Abstract
The thioredoxin system, including NADPH, thioredoxin (Trx), and thioredoxin reductase (TrxR), plays significant roles in maintaining intracellular redox homeostasis and protecting organisms against oxidative damage. In this study, the characteristics and functions of H. armigera HaTrx2 and HaTrxR1 were identified. Sequence analysis showed that HaTrx2 and HaTrxR1 were both highly conserved and shared high sequence identity with other insect counterparts. The mRNA of HaTrx2 was expressed the highest in 5th instar 96 h and was mainly detected in heads and epidermis. The expression of HaTrxR1 was highly concentrated in 5th instar 72 h and 96 h, and higher in malpighian tube, midgut and hemocyte than other examined tissues. HaTrx2 and HaTrxR1 were markedly induced by various types of stress. HaTrx2- or HaTrxR1-knockdown increased ROS production in hemocytes and also increased the lipid damage in NPV infected H. armigera larvae. Furthermore, interference with expression of HaTrx2 or HaTrxR1 transcripts in H. armigera larvae resulted in increased sensitivity to NPV infection and shortened LT50 values. Our findings indicated that HaTrx2 and HaTrxR1 contribute to the susceptibility of H. armigera to NPV and also provided the theoretical basis for the in-depth study of insect thioredoxin system.
Oxidative stress, which can damage the balance of strong oxidants and antioxidants, is a negative effect produced by reactive oxygen species (ROS) in living organisms and is one of the important factors that cause aging and disease1,2. ROS, including hydrogen peroxide, superoxide anions and hydroxyl radicals, can be induced by various adverse factors, leading to oxidative damage to biological macromolecules3. To protect against these toxic oxygen intermediates caused by ROS, oxygenic organisms have developed a battery of protective enzymes, including the antioxidase system, and non-enzymatic systems to maintain redox homeostasis by scavenging excessive ROS4,5. The thioredoxin system, comprising NADPH, thioredoxin (Trx), and thioredoxin reductase (TrxR), is one of these systems that participate in redox-regulatory processes in cells6.
The Trxs, which contain a highly conserved Cyc-Gly-Pro-Cys (CGPC) active site motif 7, are the family of small reductases (approximately 12 kDa in size) that are ubiquitously distributed from Archaea to man6. The Trxs, which were first identified in Escherichia coli as an electron donor for ribonucleotide reductase8, play numerous roles in resisting oxidative stresses, promoting cell growth, regulating cell apoptosis and transcriptional regulation in many organisms9. Oxidized Trxs present a disulfide bond, while reduced Trxs exhibit a thiol group. The reversible thiol-disulfide transformation reactions are rapid and ideally suited to regulate the functions of proteins4. It is generally believed that the antioxidant effect of Trxs are mainly manifested in two aspects: first, Trxs can serve as electron donors for peroxidases to cope with ROS and, thus, to reduce lipid peroxidation, DNA damage and protein inactivation; second, as a disulfide reductase of intracellular proteins, Trxs can reduce the disulfide bonds of many proteins (such as kinases, phosphatases and transcription factors) to restore physiological function10.
Trxs have been widely studied in mammals11,12, plants13,14, and bacteria15,16 because of their essential roles in protection against oxidative stress, whereas reports focusing on Trxs in insects are limited. In Drosophila, three Trx genes (Trx1, Trx2, and TrxT) have been identified17,18,19, and the loss of Trx-2 promoted the expression of other antioxidant genes and exacerbated oxidative stress-dependent phenotypes20. In Bombyx mori, BmTrx has been shown to protect against oxidative stress caused by extreme temperatures and microbial infection21. In Apis mellifera, three Trxs have been identified: AmTrx1, which is located in the mitochondrion, AmTrx2, which is a putative ortholog of Drosophila Trx2 and may play a vital role in redox homeostasis, and AmTrx322. In Apis cerana cerana, some Trxs, including AccTrx-like123, AccTrx224 and AccTrx125, have been demonstrated to participate in antioxidant defense. All of the above studies suggest that Trxs play a major role in maintaining redox homeostasis and resisting adverse circumstances in insects.
TrxRs are homodimeric flavoproteins that belong to the pyridine nucleotide-disulfide oxidoreductase family and can catalyze the natural substrates of thioredoxin26. There are two forms of TrxRs in different organisms: low molecular weight (MW) TrxRs, of approximately 35 kDa, which are mainly found in bacteria, plants and parasites; and high MW TrxRs, of approximately 55 kDa, which are mainly found in higher eukaryotes27. The N-terminus of mammalian TrxRs possesses a redox catalytic site structure consisting of -Cyc-Val-Asn-Gly-Cys- (CVNVGC), and the C-terminus exhibits an extension redox active site sequence of -Gly-Cys-Sec-Gly- (GCUG)28,29, while the C-terminal conserved sequence is -Cys-Cys-Ser- (CCS) in insects30. TrxRs can transfer reducing equivalents from NADPH to thioredoxin; the electron transfer path is from NADPH to FAD, then to N-terminal redox active sites, followed by the C-terminal active motifs, and finally to Trxs28. The physiological roles of TrxRs have been widely studied in mammals, including their functions in redox homeostasis and antioxidant defense31, regulating cell growth and inhibiting cell apoptosis32, and controlling early embryonic development33. There have been some reports about the use of Trx and TrxR as targets of cancer therapy34,35.
In contrast to the many studies addressing TrxRs in mammals, knowledge of TrxRs in insects is lacking. In Drosophila, two TrxRs have been identified: TrxR-1, which encodes three splice variants (one mitochondrial and two cytoplasmic forms), and TrxR-2, which encodes a protein with a potential targeting peptide36. The TrxR-1 null mutant of D. melanogaster leads to death at the end of the second larval instar37, and both cytosolic and mitochondrial TrxR-1 forms have been shown to be necessary for survival36. In Anopheles gambiae, TrxR-1, which occurs in three splice variants, shares 69% sequence identity with D. melanogaster TrxR-1 and possesses a conserved Cys-Cys active motif in its C-terminal extension30. In A. mellifera, only one TrxR gene has been identified, which exhibits two putative splice variants, but it does not appear that they encode the mitochondrial variant22. In A. cerana cerana, AccTrxR1 was shown to be induced by ultraviolet light (UV) and heat (37 °C) and to be involved in protection against oxidant stress38. In Chironomus riparius, the transcription of CrTrxR1 was found to be up-regulated after paraquat and cadmium chloride exposure and is considered to be a biomarker of oxidative stress induced by environmental contaminants39.
The cotton bollworm (Helicoverpa armigera) is one of the lepidopteran pests that cause the most damage, resulting in enormous economic losses in the cotton, corn, vegetable and other crop industries throughout Asia40. Although its population has decreased since the introduction of Bt-cotton in China in 1997, the control of this pest is a longstanding problem due to its ability to develop insecticide resistance40,41. The genes of the thioredoxin system are being considered as targets for the treatment of inflammation or cancer in humans42,43, and another antioxidant gene (thioredoxin peroxidase) was shown to be involved in resistance to the biocontrol fungus Nomuraea rileyi in Spodoptera litura44. We hypothesize that Trx and TrxR can help to resist the infection of pathogenic microorganism in insect. To elucidate the functions of thioredoxin system genes in H. armigera, we investigated their spatio-temporal distribution and evaluated their transcript levels after various types of stress treatments, including temperatures of 0 °C and 37 °C, UV, mechanical injury, E. coli exposure, Metarhizium anisopliae exposure, and nucleopolyhedrovirus (NPV) infection. Furthermore, ROS generation and lipid peroxidation in HaTrx2- or HaTrxR1-knockdown larvae and normal larvae were measured. Finally, RNA interference (RNAi) technology was used to study these two genes involved in resistance to NPV. Our results will contribute to further studies on Trx and TrxR in Insecta and will aid in the development of novel insecticides targeting Trx and TrxR.
