Abstract
Non-alcoholic fatty liver disease (NAFLD) is an emerging health concern in both developed and non-developed world, encompassing from simple steatosis to non-alcoholic steatohepatitis (NASH), cirrhosis and liver cancer. Incidence and prevalence of this disease are increasing due to the socioeconomic transition and change to harmful diet. Currently, gold standard method in NAFLD diagnosis is liver biopsy, despite complications and lack of accuracy due to sampling error. Further, pathogenesis of NAFLD is not fully understood, but is well-known that obesity, diabetes and metabolic derangements played a major role in disease development and progression. Besides, gut microbioma and host genetic and epigenetic background could explain considerable interindividual variability. Knowledge that epigenetics, heritable events not caused by changes in DNA sequence, contribute to development of diseases has been a revolution in the last few years. Recently, evidences are accumulating revealing the important role of epigenetics in NAFLD pathogenesis and in NASH genesis. Histone modifications, changes in DNA methylation and aberrant profiles or microRNAs could boost development of NAFLD and transition into clinical relevant status. PNPLA3 genotype GG has been associated with a more progressive disease and epigenetics could modulate this effect. The impact of epigenetic on NAFLD progression could deserve further applications on therapeutic targets together with future non-invasive methods useful for the diagnosis and staging of NAFLD.
Keywords: Non-alcoholic steatohepatitis, Epigenetics, Diagnosis, Treatment, Non-alcoholic fatty liver disease
Core tip: The interplay of environmental and host factors results in non-alcoholic fatty liver disease (NAFLD) development and influence its progression individually. Nevertheless, the physiopathology of this disease remains unclear, so this lack of knowledge avoids the development of new therapeutic approaches and non-invasive biomarkers. Epigenetics could be an interesting alternative to cover these issues, considering the amount of evidence accumulated in order to clarify its role on NAFLD.
EPIDEMIOLOGY
Non-alcoholic Fatty liver disease (NAFLD) is defined as an accumulation of fat in the liver in absence of significant alcohol consumption, hereditary disease or drugs[1]. It constitutes a clinicopathological disease comprising a wide spectrum of disorders, ranging from simple steatosis (SS), initially benign, to non-alcoholic steatohepatitis (NASH), accompanied by inflammation and/or hepatocellular damage. These two entities, SS and NASH, show different natural history, evolution and consequences, through necroinflammation, fibrosis, cirrhosis and even hepatocellular carcinoma. The strongest predictor of fibrosis progression in NAFLD is the presence of steatohepatitis, mainly lobular inflammation and ballooning[2].
Currently, NAFLD is considered the most common chronic liver disease in developed countries[3]. Its worldwide prevalence in general population is estimated to be around 20%-30%[4] in Western countries and 5%-18% in Asia[5], being a common and underdiagnosed condition. The reason for this variability remains unclear, but presumable genetic and epigenetics factors play an important role.
NAFLD is often associated with clinical features of metabolic syndrome, such as central obesity, insulin resistance, type 2 diabetes mellitus, arterial hypertension and dyslipidaemia[6]. Sedentary lifestyles and changes in dietary patterns are responsible for an increased prevalence of obesity and insulin resistance, leading to an increased prevalence of this disease, projected to be the top cause for liver transplantation within the next decade[7]. Furthermore, this disease is related to different systemic disorders, like cardiovascular disease[8,9].
DIAGNOSIS, MANAGEMENT AND TREATMENT
Percutaneous liver biopsy is often recommended in patients with unexplained elevated serum aminotransferases, constituting the gold standard method in the diagnosis of fibrosis and steatohepatitis. It shows inherent limitations, as high costs and associated morbidity, which can lead to major complications (i.e., bleeding, and even death)[10]. Non-invasive methods have been recently developed in order to diagnose non-alcoholic steatohepatitis, such as imaging tests (transient elastography, acoustic radiation force impulse and magnetic resonance elastography) as well as biomarkers (cytokeratin-18 and fibroblast grown factor 21).
