Abstract
Genetic variation in drug detoxification pathways may influence outcomes in pediatric acute lymphoblastic leukemia (ALL). We evaluated relapse risk and 24 variants in 17 genes in 714 patients in CCG-1961. Three TPMT and 1 MTR variant were associated with increased risks of relapse (rs4712327, OR 3.3, 95%CI 1.2–8.6; rs2842947, OR 2.7, 95%CI 1.1–6.8; rs2842935, OR 2.5, 95%CI 1.1–5.0; rs10925235, OR 4.9, 95%CI 1.1–25.1). One variant in SLC19A1 showed a protective effect (rs4819128, OR 0.5, 95%CI 0.3–0.9). Our study provides data that relapse risk in pediatric ALL is associated with germline variations in TPMT, MTR and SLC19A1.
Keywords: Acute lymphoblastic leukemia, SNPs, Relapse, Folate pathway
Highlights
-
•
Genetic variants in the folate pathway may influence ALL outcome.
-
•
We evaluated 24 SNPs in 17 genes on relapse risk in pediatric ALL.
-
•
Three TPMT variants were associated with an increased risk of relapse.
-
•
One MTR variant was positively associated with risk of relapse.
-
•
One variant in SLC19A1 showed a protective effect on relapse.
-
•
TPMT, MTR and SLC19A1 are associated with relapse risk in pediatric ALL.
1. Introduction
The introduction of risk-adapted intensified combination therapy has markedly improved outcomes for children with acute lymphoblastic leukemia (ALL). Unfortunately, disease recurrence and treatment-induced toxicity remain clinically relevant risks [1]. Candidate gene studies previously reported that polymorphisms in genes encoding for enzymes in folate metabolism and drug detoxification pathways (e.g. TPMT, MTHFR, CBS, MTR, and DHFR) may modify relapse and treatment toxicity risk [2,3]. We sought to evaluate single nucleotide polymorphism (SNPs) in these pathways to validate previous reports. We expanded our previous candidate gene genotyping efforts to include 24 SNPs in 17 genes (MTHFR, MTR, PTH1R, ABCG2, MTRR, TPMT, SOD2, ABCB1, GGH, ENOSF1, GSTP1, LRP5, NQO1, SHMT1, TYMS, SLC19A1, and CBS) in the folate and drug metabolizing pathway for their association with risk of relapse in 714 children who had been treated in the CCG-1952 study [4]. Between 1996 and 2000, a total of 2027 pediatric patients were enrolled in the CCG-1952 study. This study compared the use of intrathecal (IT) methotrexate to IT triple (ITT) chemotherapy (methotrexate, cytarabine, and hydrocortisone sodium succinate) and oral mercaptopurine to thioguanine in treating children with standard-risk ALL. Guardians provided written informed consent and permission for the use of archived samples in accordance with National Institutes of Health guidelines.
The primary outcome in the present study was ALL relapse, defined as an isolated or combined M3 (>25% blasts) disease recurrence event in the bone marrow and/or extramedullary sites after achieving initial remission. Occasionally bone marrow relapse was defined as M2 (5–25% blasts) with confirmation at relapse of the diagnostic cytogenetic clone. Isolated central nervous system relapse was defined as ≥5 white blood cells per microliter in the cerebrospinal fluid, with morphologically identifiable blasts on cytospin. Germline DNA on 714 samples was collected at remission and extracted from remission bone marrow aspirate specimens using the Qiagen Puregene Core Kit A. Genotyping was performed using either Pyrosequencing, Taqman Open Array platforms, direct sequencing, or standard restriction fragment length polymorphism. All SNPs were in Hardy–Weinberg equilibrium with a minor allele frequency (AF) exceeding 10%.
Logistic and Cox regression models were used to evaluate the differences in relapse rate and time to relapse for all SNPs, testing the effects of heterozygous (AB) and homozygous recessive (BB) patients separately. In both cases the dominant homozygous (AA) patients were chosen as the reference category. All risk estimates were adjusted for age, race, gender and immunophenotype. The difference in relapse rates among genotypes of significantly associated SNPs are visualized with Kaplan–Meier curves.
2. Results
From the 2027 patients in the overall cohort, genotyping information was available on 714 patients. Female, white, and patients with B-cell lineage progenitor ALL were slightly more represented in the genotyped samples (Table 1). There was no difference in the occurrence of relapse between genotyped and non-genotyped patients (334/2027 [16.5%] versus 112/714 [15.3%]).
Table 1.