Results
Sequence analysis of HaTrx2 and HaTrxR1
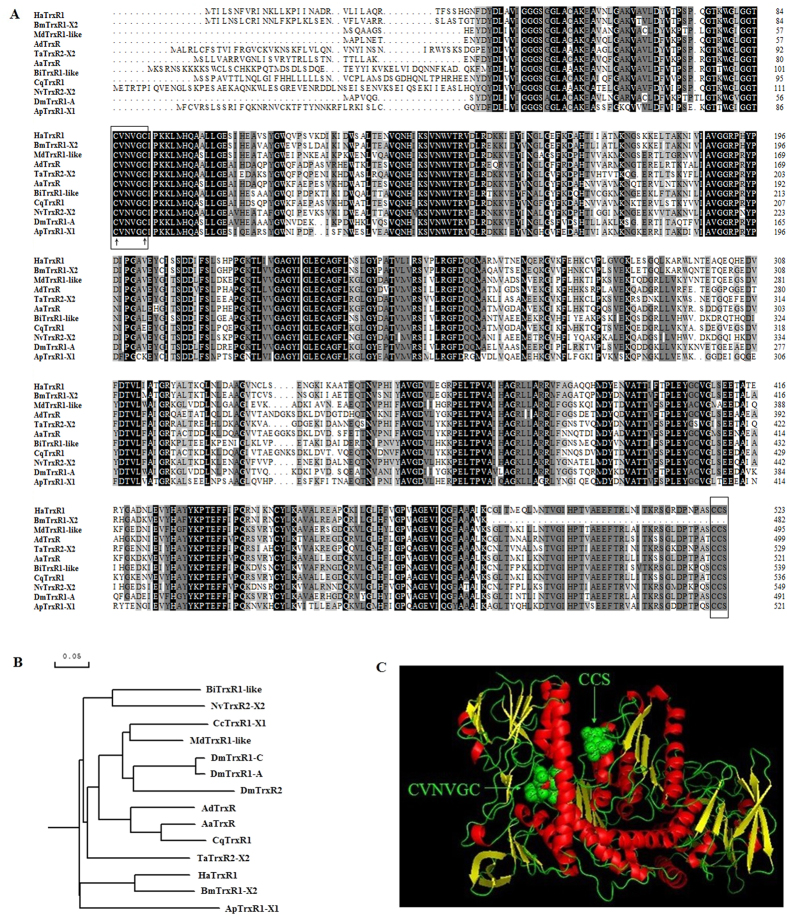
Sequence analysis showed that the full-length cDNA of HaTrx2 was 800 bp, including a 321 bp open reading frame (ORF) and encoding a deduced polypeptide of 107 amino acids with a predicted molecular weight of 12.03 kDa and a pI of 4.82. Multiple alignment analysis of the amino acid sequence showed that HaTrx2 shared high amino acid identity (61%–92%) with Trx sequences from other selected insect species. The active site sequence CGPC was found in the N-terminal portion of the HaTrx2 sequence and was highly conserved among all of the selected insect species (Fig. 1A). As shown in Fig. 1B, phylogenetic analysis revealed that HaTrx2 was most closely related to the PpTrx2 homologue (Papilio polytes, BAM19091.1) and PxTrx2 homologue (Papilio xuthus, BAM17831.1), consistent with the evolutionary relationships predicted from the multiple alignment of amino acid sequences. The potential tertiary protein structure of HaTrx2 was constructed with the SWISS-MODEL server and PyMOL-v1.3r1 software, and the cysteines (Cys32 and Cys35) in the conserved redox active motif were identified (Fig. 1C).
Figure 1. Sequence characterization of Trx from various species and the predicted tertiary structure of HaTrx2.
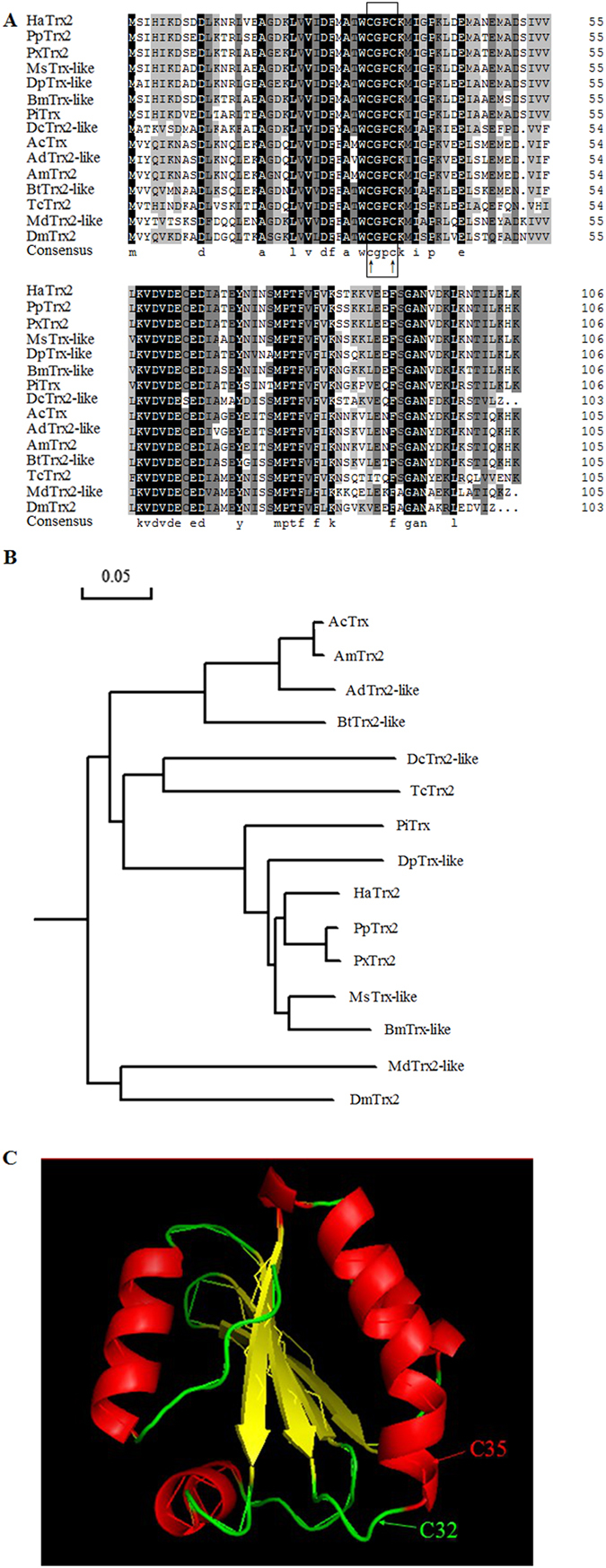
(A) Multiple alignments of the amino acid sequence of HaTrx2 with homologs from other insect species. Black represents 100% identity, gray represents 75% identity and white represents <75% identity. The conserved CGPC motif is boxed and the active sites are marked by ↑. HaTrx2 (Helicoverpa armigera, AGC39043.1), PpTrx2 (Papilio polytes, BAM19091.1), PxTrx2 (Papilio xuthus, BAM17831.1), MsTrx-like (Manduca sexta, AAF16695.1), DpTrx-like (Danaus plexippus, EHJ64037.1), BmTrx-like (Bombyx mori, NP_001091804.1), PiTrx (Plodia interpunctella, CBW45298.1), DcTrx2-like (Diaphorina citri, XP_008485323.1), AcTrx (Apis cerana, AGF33352.1), AdTrx2-like (Apis dorsata, XP_006608786.1), AmTrx2 (Apis mellifera, XP_003250408.1), BtTrx2-like (Bombus terrestris, XP_003396074.1), TcTrx2 (Tribolium castaneum, XP_967987.1), MdTrx2-like (Musca domestica, XP_005177432.1), DmTrx2 (Drosophila melanogaster, NP_523526.1). The same as below. (B) Phylogenetic tree analysis of HaTrx2 and its homologs in insects. (C) Predicted tertiary structure of HaTrx2. The SWISS-MODEL server and PyMOL software were used to construct the tertiary structure, and the active sites (Cys32 and Cys35) are marked.
The ORF of HaTrxR1 was 1572 bp, encoding a polypeptide of 523 amino acids residues with a predicted molecular weight of 57.16 kDa and a theoretical pI of 7.57. Multiple sequence alignment revealed that HaTrxR1 shared 83% identity with BmTrxR1-X2 and 64%–71% identity with TrxR sequences from other selected insect species. The active site sequence CVNVGC was found in the N-terminal portion, while CCS was found in the C-terminal portion of the HaTrxR1, and these sequences were highly conserved among the selected insect species (Fig. 2A). Phylogenetic analysis showed that HaTrxR1 was more closely related to the BmTrxR1-X2 homologue (B. mori, XP_004921588.1) than other selected species, and this result was consistent with the evolutionary relationship predicted from the multiple alignment of amino acid sequences (Fig. 2B). The tertiary protein structure of HaTrxR1 was constructed using the SWISS-MODEL server and PyMOL-v1.3r1 software, and the conserved redox active motifs (CVNVGC and CCS) were identified (Fig. 2C).
Figure 2. Sequence characterization of TrxR from various species and the predicted tertiary structure of HaTrxR1.
(A) Multiple alignments of the amino acid sequences of HaTrxR1 with homologs from other insect species. Black represents 100% identity, gray represents 75% identity and white represents <75% identity. The conserved CVNVGC motif and CCS motif are boxed and the active sites are marked by ↑. HaTrxR1 (Helicoverpa armigera, KM658552), BmTrxR1-X2 (Bombyx mori, XP_004921588.1), MdTrxR1-like (Musca domestica, NP_001273801.1), AdTrxR (Anopheles darlingi, ETN61621.1), TaTrxR2-X2 (Tribolium castaneum, XP_008191173.1), AaTrxR (Aedes aegypti, XP_001662666.1), BiTrxR1-like (Bombus impatiens, XP_003485381.1), CqTrxR1 (Culex quinquefasciatus, XP_001847793.1), NvTrxR2-X2 (Nasonia vitripennis, XP_008202131.1), DmTrxR1-A (Drosophila melanogaster, NP_727252.1), ApTrxR1-X1 (Acyrthosiphon pisum, XP_008179444.1). The same as below. (B) Phylogenetic tree analysis of HaTrxR1 and its homologs in insects. CcTrxR1-X1 (Ceratitis capitata, XP_004518825.1), DmTrxR1-C (Drosophila melanogaster, NP_727252.1), DmTrxR2 (Drosophila melanogaster, NP_524216.1). (C) Predicted tertiary structure of HaTrxR1. SWISS-MODEL server and PyMOL software were used to build the tertiary structure and the conserved motifs (CVNVGC and CCS) were marked.