NAFLD pathogenesis can be resumed as the excessive accumulation of fat in hepatocytes, leading to increased intracellular vacuoles of fat, lack of capacity for mitochondrial beta-oxidation, oxidative stress, pro-inflammatory mechanisms and hepatocellular apoptosis[11,12]. Since pharmacological treatment for NAFLD remains ineffective[13], the first-line approach for these patients is the reduction on total body weight achieved by decrease of energy consumption and increase of exercise.
EPIGENETIC MECHANISMS
Historically, the term of epigenetic was coined by Conrad Waddington in the 1940s as the branch of biology which study the causal interactions between genes and their products which bring the phenotype into being[14]. Currently, epigenetic modification is defined as phenotypic changes in gene expression that can be inherited through mitosis and/or meiosis caused by an adaptive mechanism unrelated to alteration of primary DNA sequences[15,16].
Epigenetic phenomena are heritable adaptive mechanisms considered reversible, since they are being modulated by environmental stimuli. Disruption of the balance could lead to the development of serious disorders. The most described epigenetic modifications include: (1) histone modification; (2) DNA methylation; (3) microRNAs; and (4) chromatin remodelling. To date, the most intensively studied epigenetic mechanisms in NAFLD are DNA methylation and microRNAs (Figure 1).
Figure 1.
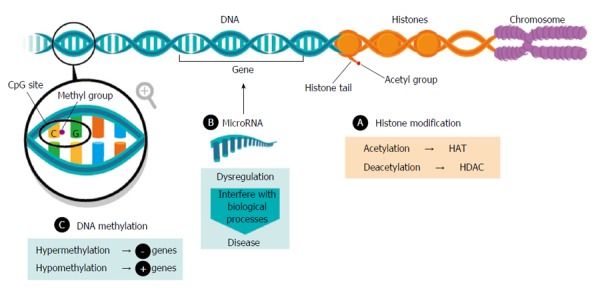
Epigenetic phenomena. HAT: Histone acetyltransferase; HDAC: Histone deacetylase.
HISTONE MODIFICATION
Modifications of the amino-terminal tails histones constitute an important determinant of chromatin structure and gene expression[17]. Aberrant histone modifications promote the development of insulin resistance and consequently, NAFLD[18]. Among the most common modifications acetylation has been reported, associated with gene transcription activation and catalysed by histone acetyltransferase (HAT) and deacetylation, involved in gene repression, and catalysed by histone deacetylase (HDAC).
So far, the most important findings in NAFLD have been described in mice. Imbalance between HAT and HDAC influences gene expression profile on NAFLD, resulting on liver injury[19]. p300 is a transcriptional coactivator that belongs to the HAT family. It has been identified as a key upstream regulator of carbohydrate-responsive element-binding protein activity, an activator of glycolytic and lipogenic genes, so inhibition of its activity may be beneficial for treating hepatic steatosis[20]. Several HDCA also play important roles on NAFLD. HDAC3 displays a circadian rhythm in mouse liver controlling hepatic lipogenesis, and disruption of this mechanism exacerbates metabolic diseases, including obesity and diabetes. Its depletion reroutes metabolic precursors toward lipid synthesis and storage in lipid droplets[21,22]. Sirtuins (SIRT) are master metabolic regulators with protective roles against obesity, glucose and lipid metabolism. Activation of SIRT1 shows potential against NAFLD-related physiological mechanisms, and it has been found increased plasma levels in NAFLD obese patients[23]. SIRT1 could play a dual role, acting as a potential therapeutic target and a noninvasive biomarker on NAFLD[11].
DNA METHYLATION
Many methylations occur in the liver, and hepatic steatosis is often view from the standpoint of the deregulation of one-carbon metabolism, being related to folate deficiency[24]. DNA methylation is the addition of a methyl group on cytosine with guanine as the next nucleotide, also known as CpG site[25].