Patient characteristics of the overall and genotyped sub-cohort of the CCG-1952 study.
| CCG-1952 |
Overall |
Genotyped |
P | |||
|---|---|---|---|---|---|---|
| Characteristics | N | % | N | % | ||
| n | 2027 | 100.0 | 714 | 100.0 | ||
| Age | 1–1.99 | 165 | 8.1 | 62 | 8.7 | 0.78 |
| 2–5.99 | 1402 | 69.2 | 493 | 69.0 | ||
| 6–9.99 | 460 | 22.7 | 159 | 22.3 | ||
| Sex | Male | 1129 | 55.7 | 375 | 52.5 | 0.03 |
| Female | 898 | 44.3 | 339 | 47.5 | ||
| Race/Ethnicity | White | 1376 | 68.2 | 537 | 75.2 | <0.01 |
| Black | 72 | 3.6 | 23 | 3.2 | ||
| Hispanic | 444 | 22.0 | 98 | 13.7 | ||
| Other | 125 | 6.2 | 56 | 7.8 | ||
| Immunophenotype | B-cell | 1802 | 93.3 | 577 | 95.8 | <0.01 |
| T-cell | 130 | 6.7 | 25 | 4.2 | ||
| Relapse | No | 1693 | 83.5 | 602 | 84.3 | 0.47 |
| Yes | 334 | 16.5 | 112 | 15.7 | ||
Total summations of less than 2027 patients in the overall cohort, or less than 714 patients in the genotyped sub-cohort, are due to missing observations.
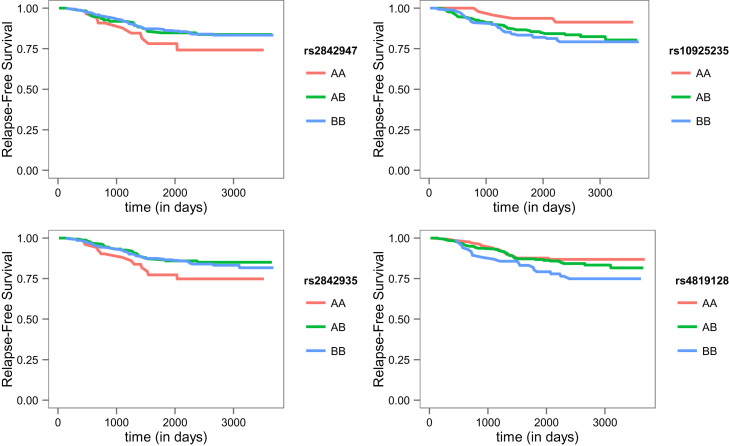
Genotype association results are presented in Table 2. A subset of SNPs in the TPMT gene, specifically variants in rs4712327, rs2842947, and rs2842935, were associated with increased risks of relapse (OR 3.3, 95%CI 1.2–8.6; OR 2.7, 95%CI 1.1–6.8; OR 2.5, 95%CI 1.1–5.0, respectively). These three SNPs were in high linkage disequilibrium (LD) (min. D′=0.77, 1000 Genomes Project). Additionally, one SNP in the MTR gene, rs10925235, was associated with relapse (OR 4.9, 95%CI 1.1–25.1) (Fig. 1). SLC19A1 variant rs4819128 showed a significant protective effect on relapse both in heterozygote (OR 0.5, 95%CI 0.3–0.9) and recessive homozygote (OR 0.4, 95%CI 0.2–1.0) patients. Borderline significant protective effects were observed for two SNPs upstream of CBS: rs466791 (OR 0.6, 95%CI 0.3–1.2) and rs719038 (OR 0.4, 95%CI 0.1–1.2). A borderline significant association between the MTHFR C677T polymorphism and relapse-free survival was observed; both heterozygote and homozygote patients showed a >2.5 fold increased risk for relapse.
Table 2.
Odds ratios and hazard ratios of SNPs on ALL relapse.