Temporal and spatial expression profiles of HaTrx2 and HaTrxR1
To determine the transcription profile of HaTrx2 in different developmental stages and larval tissues in H. armigera, qRT-PCR was carried out using total RNA prepared from the above collected samples. Standard curves for the primers were generated before formal experiments. The correlation coefficients (R2) of the four genes (HaTrx2, HaTrxR1, RPS15, and RPL32) were greater than 0.99, and the amplification efficiencies of the primers were 98.06%, 103.97%, 105.20%, and 98.70%, respectively (Figure S1). The HaTrx2 transcript showed ubiquitous expression in all developmental stages, mainly being expressed in the 96 h larvae of the 5th instar (Fig. 3A). The spatial expression profiles revealed that the HaTrx2 gene could be detected in all of the investigated tissues, and the expression levels were higher in the head, epidermis, midgut and Malpighian tubules than other tissues (Fig. 3B).
Figure 3. Expression profile of HaTrx2 and HaTrxR1 in different developmental stages and different larval tissues.
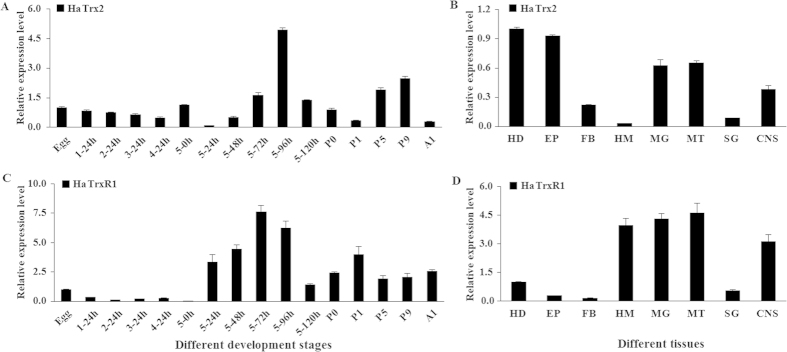
(A) Transcript levels of HaTrx2 during different developmental stages. 1–24h, 24 h larvae of 1st instar; 2–24 h, 24 h larvae of 2nd instar; 3–24 h, 24 h larvae of 3rd instar; 4–24 h, 24 h larvae of 4th instar; 5–0 h, 5–24 h, 5–48 h, 5–72 h, 5–96 h, and 5–120 h stand for 5th instar larvae at 0, 24, 48, 72, 96, and 120 h, respectively; P0, P1, P5, and P9 stand for 0, 1, 5, and 9 day pupae, respectively; A1, 1 day adults. The same as below. (B) Transcript levels of HaTrx2 in the tissues of 5th instar 24 h larvae. HD, heads; EP, epidermis; FB, fat body; HM, hemolymph; MG, midgut; MT, malpighian tubule; SG, salivary glands; CNS, central nervous system. The same as below. (C) Transcript levels of HaTrxR1 during different developmental stages. (D) Transcript levels of HaTrxR1 in the tissues of 5th instar 24 h larvae. The data represent the mean ± standard deviation (SD) from 3 biological samples.
The qRT-PCR results showed that HaTrxR1 was mainly expressed in 24 h, 48 h, 72 h, and 96 h larvae of the 5th instar and the first-day pupae, with relatively lower expression being observed in other larval stages (Fig. 3C). The obtained spatial expression profiles showed that the HaTrxR1 gene was mainly expressed in the hemocytes, midgut, Malpighian tubules, and CNS (Fig. 3D).
The response of the expression profiles of HaTrx2 and HaTrxR1 to various types of adversity
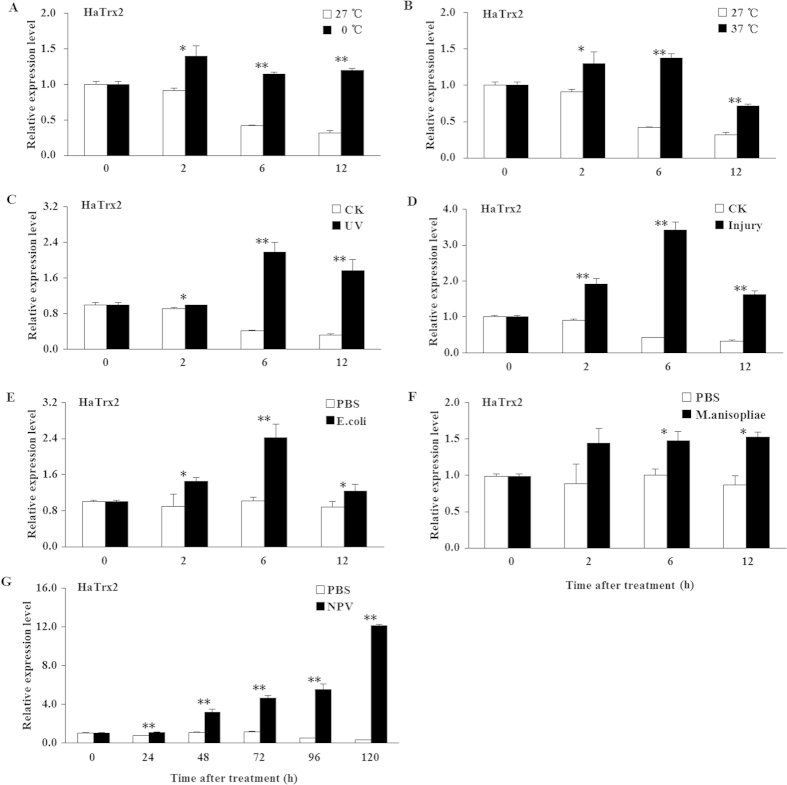
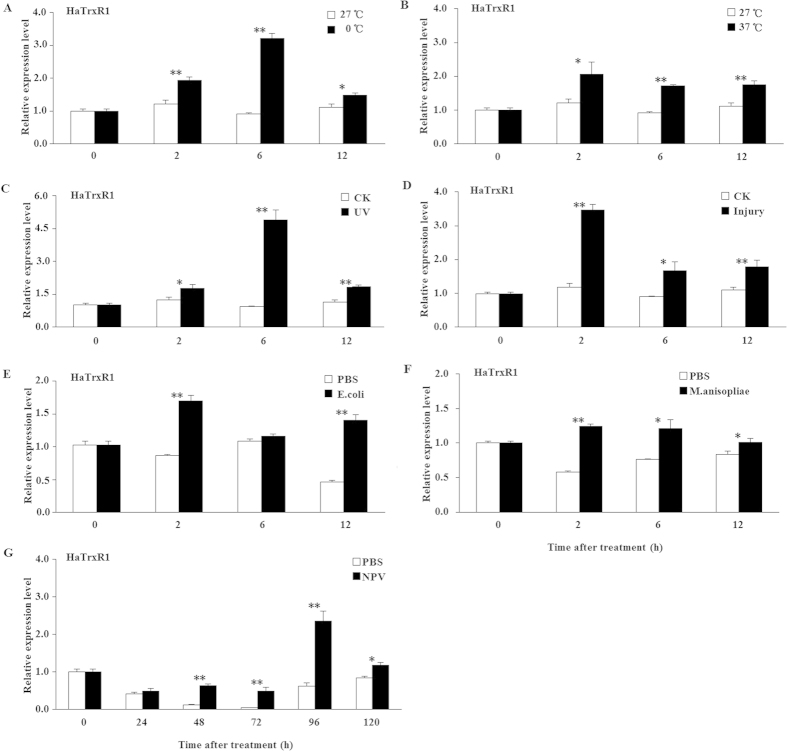
To study the effect of various adverse stresses on HaTrx2 and HaTrxR1 transcription, larvae were challenged with low temperature, high temperature, UV light, mechanical injury, E. coli exposure, M. anisopliae exposure, and NPV infection. As shown in Fig. 4, the transcription of HaTrx2 was significantly induced by the 0 °C, 37 °C, UV, mechanical injury, and E. coli exposure treatments at 2 h, 6 h, and 12 h, in addition to being increased at 6 h and 12 h after M. anisopliae exposure treatment and being markedly up-regulated at 24 h, 48 h, 72 h, 96 h, and 120 h after NPV infection. For the HaTrxR1 transcript, we observed a similar tendency (Fig. 5). HaTrxR1 transcription was markedly up-regulated at 2 h, 6 h, and 12 h after the 0 °C, 37 °C, UV, mechanical injury, M. anisopliae exposure treatments, in addition to being significantly up-regulated at 2 h and 12 h after E. coli exposure and being markedly increased at 48 h, 72 h, 96 h, and 120 h after NPV infection (Fig. 5). Taken together, all of the above results suggested that HaTrx2 and HaTrxR1 may play an important role in protection against the oxidative stress caused by various types of adversity, and especially NPV infection.