DNA methylation plays a central role in the regulation of gene expression, representing a level of epigenetic regulation, which is commonly associated with transcriptional repression and chromatin accessibility. Aberrant DNA methylation patterns of genomic stability affect cell homeostasis, such as hypermethylation, associated with gene repression, and hypomethylation, related to gene activation. In mice, kick-off of steatosis is accompanied by alterations in DNA methyltranferases (DNMT) expression in the liver[26]. In humans, hepatic DNMT was found higher in NASH than SS patients and significantly associated with NAS Score[27]. Abnormal DNA methylation is the hallmark of carcinogenesis; this process has been studied in hepatocellular carcinoma (HCC)[28], more specifically in NAFLD-HCC. Metabolites derived from metabolic syndrome, such as insulin, glucose and lipids could perturb epigenetic gene regulation leading to a pro-inflammatory status and disturbing metabolic pathways[29]. Furthermore, DNA methylation signatures can be remodelled by transcriptional factors, so it has also being evaluated after bariatric surgery and the massive loss weight that entails, suggesting that NAFLD-associated methylation changes could be partially reversible[30]. It has been reported that functionally relevant differences in methylation could distinguish between mild and advanced NAFLD in 100 human frozen liver biopsies. Moreover, in patients with advanced vs mild NAFLD, 69247 differentially methylated CpG sites (78% hypomethylated, 24% hypermethylated) were described. These findings indicate that differential methylation contributes to differences in expression[31].
MICRORNAS
MicroRNAs (miRNAs) are receiving growing attention because they are commonly deregulated in pathological situations, being the most extensively investigated epigenetic mechanism in NAFLD. MiRNAs constitute a class of small, single-stranded non-coding RNA highly conserved, acting as regulators of gene expression and protein translation. They can interfere in each single aspect of cellular activity, such as differentiation and development, proliferation, metabolism, apoptosis and tumorigenesis[32]. A single miRNA could target multiple genes (multiplicity) and multiple miRNAs could target a just one gene (cooperativity). Taking into account its large potential roles on carcinogenesis, miRNAs could also be categorized as oncogenes (onco-miRNAs) or tumour suppressors[33]. It has been shown their stability in serum, plasma, urine and saliva. Circulating miRNAs, protected from degradation by RNAses contained in body fluids, are currently extensively studied for noninvasive diagnosis of a sort of liver diseases[34], including NAFLD[35]. Thereof, it has been identified several miRNAs in serum/plasma of NAFLD patients that show diagnostic potential for defining different phenotypes of this disease, from simple steatosis to NASH, going through fibrosis[36].
Actually, the major importance or miRNAs on NAFLD stands on the discrimination of NASH and the diagnosis of HCC. In this sense miR-122, the most expressed miRNA in human liver has been reported significantly under-expressed in NASH[37], acting as a tumor-supressor in the liver[29,38]. It has been proposed as a potential therapeutic target in the treatment of hypercholesterolemia[39] or different dyslipidaemia[40].
Besides miR-122, other miRNAs have been demonstrated to be involved in NAFLD. It has been reported a link between liver cell apoptosis, miR-34a/SIRT1/p53 signalling and NAFLD severity[41], and recently, miR-21 seems to regulate triglyceride and cholesterol metabolism in vivo and in vitro[42]. Moreover, mir-24 is robustly induced in fatty acids treated human hepatocytes, HepG2 cells and high-fat diet mice, revealing the potential role of miR-24 inhibitor as a promising therapeutic target for NAFLD[43]. miR-33a and miR-33b also inhibit genes involved in fatty acids metabolism and insulin signalling in hepatocytes, regulating pathways controlling three risk factors of metabolic syndrome, HDL levels, triglycerides, and insulin[44]. The miR-200 family and others, like miR-155, are also related with NAFLD[33] (Table 1).
Table 1.
Dysregulated miRNAs in non-alcoholic fatty liver disease
| miR | Modulation | Experimental model |
| miR-15b[45] | Upregulation | In vitro/rats/human |
| miR-34a[46] | Upregulation | Human |
| miR-221[47] | Upregulation | In vitro |
| miR-222[47] | Upregulation | In vitro |
| miR-155[48] | Downregulation | Mice |
| miR-198[37] | Downregulation | Human |
| miR-451[37] | Downregulation | Human |
| miR-146b[37] | Upregulation | Human |
Besides, there are still some barriers to the therapeutic use of miRNAs. MiRNAs can be degraded by endogenous RNAses and affect several pathways in different organs, so caution is needed to avoid undesirable adverse effects.