| Gene | Allele | Size | Relapse | Remission | Heterozygote | Homozygote | Heterozygote | Homozygote | |
|---|---|---|---|---|---|---|---|---|---|
| SNP rs# | A | B | n | BB/AB/AA | BB/AB/AA | OR (95%CI) | OR (95%CI) | HR (95%CI) | HR (95%CI) |
| MTHFR | |||||||||
| rs1801131 | A | C | 637 | 3/49/47 | 62/219/257 | 3.2 (0.9–11) | 2.6 (0.8–8.9) | 2.9 (0.9−9.4) | 2.5 (0.8−8.2) |
| rs1801133 | C | T | 525 | 17/30/37 | 51/193/197 | 0.7 (0.4–1.2) | 1.4 (0.6–3.0) | 0.7 (0.4−1.2) | 1.4 (0.7−2.7) |
| MTR | |||||||||
| rs10925235 | C | T | 417 | 4/32/34 | 49/160/138 | 4.1 (0.9–18) | 4.9 (1.1–22) | 3.7 (0.9−15.7) | 4.1 (1.0−17.4) |
| rs1805087 | A | G | 478 | 4/19/49 | 16/100/290 | 1.3 (0.7–2.4) | 1.8 (0.6–5.9) | 1.2 (0.7−2.1) | 1.5 (0.5−4.4) |
| PTH1R | |||||||||
| rs1138518 | C | T | 305 | 5/22/15 | 36/120/107 | 1.4 (0.4–4.6) | 1.0 (0.3–3.5) | 1.3 (0.4−3.9) | 1 (0.3−3.2) |
| ABCG2 | |||||||||
| rs2231142 | G | T | 327 | 1/11/34 | 2/45/234 | 2.1 (0.9–4.9) | 5.5 (0.5–68) | 1.9 (0.9−4.1) | 4.3 (0.6−32.2) |
| MTRR | |||||||||
| rs1532268 | C | T | 393 | 3/32/29 | 38/132/159 | 4.0 (0.9–18) | 2.6 (0.6–12) | 3.5 (0.8–15.0) | 2.7 (0.6−11.4) |
| rs162036 | A | G | 415 | 2/21/47 | 13/100/232 | 1.0 (0.5–1.9) | 0.3 (0.0–2.6) | 1.1 (0.6−1.9) | 0.4 (0.1−2.8) |
| TPMT | |||||||||
| rs2842947 | C | T | 484 | 50/19/8 | 272/110/25 | 1.5 (0.8–2.8) | 2.7 (1.1–6.8) | 1.6 (0.9−2.8) | 2.2 (1.0−4.8) |
| TPMT | |||||||||
| rs4712327 | G | A | 377 | 8/15/38 | 21/85/210 | 1.2 (0.6–2.4) | 3.3 (1.2–8.6) | 1.2 (0.6−2.4) | 2.4 (1.1−5.3) |
| rs2842935 | C | T | 487 | 43/23/12 | 225/145/39 | 2.5 (1.0–5.0) | 2.5 (1.1–5.0) | 0.5 (0.2–1.0) | 0.5 (0.2−0.9) |
| SOD2 | |||||||||
| rs4880 | A | G | 320 | 16/25/12 | 76/138/53 | 1.1 (0.5–2.5) | 1.1 (0.4–2.8) | 1 (0.5–2.2) | 1 (0.4–2.4) |
| ABCB1 | |||||||||
| rs1045642 | G | A | 339 | 7/25/15 | 68/151/73 | 0.9 (0.4–2.1) | 0.4 (0.1–1.3) | 1 (0.5–1.9) | 0.4 (0.2–1.2) |
| GGH | |||||||||
| rs11545078 | G | A | 337 | 2/9/35 | 16/43/232 | 1.7 (0.7–4.1) | 1.1 (0.2–5.1) | 1.5 (0.7–3.3) | 1 (0.2–4.3) |
| GSTP1 | |||||||||
| rs1695 | A | G | 328 | 6/24/17 | 34/118/129 | 1.9 (0.9–4.1) | 1.2 (0.4–3.9) | 1.6 (0.8–3.3) | 1.4 (0.5–3.9) |
| rs1138272 | C | T | 320 | 1/16/25 | 1/101/176 | 0.2 (0.0–3.6) | 0.1 (0.0–2.3) | 0.3 (0.0–2.6) | 0.2 (0.0–1.6) |
| LRP5 | |||||||||
| rs2306862 | C | T | 345 | 4/14/28 | 7/74/218 | 0.7 (0.1–4.4) | 0.6 (0.1–3.5) | 0.5 (0.1–2.3) | 0.4 (0.1–1.8) |
| NQO1 | |||||||||
| rs1800566 | G | A | 331 | 3/9/32 | 22/102/163 | 0.5 (0.2–1.1) | 0.2 (0.0–1.8) | 0.5 (0.2–1.2) | 0.3 (0–2.1) |
| SHMT1 | |||||||||
| rs2273026 | C | T | 461 | 1/13/61 | 5/94/287 | 0.7 (0.3–1.4) | 0.8 (0.1–7.1) | 0.7 (0.3–1.3) | 0.8 (0.1–5.9) |
| TYMS | |||||||||
| rs2853533 | C | G | 312 | 25/17/4 | 168/79/19 | 1.2 (0.3–5.2) | 0.7 (0.2–2.8) | 1.3 (0.4–4.4) | 0.7 (0.2–2.5) |
| ENOSF1 | |||||||||
| rs3744962 | A | G | 422 | 0/11/61 | 4/77/269 | 0.6 (0.3–1.4) | na | 0.7 (0.3–1.4) | na |
| SLC19A1 | |||||||||
| rs4819128 | C | T | 478 | 17/40/20 | 116/221/64 | 0.5 (0.3–0.9) | 0.4 (0.2–1.0) | 0.6 (0.3–1.0) | 0.5 (0.3–1.0) |
| CBS | |||||||||
| rs466791 | C | T | 411 | 0/14/55 | 8/92/242 | 0.6 (0.3–1.2) | na | 0.6 (0.3–1.1) | na |
| rs719038 | A | G | 398 | 9/28/23 | 23/158/157 | 0.4 (0.1–1.2) | 0.4 (0.1–1.1) | 0.5 (0.2–1.1) | 0.4 (0.2–1.0) |
Single nucleotide polymorphisms (SNPs) are grouped by gene in relapsed and unaffected patients, together with odds ratios (OR), 95% confidence intervals (CI) and p-values (P). All estimates have been adjusted for the potential covarying factors of: age, race, gender, and immunophenotype. BB, recessive homozygote; AB, heterozygote; AA, dominant homozygote.