Figure 4. Expression profiles of HaTrx2 under different abiotic and biotic stresses.
Total RNA was harvested from H. armigera samples under different stress challenges, including low temperature (4 °C), high temperature (37 °C), UV light, mechanical injury, E. coli, M. anisopliae, and NPV infection, and then subjected to real-time PCR analysis. The data represent the mean ± SD of 3 biological samples. *0.01 < P < 0.05; **P < 0.01.
Figure 5. Expression profiles of HaTrxR1 under different abiotic and biotic stresses.
Total RNA was harvested from H. armigera samples under different stress challenges, including low temperature (4 °C), high temperature (37 °C), UV light, mechanical injury, E. coli, M. anisopliae, and NPV infection, and then subjected to real-time PCR analysis. The data represent the mean ± SD of 3 biological samples. *0.01 < P < 0.05; **P < 0.01.
ROS generation and lipid peroxidation in HaTrx2- or HaTrxR1-knockdown larvae and normal larvae
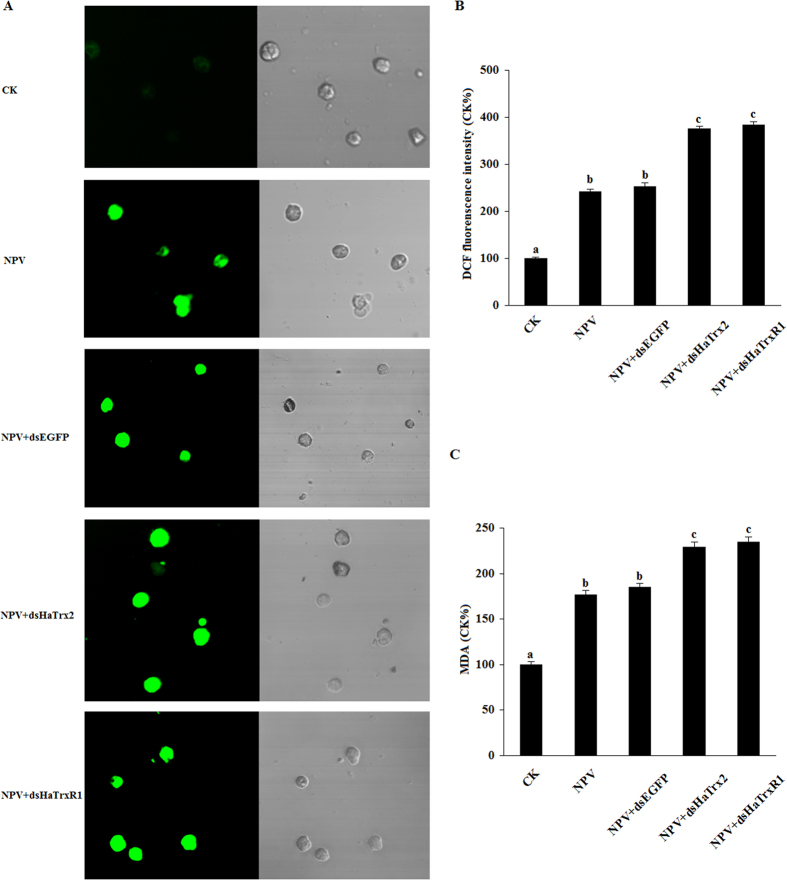
To confirm HaTrx2 and HaTrxR1 play vital roles in protecting H. armigera against oxidative damage caused by NPV infection, ROS generation was determined in HaTrx2- or HaTrxR1-knockdown larvae and normal larvae. As shown in Fig. 6A,B, the fluorescence intensity of larvae hemocytes in the NPV + dsHaTrx2 or NPV + dsHaTrxR1 group were both stronger than that in the NPV+dsEGFP or NPV groups.
Figure 6. Stimulatory effect of HaTrx2- or HaTrxR1-knockdown on ROS levels in hemocytes of NPV infected H. armigera larvae.
(A) Effects of HaTrx2- or HaTrxR1-knockdown on ROS generation in hemocytes. (B) The relevant DCF-fluorescent intensity quantification is shown as a percentage of the values found in larvae hemocytes of CK group. (C) The results of MDA generation and lipid peroxidation in H. armigera hemolymph after HaTrx2- or HaTrxR1-knockdown. The data represent the mean ± SD of 3 biological samples. a & b: signification difference, P < 0.05.
As ROS damage also caused lipid peroxidation in living organism. We measured the concentration of a terminal product (malonyl dialdehyde, MDA) of lipid peroxidation in hemocytes after HaTrx2- or HaTrxR1-knockdown to confirm HaTrx2 and HaTrxR1 play vital roles in protecting H. armigera against oxidative damage caused by NPV infection. The results showed that MDA levels were markedly increased after HaTrx2- or HaTrxR1-knockdown compared to EGFP dsRNA injection or NPV infection (Fig. 6C).
RNA interference and survival assay
To further confirm the functions of HaTrx2 and HaTrxR1, the adverse stress of NPV infection was chosen because the expression of these two genes was increased to a greater extent by NPV infection than the other selected adverse stresses (Fig. 4 and 5).
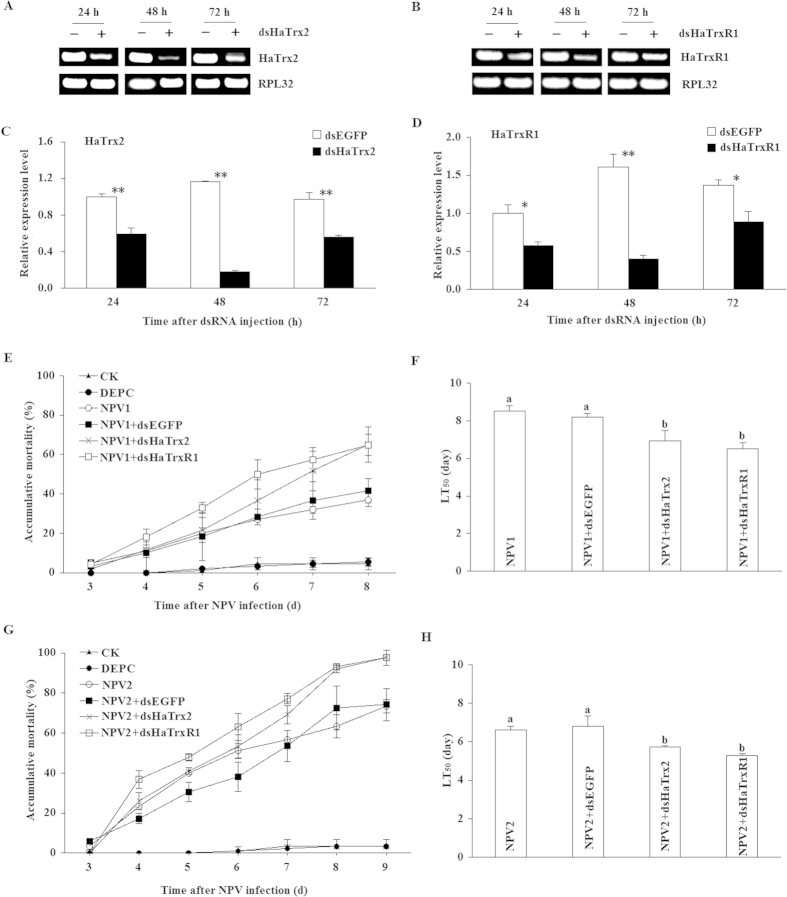
The results of agarose gel electrophoresis and real-time PCR analyses showed that the transcripts of HaTrx2 and HaTrxR1 were significantly decreased at 24 h, 48 h, and 72 h after HaTrx2 and HaTrxR1 dsRNA injection compared with EGFP dsRNA injection (Fig. 7A,B). HaTrx2 expression was decreased by 40.77%, 84.43%, and 39.89% (Fig. 7C), while HaTrxR1 expression was decreased 42.54%, 77.81%, and 35.06% (Fig. 7D) at 24 h, 48 h, and 72 h after the injection of HaTrx2 or HaTrxR1 dsRNA, respectively, compared with EGFP dsRNA injection.
Figure 7. RNAi efficiency of HaTrx2 or HaTrxR1 dsRNA and effects of HaTrx2- or HaTrxR1-knockdown on the susceptibility of H. armigera larvae to NPV infection.