POTENTIAL ROLE OF EPIGENETICS ON NAFLD MANAGEMENT
NAFLD is a complex disease trait where interpatient genetic[49] and epigenetic variations and environmental factors are combined to define development and disease progression. Upsetting the balance of cellular homeostasis, either modifying some of these mechanisms, could trigger disease development. Since epigenetic phenomena are reversible, novel therapies intended to modulate epigenetic abnormalities are trend. Altered epigenetics patterns could distinguish between NAFLD stages, but a better understanding of the molecular mechanisms is mandatory to identify reliable biomarkers and effective treatments. Among epigenetic mechanisms, miRNAs occupy a top position, because their disturbances present potential prognostic and diagnostic, and the ability to be therapeutic targets. Further research is needed to increase knowledge of the role that epigenetics mechanisms could play in determining most aggressive phenotypes of NAFLD. This could lead to disease stratification, from simple steatosis to non-alcoholic steatohepatitis, in order to target therapies, providing new tracks in NAFLD pathogenesis.
Footnotes
P- Reviewer: Jin B, Procopet B S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
Supported by European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement, No. HEALTH-F2-2009-241762 for the project FLIP; and Consejería de Salud de la Junta de Andalucía, No. PI-0488/2012.
Conflict-of-interest statement: The authors affirm there is no conflict-of-interest to declare.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: January 28, 2015
First decision: March 6, 2015
Article in press: October 19, 2015
References
- 1.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 2.Farrell GC, van Rooyen D, Gan L, Chitturi S. NASH is an Inflammatory Disorder: Pathogenic, Prognostic and Therapeutic Implications. Gut Liver. 2012;6:149–171. doi: 10.5009/gnl.2012.6.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 4.Bellentani S, Scaglioni F, Marino M, Bedogni G. Epidemiology of non-alcoholic fatty liver disease. Dig Dis. 2010;28:155–161. doi: 10.1159/000282080. [DOI] [PubMed] [Google Scholar]
- 5.Masarone M, Federico A, Abenavoli L, Loguercio C, Persico M. Non alcoholic fatty liver: epidemiology and natural history. Rev Recent Clin Trials. 2014;9:126–133. doi: 10.2174/1574887109666141216111143. [DOI] [PubMed] [Google Scholar]
- 6.Cordero P, Campion J, Milagro FI, Martinez JA. Transcriptomic and epigenetic changes in early liver steatosis associated to obesity: effect of dietary methyl donor supplementation. Mol Genet Metab. 2013;110:388–395. doi: 10.1016/j.ymgme.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–344. doi: 10.1038/nrgastro.2013.41. [DOI] [PubMed] [Google Scholar]
- 8.Ampuero J, Romero-Gómez M. [Influence of non-alcoholic fatty liver disease on cardiovascular disease] Gastroenterol Hepatol. 2012;35:585–593. doi: 10.1016/j.gastrohep.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Ampuero J, Gallego-Durán R, Romero-Gómez M. Association of NAFLD with subclinical atherosclerosis and coronary-artery disease: meta-analysis. Rev Esp Enferm Dig. 2015;107:10–16. [PubMed] [Google Scholar]
- 10.Piccinino F, Sagnelli E, Pasquale G, Giusti G. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J Hepatol. 1986;2:165–173. doi: 10.1016/s0168-8278(86)80075-7. [DOI] [PubMed] [Google Scholar]
- 11.Colak Y, Yesil A, Mutlu HH, Caklili OT, Ulasoglu C, Senates E, Takir M, Kostek O, Yilmaz Y, Yilmaz Enc F, et al. A potential treatment of non-alcoholic fatty liver disease with SIRT1 activators. J Gastrointestin Liver Dis. 2014;23:311–319. doi: 10.15403/jgld.2014.1121.233.yck. [DOI] [PubMed] [Google Scholar]
- 12.Gallego-Durán R, Ampuero J, Funuyet J, Romero-Gómez M. [Alcoholic and non-alcoholic steatohepatitis: who is affected and what can we do for them?] Gastroenterol Hepatol. 2013;36:587–596. doi: 10.1016/j.gastrohep.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Musso G, Gambino R, Cassader M, Pagano G. A meta-analysis of randomized trials for the treatment of nonalcoholic fatty liver disease. Hepatology. 2010;52:79–104. doi: 10.1002/hep.23623. [DOI] [PubMed] [Google Scholar]
- 14.Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann N Y Acad Sci. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- 15.Li YY. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6546–6551. doi: 10.3748/wjg.v18.i45.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi SW, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Friso S, Choi SW. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients. 2014;6:3303–3325. doi: 10.3390/nu6083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bricambert J, Miranda J, Benhamed F, Girard J, Postic C, Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120:4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariani S, Fiore D, Basciani S, Persichetti A, Contini S, Lubrano C, Salvatori L, Lenzi A, Gnessi L. Plasma levels of SIRT1 associate with non-alcoholic fatty liver disease in obese patients. Endocrine. 2015;49:711–716. doi: 10.1007/s12020-014-0465-x. [DOI] [PubMed] [Google Scholar]
- 24.da Silva RP, Kelly KB, Al Rajabi A, Jacobs RL. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40:277–283. doi: 10.1002/biof.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5’ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 26.Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA, Latendresse JR, Rusyn I, Beland FA. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176–186. doi: 10.1016/j.jhep.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 28.Ammerpohl O, Pratschke J, Schafmayer C, Haake A, Faber W, von Kampen O, Brosch M, Sipos B, von Schönfels W, Balschun K, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130:1319–1328. doi: 10.1002/ijc.26136. [DOI] [PubMed] [Google Scholar]
- 29.Tian Y, Wong VW, Chan HL, Cheng AS. Epigenetic regulation of hepatocellular carcinoma in non-alcoholic fatty liver disease. Semin Cancer Biol. 2013;23:471–482. doi: 10.1016/j.semcancer.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 33.Gori M, Arciello M, Balsano C. MicroRNAs in nonalcoholic fatty liver disease: novel biomarkers and prognostic tools during the transition from steatosis to hepatocarcinoma. Biomed Res Int. 2014;2014:741465. doi: 10.1155/2014/741465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Huang C, Zhang SP, Sun X, Long XR, Li J. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268–2272. doi: 10.1016/j.cellsig.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 35.Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, Gursoy S, Guven K, Ozbakır O, Dundar M, et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol. 2014;6:613–620. doi: 10.4254/wjh.v6.i8.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaki Y, Saito Y, Takasugi A, Toshimitsu K, Yamada S, Muramatsu T, Kimura M, Sugiyama K, Suzuki H, Arai E, et al. Silencing of microRNA-122 is an early event during hepatocarcinogenesis from non-alcoholic steatohepatitis. Cancer Sci. 2014;105:1254–1260. doi: 10.1111/cas.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 40.Sacco J, Adeli K. MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Curr Opin Lipidol. 2012;23:220–225. doi: 10.1097/MOL.0b013e3283534c9f. [DOI] [PubMed] [Google Scholar]
- 41.Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol. 2013;58:119–125. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Sun C, Huang F, Liu X, Xiao X, Yang M, Hu G, Liu H, Liao L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med. 2015;35:847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 43.Ng R, Wu H, Xiao H, Chen X, Willenbring H, Steer CJ, Song G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology. 2014;60:554–564. doi: 10.1002/hep.27153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Cheng X, Lu Z, Wang J, Chen H, Fan W, Gao X, Lu D. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Res Clin Pract. 2013;99:327–334. doi: 10.1016/j.diabres.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 46.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 48.Miller AM, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA, Fernández-Hernando C, McInnes IB, Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anstee QM, Day CP. The genetics of NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10:645–655. doi: 10.1038/nrgastro.2013.182. [DOI] [PubMed] [Google Scholar]