(A–D) RNAi-induced reduction of HaTrx2 and HaTrxR1 transcription. First, 4th instar first-day larvae were fed with an artificial diet contaminated with 10 μL of NPV (1 × 106 PIB/ml). At 48 h after NPV infection (at the concentration of 1 × 106 PIB/ml or 1 × 107 PIB/ml), a total of 15 μg of the dsRNA of HaTrx2, HaTrxR1, or EGFP was injected into each treatment larva. The samples of H. armigera larvae were collected 24, 48, and 72 h after dsRNA injection and then subjected to total RNA extraction, cDNA synthesis, and real-time PCR analysis. (E) Cumulative mortality of NPV infected larvae at the concentration of 1 × 106 PIB/ml (NPV1) after HaTrx2- or HaTrxR1-knockdown. (F) LT50 values in the bioassay of NPV infected larvae at the concentration of 1 × 106 PIB/ml after HaTrx2- or HaTrxR1-knockdown. (G) Cumulative mortality of NPV infected larvae at the concentration of 1 × 107 PIB/ml (NPV2) after HaTrx2- or HaTrxR1-knockdown. (H) LT50 values in the bioassay of NPV infected larvae at the concentration of 1 × 107 PIB/ml after HaTrx2- or HaTrxR1-knockdown. The data represent the mean ± SD of 3 biological samples. *0.01 < P < 0.05; **P < 0.01. a & b: signification difference, P < 0.05.
qRT-PCR results also showed that expression level of HaTrxR1 increased significantly after HaTrx2 knockdown, however, HaTrx2 expression level remained unchanged after HaTrxR1 knockdown (Figure S2). To determine how is the NPV infection going on at 48 h post infection and after 48 h of dsRNA injection, qRT-PCR was used to quantify the virus gDNA abundance. As shown in Figure S3A, viral gDNA level at 48 h after NPV infection increased about 300 times than 0 h or 24 h after NPV infection. HaTrx2- or HaTrxR1-knockdown also obviously promoted viral gDNA levels compared to EGFP dsRNA injection at 48 h (Figure S3B).
At 48 h after NPV inoculation, HaTrx2 dsRNA, HaTrxR1 dsRNA, or EGFP dsRNA was injected into the larvae. The overall trend was that injection of HaTrx2 dsRNA or HaTrxR1 dsRNA accelerated the mortality of larvae infected with NPV at the both concentrations of 1.0 × 106 PIB/mL (NPV1) and 1.0 × 107 PIB/mL (NPV2) (Fig. 7E,G). When larvae infected with NPV at the concentration of 1.0 × 106 PIB/mL, the LT50 (time required to reached 50% mortality) of NPV1 (8.54 d) and NPV1 + dsEGFP (8.20 d) larvae was significantly higher than in NPV1 + dsHaTrx2 (6.93 d) or NPV1 + dsHaTrxR1 (6.50 d) larvae (P < 0.05, Fig. 7F). When larvae infected with NPV at the concentration of 1.0 × 107 PIB/mL, the LT50 of NPV2 (6.62 d) and NPV2 + dsEGFP (6.79 d) larvae was significantly higher than in NPV2 + dsHaTrx2 (5.73 d) or NPV2 + dsHaTrxR1 (5.27 d) larvae (P < 0.05, Fig. 7H). These results suggested that HaTrx2 and HaTrxR1 are required for resistance against NPV infection in H. armigera larvae.
Discussion
Many studies addressing the functions of the thioredoxin system, which are involved in regulating cellular redox homeostasis and resisting oxidative stress caused by adversity, have been conducted in mammals43,45 and some model insect species21,24,30,38. However, research on Trx and TrxR in the lepidopteran pest H. armigera is lacking. In this study, HaTrx2 and HaTrxR1 were identified and characterized in the larvae of H. armigera. Sequence analysis suggested that HaTrx2 shared high amino acid identity (61%–92%) with other insect counterparts, and all of these proteins contained the highly conserved CGPC active-site motif, which is essential for their catalytic activity7. Multiple alignment and phylogenetic analysis revealed that HaTrxR1 shared 64%–83% sequence identity with other insect species, including the important active site sequence CVNVGC in the N-terminal portion and the CCS motif in the C-terminal extension (Fig. 2A)30. These results demonstrated that both HaTrx2 and HaTrxR1 possessed redox active sites and belonged to the typical Trx and TrxR families, respectively, and they might be involved in resistance to adversity.
The changes in HaTrx2 and HaTrxR1 transcription observed at different developmental stages showed that these two genes were mainly expressed in 5th instar and pupal stage. The obtained spatial expression profiles revealed that the HaTrx2 gene was expressed at higher levels in the head, epidermis, midgut and Malpighian tubules than other tissues (Fig. 3B), suggested that it may play vital roles in antioxidant defense in these tissues, which are central organs in metabolism and detoxification. The expression of Trx in larval tissues appears to show a species-dependent pattern: BmTrx is mainly expressed in the fat body and silk gland21; AccTrx1 and AccTrx-like1 exhibit higher expression in the epidermis than in other tissues23,25; and AccTrx2 is expressed at higher levels in the brain and midgut24. However, the HaTrxR gene is mainly expressed in the hemocytes, midgut, Malpighian tubules, and CNS (Fig. 3D), implying that it may mainly play crucial roles in these tissues with antioxidant functions.
It has been reported that adverse environmental factors, such as pesticides, heavy metals, UV radiation, and abnormal temperatures can lead to oxidative damage to living organisms46. A majority of antioxidant enzymes, such as peroxidases and catalases, play significant roles in the scavenging or quenching of oxidants and, thus, constitute a primary short-term line of defense. In previous studies, the expression of BmTrx in the fat body of B. mori larvae was shown to be greatly increased after treatment with H2O2, paraquat, low or high temperatures, or microorganism (bacterium, fungus, and NPV) infection21; AccTrx-like1 was found to be up-regulated by treatment with H2O2 or temperatures of 4, 15, and 25 °C23; AccTrx2 was shown to be stimulated by treatment with H2O2, temperatures of 4, 16, and 25 °C, acaricide, cyhalothrin, phoxim, paraquat, and HgCl224; and AccTrx1 was found to be induced by treatment with H2O2, temperatures of 4, 16, and 42 °C and pesticides (acaricide, phoxim, cyhalothrin, and paraquat)25, suggesting that Trx may play important roles in protection against oxidative stress caused by an adverse environment. The TrxR gene of A. gambiae was also shown to be induced by injury, bacterial challenge, and malaria infection47. In the present study, the transcripts of HaTrx2 and HaTrxR1 were significantly induced by various types of adversity, including low temperature, high temperature, UV light, mechanical injury, E. coli exposure, M. anisopliae exposure, and NPV infection, which suggests that HaTrx2 and HaTrxR1 may participate in resistance to these adverse conditions. The possible mechanism underlying Trx and TrxR involvement in antioxidant defense may be elucidated as follows: ROS are first formed under adverse stress and then act on cellular biomacromolecules that are susceptible to oxidation stress by disrupting intracellular redox homeostasis, and Trx and TrxR may be play crucial roles in the removal of excessive ROS to protect organisms48.
In this study, to confirm the role of HaTrx2 and HaTrxR1 in the removal of excessive ROS caused by NPV infection to protect H. armigera larvae, the expression of HaTrx2 and HaTrxR1 was successfully knockdown with the injection of the related dsRNA, as examined by semi-quantitative RT-PCR and qRT-PCR. Further study confirmed that HaTrx2- or HaTrxR1-knockdown increased ROS production in hemocytes and also increased the lipid damage in NPV infected H. armigera larvae. Together, these results indicated that HaTrx2 and HaTrxR1 may participate in the removal of excessive ROS caused by NPV infection in H. armigera. However, further study to provide in-depth confirmation is warranted.
In S. litura, larval mortality was accelerated after knockdown of the antioxidant gene SlTpx through dsRNA interference in the presence of N. rileyi infection, suggesting that SlTpx plays a vital role in resisting oxidative damage caused by N. rileyi infection44. In D. melanogaster, Drosophila cells become susceptible to H2O2 treatment after knockdown of the Tpx transcript through RNAi49. Here, the expression of HaTrx2 and HaTrxR1 was found to be significantly stimulated by NPV infection, which is widely applied in the management of the pest H. armigera due to its strong pathogenicity. In a further experiment, knockdown of HaTrx2 or HaTrxR1 transcripts resulted in increased sensitivity to NPV infection and shortened LT50 values. All of these observations indicated that expression of HaTrx2 and HaTrxR1 is essential in defense against NPV infection in H. armigera larvae.
In conclusion, we have characterized two typical thioredoxin system genes from H. armigera, and determined the temporal and spatial expression profiles of HaTrx2 and HaTrxR1. The transcription of HaTrx2 and HaTrxR1 was induced by various types of adversity (low temperature, high temperature, UV light, mechanical injury, E. coli exposure, M. anisopliae exposure, and NPV infection), suggesting HaTrx2 and HaTrxR1 play important roles in resistance to various types of adversity. HaTrx2- or HaTrxR1-knockdown increased ROS production in hemocytes and also increased the lipid damage in NPV infected H. armigera larvae. RNAi experiments further confirmed that HaTrx2 and HaTrxR1 are involved in resistance to NPV infection. These observations provide powerful evidence demonstrating that HaTrx2 and HaTrxR1 play vital roles in protecting H. armigera against oxidative damage and enrich our knowledge of the thioredoxin system in insects. Therefore, the development of novel chemicals and microbial pesticides targeting HaTrx2 or HaTrxR1 for H. armigera control will require further in-depth research.
Methods
Insect maintenance and H. armigera NPV
H. armigera were reared in our laboratory with an artificial diet50 at a constant temperature of 26 ± 1 °C, under 75 ± 10% RH and a 16 L: 8 D light regime. The larvae were individually reared in separate glass tubes (5.5 cm in length × 2.0 cm in diameter) after the 3rd instar stage to prevent cannibalism.
Raw powder of H. armigera NPV (5 × 1011 PIB/g) was bought from the Henan Jiyuan Baiyun Industry Co., Ltd (China) and stored at 4 °C for later use.
Sequence analysis of HaTrx2 and HaTrxR1
The GenBank accession number of HaTrx2 and HaTrxR1 were JQ744277.1 and KM658552. The physicochemical properties of HaTrx2 and HaTrxR1 were analyzed using the online bioinformatics ProtParam tool (http://web.expasy.org/protparam/). Homologous protein sequences of Trxs and TrxRs from various species were obtained from the NCBI database and aligned using DNAman6.0.3 software. Phylogenetic analysis was carried out using MEGA5.10 software. Finally, the tertiary protein structures of HaTrx2 and HaTrxR1 were predicted with the online server SWISS-MODEL and were modified with PyMOL-v1.3r1 software51.
Developmental analysis and tissue distribution of HaTrx2 and HaTrxR1
To examine the temporal expression profiles of HaTrx2 and HaTrxR1, H. armigera samples were collected at different developmental stages, including eggs; 24 h larvae of the first, second, third, and 4th instar; 0, 24, 48, 72, 96, and 120 h larvae of the 5th instar; 0, 1, 3, 5 and 9 day pupae; and 1 day adults (equal numbers of females and males). To analyze the spatial expression patterns of HaTrx2 and HaTrxR1, the tissues of the 5th instar 48 h larvae were collected, including the head, epidermis, fat body, hemocytes, midgut, Malpighian tubules, salivary glands, and central nervous system (CNS)52. Each sample was repeated three times and immediately stored at −80 °C for total RNA extraction.
Effect of different types of stress on the expression of HaTrx2 and HaTrxR1
For the temperature treatments, 0 °C (low temperature) and 37 °C (high temperature) were chosen21,53. The first-day larvae of the 5th instar were held for 12 h under the two temperatures, while the controls were maintained at 27 °C (normal temperature)21. In the UV treatment, the first-day larvae of the 5th instar were irradiated with 300 nm wavelength light, and the control larvae were kept under normal light for 12 h53. In the mechanical injury experiment, each larva was impaled 10 times with an insect pin (30 × 0.5 mm), and normal larvae were used as controls. In the E. coli infection treatment, E. coli cells were diluted in PBS and subsequently injected into the abdomens of first-day larvae of the 5th instar with a syringe, injecting 10 μL of 1.0 × 105 E. coli cells per larva21. Control larvae were injected with an equal volume of PBS (10 μL/larva). For challenge by M. anisopliae, M. anisopliae was first inoculated on potato dextrose agar plates and incubated at 26 °C for 7–10 days. The produced conidia were then scraped and diluted with sterile water containing 0.1% Tween−80 to 1.9 × 108 conidia/μL, which has been reported as the LC50 concentration of H. armigera54. The first-day larvae of the 5th instar were injected with 5 μL of the diluted M. anisopliae suspension, and control larvae were injected with an equal volume of PBS (5 μL/larva)55. The treatment and control larvae from each group were collected at 0, 2, 6, and 12 h after treatment, then immediately stored at −80 °C for further total RNA extraction. For the virus challenge, the first-day larvae of the 4th instar were inoculated with 10 μL of NPV at a concentration of 1.0 × 106 PIB/mL per larva, and control larvae were inoculated with 10 μL of sterile water. The treatment and control larvae were collected after 0, 24, 48, 72, 96, and 120 h and then immediately stored at −80 °C for later total RNA extraction46. At least three independent biological replications were carried out in each of the adverse condition experiments, and at least 15 larvae were used in both the control and treatment replications.
Primer design
The primers of HaTrx2 and HaTrxR1 used for RT-PCR, real-time PCR, and dsRNA synthesis were designed with DNAClub software according to their sequences. The H. armigera ribosomal proteins S15 (RPS15) and L32 (RPL32) were used as internal controls for real-time PCR normalization. All of the primers were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China) (Table 1).
Table 1. Primers used in the current study.
| Gene name (Abbreviation) | Description | Sequence (5′–3′) | Product length (bp) |
|---|---|---|---|
| HaTrx2-qF | Real-time PCR primer, forward | GTCGATCCACATCAAGGAC | 94 |
| HaTrx2-qR | Real-time PCR primer, reverse | GCACCAAGTGGCCATGAAG | |
| HaTrx2-RNAiF1 | The first step PCR primer of dsRNA synthesis, forward | GGACCCTGCAAGATGATCG | 213 |
| HaTrx2-RNAiR1 | The first step PCR primer of dsRNA synthesis, reverse | CAGGATAGTGTTCCTCAG | |
| HaTrx2-RNAiF2 | The second step PCR primer of dsRNA synthesis, forward | GATCACTAATACGACTCACTATAGGGAGAGGACCCTGCAAGATGATCG | 271 |
| HaTrx2-RNAiR2 | The second step PCR primer of dsRNA synthesis, reverse | GATCACTAATACGACTCACTATAGGGAGACAGGATAGTGTTCCTCAG | |
| HaTrxR1-qF | Real-time PCR primer, forward | CGAGGTCATACAGGGCTTC | 125 |
| HaTrxR1-qR | Real-time PCR primer, reverse | TGCGCTTGGTGATGTTGAGG | |
| HaTrxR1-RNAiF1 | The first step PCR primer of dsRNA synthesis, forward | GATGCTCATACGATCATCGC | 391 |
| HaTrxR1-RNAiR1 | The first step PCR primer of dsRNA synthesis, reverse | CCTTGAGCTGTCCTGACTC | |
| HaTrxR1-RNAiF2 | The second step PCR primer of dsRNA synthesis, forward | GATCACTAATACGACTCACTATAGGGAGAGATGCTCATACGATCATCGC | 449 |
| HaTrxR1-RNAiR2 | The second step PCR primer of dsRNA synthesis, reverse | GATCACTAATACGACTCACTATAGGGAGACCTTGAGCTGTCCTGACTC | |
| EGFP-RNAiF1 | The first step PCR primer of dsRNA synthesis, forward | CCTGAAGTTCATCTGCACCAC | 538 |
| EGFP-RNAiR1 | The first step PCR primer of dsRNA synthesis, reverse | CTCCAGCAGGACCATGTGATC | |
| EGFP-RNAiF2 | The second step PCR primer of dsRNA synthesis, forward | GATCACTAATACGACTCACTATAGGGAGACCTGAAGTTCATCTGCACCAC | 596 |
| EGFP-RNAiR2 | The second step PCR primer of dsRNA synthesis, reverse | GATCACTAATACGACTCACTATAGGGAGACTCCAGCAGGACCATGTGATC | |
| Poly-qF | Real-time PCR primer, forward | CAAACCGAACCGTTGTTACC | 171 |
| Poly-qR | Real-time PCR primer, reverse | TGCAAGTTCATAACGGGAC | |
| ACT-qF | Real-time PCR primer, forward | GACGGTCAGGTCATCACCATC | 151 |
| ACT-qR | Real-time PCR primer, reverse | ACAGGTCCTTACGGATGTCA | |
| RPS15-qF | Real-time PCR primer, forward | CTGAGGTCGATGAAACTCTC | 107 |
| RPS15-qR | Real-time PCR primer, reverse | CTCCATGAGTTGCTCATTG | |
| RPL32-qF | Real-time PCR primer, forward | CATCAATCGGATCGCTATG | 152 |
| RPL32-qR | Real-time PCR primer, reverse | CCATTGGGTAGCATGTGAC |
Total RNA extraction, cDNA synthesis, and real-time PCR amplification
Total RNA was extracted from the above samples using the TRIzol reagent (Invitrogen, USA) following the manufacturer’s protocols. The purity and concentration of the RNA samples were determined three times with an ultraviolet spectrophotometer (Abs260) to reduce deviation. First-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA following the instruction manual of the PrimeScript RT reagent kit with gDNA Eraser (Takara, Kyoto, Japan) and immediately stored at −80 °C for later use. The cDNA samples were evaluated in triplicate.
qRT-PCR was performed using SYBR green supermix (TaKaRa) in a Bio-Rad CFX ConnectTM Real-Time PCR System (Bio-Rad, USA) to determine the gene expression levels. The real-time PCR amplification conditions for HaTrx2, HaTrxR1, RPS15, and RPL32 are listed in Table S1. The reliability of the qRT-PCR results was confirmed through standard curve and melting curve analyses. Standard curves were generated using 10-fold dilution series of cDNA as a template for each treatment, employing a linear regression model (Figure S1s)56. The efficiencies (E) of the primers used for qRT-PCR were calculated according to the equation: E = (10[−1/slope]−1) × 100%57. The specificity of the amplified product was further confirmed through melting curve analysis from 65 °C to 95 °C and agarose gel electrophoresis. The mRNA expression of target genes was quantified using the comparative CT (cross threshold) method58. The CT value of the reference gene was subtracted from the CT value of the target gene to obtain ΔCT. The normalized fold changes of target gene mRNA expression were expressed as 2−ΔΔCT, where ΔΔCT is equal to ΔCTtreated sample –ΔCTcontrol.
Synthesis of dsRNA and detecting of RNAi efficiency
To synthesize the dsRNAs, gene-specific primers containing a T7 polymerase promoter sequence were used to amplify the target sequences via reverse transcription-PCR (RT-PCR) (Table 1). The applied RT-PCR amplification conditions are listed in Table S1. The MEGAscript RNAi kit (Ambion) was employed to synthesize the dsRNAs according to the manufacturer’s instructions. DNase and ribonuclease (RNase) were used to remove the template DNA and single-stranded RNA from the transcription reaction. dsRNAs were purified with MEGAclear columns (Ambion) and eluted with diethyl pyrocarbonate (DEPC)-treated nuclease-free water. The purity and concentration of dsRNA were then measured via ultraviolet spectrophotometry (Table S2) and gel electrophoresis. As a negative control, dsRNA of enhanced green fluorescent protein (EGFP) was also synthesized.
To evaluate the effects of RNAi on the gene expression, the first day larvae of 4th instar which inoculated with NPV (total quantity of 104 PIB per larva) at 48 h were injected with 15 μg dsRNA of HaTrx2, HaTrxR1, or EGFP, respectively. The whole-body samples were collected at 24, 48, and 72 h after dsRNA injection, and then used for total RNA extraction and real-time PCR analysis. To determine the relation between HaTrx2 and HaTrxR1, HaTrxR1 expression was measured after HaTrx2 dsRNA injection at 48 h and HaTrx2 expression was also measured after HaTrxR1 dsRNA injection at 48 h.
Measurement of ROS production and lipid peroxidation
To study the effect of RNAi on ROS generation in hemocytes, newly molted 4th instar larvae were fed the NPV-contaminated diet (10 μL of NPV at the concentration of 1.0 × 106 PIB/mL per larva) as the method of NPV challenge. After NPV infection 48 h, the infected larvae were injected with the dsRNA of EGFP, HaTrx2, or HaTrxR1, respectively. After dsRNA injection 48 h, hemolymph of each groups were collected from H. armigera and centrifuged immediately at 4000 × g at 4 °C for 10 min to isolate hemocytes. Then, ROS production was measured with Reactive Oxygen Species Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Hemocytes were incubated with DCFH-DA (2, 7-dichlorofuorescin diacetate) at a final concentration of 10 μM for 20 min. Hemocyte morphology was observed using a OLYMPUS BX61 (Olympus, Tokyo, Japan) laser scanning confocal microscope. ROS production in hemocytes was measured fluorometrically at excitation and emission wavelengths of 488 and 525 nm, respectively.
Usually, ROS damage caused lipid peroxidation in the organism. As a terminal product of lipid peroxidation, MDA was measured to evaluate the degree of lipid peroxidation in the hemolymph using MDA Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
qRT-PCR analysis of virus gDNA abundance at 48 h post NPV infection and after 48 h of dsRNA injection
qRT-PCR was used to quantify the virus abundance in NPV infected larvae at 48 h post NPV infection and after 48 h of dsRNA injection using specific primers to the polyhedrin gene (Accession no. NC_002654.2) of HaSNPV. The first-day larvae of the 4th instar were inoculated with 10 μL of NPV at a concentration of 1.0 × 106 PIB/mL per larva as the above method. The samples of whole body were collected at 0, 24, and 48 h after NPV infection, respectively. After 48 h of NPV infection, dsRNA of EGFP, HaTrx2, or HaTrxR1 were injected to the NPV infected larvae, respectively. The sample of each treatment were collected at 48 h after dsRNA injection. The genomic DNA was extracted and used in qRT-PCR technique as described59. H. armigera actin gene (Accession no. HM629437.1) was used as the housekeeping gene for normalization the data of virus gDNA quantification.
RNA interference and survival assay
To determine the effects of HaTrx2- or HaTrxR1-knockdown on the susceptibility of H. armigera larvae to NPV infection, two inoculation doses (10 μL of NPV at the concentration of 1.0 × 106 PIB/mL or 1.0 × 107 PIB/mL per larva) were selected60.
The first-day larvae of the 4th instar were inoculated with 10 μL of NPV at a concentration of 1.0 × 106 PIB/mL per larva (the group of “NPV1”) according to the above method (for the virus challenge). Control larvae were inoculated with 10 μL of sterile water (the “CK” group) or injected with 10 μL DEPC solution (the “DEPC” group). At 48 h after NPV inoculation, 15 μg of HaTrx2 dsRNA (the “NPV1 + dsHaTrx2” group), 15 μg of HaTrxR1 dsRNA (the “NPV1 + dsHaTrxR1” group), or 15 μg of EGFP dsRNA (the “NPV1 + dsEGFP” group) was injected into the proleg of each H. armigera larva using a capillary microsyringe. As the above method, The first-day larvae of the 4th instar were inoculated with 10 μL of NPV at a concentration of 1.0 × 107 PIB/mL per larva (the group of “NPV2”). At 48 h after NPV infection, 15 μg of HaTrx2 dsRNA (the “NPV2 + dsHaTrx2” group), 15 μg of HaTrxR1 dsRNA (the “NPV2 + dsHaTrxR1” group), or 15 μg of EGFP dsRNA (the “NPV2 + dsEGFP” group) was injected into the proleg of each H. armigera larva. The number of dead larvae was observed and recorded in each group until the larvae pupated. At least 30 larvae were included in each replicate, and every treatment was replicated three times.
Statistical analysis
The real-time PCR experiments and RNAi experiments were both carried out with three independent replications, and the results are presented as the means ± standard deviation (SD). Statistically significant differences in gene expression observed in the real-time PCR assays are denoted by *(0.01 < p < 0.05) and **(p < 0.01), as obtained through pair-wise Student’s t-test analysis. The mortality rate was analyzed using ANOVA followed by Turkey’s HSD multiple comparison test in SPSS 17.0 software to detect statistically significant differences between different groups (p < 0.05).
Additional Information
How to cite this article: Zhang, S. et al. Sequence analysis, expression profiles and function of thioredoxin 2 and thioredoxin reductase 1 in resistance to nucleopolyhedrovirus in Helicoverpa armigera. Sci. Rep. 5, 15531; doi: 10.1038/srep15531 (2015).
Supplementary Material
Acknowledgments
This study was supported by a grant from the Major State Basic Research Development Program of China (973 Program) (No. 2012CB114103). The authors thank Lihua Liang of IPM laboratory in China Agricultural University for providing the Helicoverpa armigera larvae.
Footnotes
Author Contributions S.Z. and X.L. conceived and designed the experiments, S.Z. Performed the experiments, S.Z. and X.N. analyzed the data, Z.L., X.N., F.W., Z.S. and B.Z. contributed reagents/materials/analysis tools, S.Z., Z.L., X.N., Q.Z. and X.L. wrote the paper.
References
- Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 82(1), 47–95 (2002). [DOI] [PubMed] [Google Scholar]
- Gertz M., Fischer F., Leipelt M., Wolters D. & Steegborn C. Identification of Peroxiredoxin 1 as a novel interaction partner for the lifespan regulator protein p66Shc. Aging (Albany NY) 1(2), 254–265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418 (2003). [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264(24), 13963–13966 (1989). [PubMed] [Google Scholar]
- Kobayashi-Miura M. et al. Oxygen sensing and redox signaling: the role of thioredoxin in embryonic development and cardiac diseases. Am. J. Physiol. Heart. Circ. Physiol. 292(5), H2040–2050 (2007). [DOI] [PubMed] [Google Scholar]
- Lu J. & Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 66, 75–87 (2014). [DOI] [PubMed] [Google Scholar]
- Powis G. & Montfort W. R. Properties and biological activities of thioredoxins. Annu. Rev. Pharmacol. Toxicol. 41, 261–295 (2001). [DOI] [PubMed] [Google Scholar]
- Laurent T. C., Moore E. C. & Reichard P. Enzymatic synthesis of deoxyribonucleotides. Iv. isolation and characterization of thioredoxin, the hydrogen donor from Escherichia Coli B. J. Biol. Chem. 239, 3436–3444 (1964). [PubMed] [Google Scholar]
- Arner E. S. & Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267(20), 6102–6109 (2000). [DOI] [PubMed] [Google Scholar]
- Kalinina E. V., Chernov N. N. & Saprin A. N. Involvement of thio-, peroxi-, and glutaredoxins in cellular redox-dependent processes. Biochemistry (Mosc) 73(13), 1493–1510 (2008). [DOI] [PubMed] [Google Scholar]
- Lee S., Kim S. M. & Lee R. T. Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance. Antioxid Redox Signal 18(10), 1165–1207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai J. & Jones D. P. Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett. 580(28-29), 6596–6602 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y. et al. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106(9), 3615–3620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann P. & Jacquot J. P. Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371–400 (2000). [DOI] [PubMed] [Google Scholar]
- Mishra S. & Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525(2), 145–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serata M., Iino T., Yasuda E. & Sako T. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology 158 (Pt 4), 953–962 (2012). [DOI] [PubMed] [Google Scholar]
- Kanzok S. M. et al. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science 291(5504), 643–646 (2001). [DOI] [PubMed] [Google Scholar]
- Bauer H., Kanzok S. M. & Schirmer R. H. Thioredoxin-2 but not thioredoxin-1 is a substrate of thioredoxin peroxidase-1 from Drosophila melanogaster: isolation and characterization of a second thioredoxin in D. Melanogaster and evidence for distinct biological functions of Trx-1 and Trx-2. J. Biol. Chem. 277(20), 17457–17463 (2002). [DOI] [PubMed] [Google Scholar]
- Svensson M. J., Chen J. D., Pirrotta V. & Larsson J. The ThioredoxinT and deadhead gene pair encode testis- and ovary-specific thioredoxins in Drosophila melanogaster. Chromosoma 112(3), 133–143 (2003). [DOI] [PubMed] [Google Scholar]
- Tsuda M. et al. Loss of Trx-2 enhances oxidative stress-dependent phenotypes in Drosophila. FEBS. Lett. 584(15), 3398–3401 (2010). [DOI] [PubMed] [Google Scholar]
- Kim Y. J. et al. Thioredoxin from the silkworm, Bombyx mori: cDNA sequence, expression, and functional characterization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 147(3), 574–581 (2007). [DOI] [PubMed] [Google Scholar]
- Corona M. & Robinson G. E. Genes of the antioxidant system of the honey bee: annotation and phylogeny. Insect Mol. Biol. 15(5), 687–701 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W. J. et al. Identification and antioxidant characterisation of thioredoxin-like1 from Apis cerana cerana. Apidologie 43, 737–752. (2012). [Google Scholar]
- Yao P. et al. Molecular cloning, expression and antioxidant characterisation of a typical thioredoxin gene (AccTrx2) in Apis cerana cerana. Gene 527(1), 33–41 (2013). [DOI] [PubMed] [Google Scholar]
- Yao P. et al. Glutaredoxin 1, glutaredoxin 2, thioredoxin 1, and thioredoxin peroxidase 3 play important roles in antioxidant defense in Apis cerana cerana. Free Radic. Biol. Med. 68, 335–346 (2014). [DOI] [PubMed] [Google Scholar]
- Argyrou A. & Blanchard J. S. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78, 89–142 (2004). [DOI] [PubMed] [Google Scholar]
- Williams C. H. et al. Thioredoxin reductase two modes of catalysis have evolved. Eur. J. Biochem. 267(20), 6110–6117 (2000). [DOI] [PubMed] [Google Scholar]
- Zhong L., Arner E. S. & Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc. Natl. Acad. Sci. USA 97(11), 5854–5859 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Sandalova T., Lindqvist Y. & Arner E. S. Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1. J. Biol. Chem. 284(6), 3998–4008 (2009). [DOI] [PubMed] [Google Scholar]
- Bauer H. et al. Thioredoxin reductase from the malaria mosquito Anopheles gambiae. Eur. J. Biochem. 270(21), 4272–4281 (2003). [DOI] [PubMed] [Google Scholar]
- Xia L. et al. The mammalian cytosolic selenoenzyme thioredoxin reductase reduces ubiquinone. A novel mechanism for defense against oxidative stress. J. Biol. Chem. 278(4), 2141–2146 (2003). [DOI] [PubMed] [Google Scholar]
- Sakurai A. et al. Overexpression of thioredoxin reductase 1 regulates NF-kappa B activation. J. Cell Physiol. 198(1), 22–30 (2004). [DOI] [PubMed] [Google Scholar]
- Conrad M. Transgenic mouse models for the vital selenoenzymes cytosolic thioredoxin reductase, mitochondrial thioredoxin reductase and glutathione peroxidase 4. Biochim. Biophys. Acta. 1790(11), 1575–1585 (2009). [DOI] [PubMed] [Google Scholar]
- Pennington J. D. et al. Thioredoxin and thioredoxin reductase as redox-sensitive molecular targets for cancer therapy. Curr. Pharm. Des. 13(33), 3368–3377 (2007). [PubMed] [Google Scholar]
- Prast-Nielsen S., Cebula M., Pader I. & Arner E. S. Noble metal targeting of thioredoxin reductase–covalent complexes with thioredoxin and thioredoxin-related protein of 14 kDa triggered by cisplatin. Free Radic. Biol. Med. 49(11), 1765–1778 (2010). [DOI] [PubMed] [Google Scholar]
- Missirlis F. et al. Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J. Biol. Chem. 277(13), 11521–11526 (2002). [DOI] [PubMed] [Google Scholar]
- Missirlis F., Phillips J. P. & Jackle H. Cooperative action of antioxidant defense systems in Drosophila. Curr. Biol. 11(16), 1272–1277 (2001). [DOI] [PubMed] [Google Scholar]
- Yang H., Kang M., Guo X. & Xu B. Cloning, structural features, and expression analysis of the gene encoding thioredoxin reductase 1 from Apis cerana cerana. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 156(3), 229–236 (2010). [DOI] [PubMed] [Google Scholar]
- Nair P. M. & Choi J. Characterization and transcriptional regulation of thioredoxin reductase 1 on exposure to oxidative stress inducing environmental pollutants in Chironomus riparius. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 161(2), 134–139 (2012). [DOI] [PubMed] [Google Scholar]
- Wu K. M., Lu Y. H., Feng H. Q., Jiang Y. Y. & Zhao J. Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321(5896), 1676–1678 (2008). [DOI] [PubMed] [Google Scholar]
- Lu Y., Wu K., Jiang Y., Guo Y. & Desneux N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 487(7407), 362–365 (2012). [DOI] [PubMed] [Google Scholar]
- Arner E. S. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim. Biophys. Acta. 1790(6), 495–526 (2009). [DOI] [PubMed] [Google Scholar]
- Holmgren A. & Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem. Biophys. Res. Commun. 396(1), 120–124 (2010). [DOI] [PubMed] [Google Scholar]
- Chen H. et al. Thioredoxin peroxidase gene is involved in resistance to biocontrol fungus Nomuraea rileyi in Spodoptera litura: gene cloning, expression, localization and function. Dev. Comp. Immunol. 44(1), 76–85 (2014). [DOI] [PubMed] [Google Scholar]
- Myers J. M., Antholine W. E. & Myers C. R. Hexavalent chromium causes the oxidation of thioredoxin in human bronchial epithelial cells. Toxicology 246(2-3), 222–233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak V. I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 101(1), 13–30 (2011). [DOI] [PubMed] [Google Scholar]
- Dimopoulos G. et al. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc. Natl. Acad. Sci. USA 99(13), 8814–8819 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel-Harel O. & Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54, 439–461 (2000). [DOI] [PubMed] [Google Scholar]
- Radyuk S. N., Sohal R. S. & Orr W. C. Thioredoxin peroxidases can foster cytoprotection or cell death in response to different stressors: over- and under-expression of thioredoxin peroxidase in Drosophila cells. Biochem. J. 371 (Pt 3), 743–752 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.J. & G, P. A new and practical artificial diet for the cotton bollworm. Entomol. Sinica. 14, 227–282 (1997). [Google Scholar]
- Li Z. et al. Cloning and characterization of a 2-cys peroxiredoxin in the pine wood nematode, Bursaphelenchus xylophilus, a putative genetic factor facilitating the infestation. Int. J. Biol. Sci. 7(6), 823–836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. et al. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555(2), 393–402 (2015). [DOI] [PubMed] [Google Scholar]
- An S., Zhang Y., Wang T., Luo M. & Li C. Molecular characterization of glutaredoxin 2 from Ostrinia furnacalis. Integr. Zool. 8 Suppl 1, 30–38 (2013). [DOI] [PubMed] [Google Scholar]
- Lawo N. C. et al. Effectiveness of Bacillus thuringiensis-transgenic chickpeas and the entomopathogenic fungus Metarhizium anisopliae in controlling Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 74(14), 4381–4389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhou F., Chu Y., Zhao Z. & An C. Identification and expression profile analysis of antimicrobial peptide/protein in Asian corn borer, Ostrinia furnacalis (Guenee). Int. J. Biol. Sci. 9(9), 1004–1012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C. & Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26(6), 509–515 (2004). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29(9), e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4), 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Jayachandran B., Hussain M. & Asgari S. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86(24), 13729–13734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. et al. Effects of nucleopolyhedrovirus infection on the development of Helicoverpa armigera (Lepidoptera: Noctuidae) and expression of its 20-hydroxyecdysone- and juvenile hormone-related genes. Florida Entomologist 98(2), 681–689 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.