Fig. 1.
Relapse-free survival of selected SNPs in pediatric ALL. Kaplan–Meier curves for relapse-free survival stratified by SNP genotype, e.g. dominant homozygote (AA), heterozygote (AB) and recessive homozygote (BB).
3. Discussion
Our results confirm that germline variations in the TPMT gene are associated with treatment outcome in children with ALL [5]. The ability of TPMT to catalyze S-methylation and inactivate 6-mercaptopurine makes it a major determinant of 6-mercaptopurine toxicity. Furthermore, reduced TPMT activity is associated with both methotrexate availability and occurrence of minimal residual disease after induction chemotherapy [6]. While these intronic SNPs may have an unknown regulatory function, it is more likely that the observed effects of intronic SNPs are due to a nearby exonic TPMT variant. All three intronic SNPs are in close LD with the TPMT*2 variant (min. D′=0.67).
The MTHFR gene modulates methotrexate toxicity and is another well-investigated example of pharmacogenetics in pediatric ALL treatment-related outcome [3,7,8]. The observed borderline statistical significance may be a sample size issue as our study contains merely 3 relapse patients with the MTHFR 677TT genotype.
Previously Sepe et al. observed a significant association between the MTRR G66A (rs1801394) variant allele and relapse risk among 557 patients in the predecessor COG study, CCG-1891 [3]. Another study reported a relationship between the MTRR G66A variant and in vitro methotrexate sensitivity among 119 patients but did not investigate treatment outcome [9]. In the current study, the MTRR variant rs1532268 was borderline associated with relapse risk and is in moderate LD with MTRR G66A (D′=0.37), whereas rs162036 which is in high LD with MTRR G66A (D′=1.00) was not associated with relapse. Since the recessive homozygous variant of MTR G66A was not observed in our study, we believe we are underpowered to detect associations in this SNP.
In our data, an intronic MTR variant (rs10925235) was highly associated with ALL treatment outcome whereas the known missense mutation in MTR A2756G (rs1805087) was not. The two SNPs are in perfect LD and point towards the same direction of association, however the intronic variant has a higher allele frequency than the exonic variant (AF=0.35 in rs10925235, AF=0.21 in rs1805087). The gain in statistical power from the higher allele frequency may explain why significance was reached by this intronic variant and not in the missense MTR A2756G mutation.
Our finding that multiple SNPs upstream of CBS (rs719038, rs466791) may be associated with the response to anti-leukemic therapy in childhood ALL has not been previously reported. Within CBS, two consecutive SNPs showed a consistent trend towards a protective effect for relapse, although the trend was not statistically significant.
Previous studies show varied results on the role of GSPT1 in ALL treatment response, ranging from a protective [2,10] to a risk association between the GSTP1 Val105/Val105 (rs1695) genotype and relapse [3]. Our data suggest an increased risk, although not statistically significant (OR for heterozygotes of 1.9, 95%CI 0.9–4.1) and indicate that GSTP1 variants might have a modest effect on the modification of relapse risk.
Various observational studies have highlighted the importance of the folate pathway in chemotherapeutic drug response, but the specific results across these studies have been inconsistent [1]. The current study provides additional data to support the hypothesis that the risk of relapse after pediatric ALL treatment is associated with germline variation in genes (e.g. TPMT, MTHFR and MTR) that encode for enzymes that metabolize chemotherapeutic agents. Additional work is needed to refine these findings and to determine which germline variants beyond TPMT have pharmacogenomic potential for pediatric ALL treatment.
Conflict of interest statement
There are no conflicts of interests.
Authors' contributions
The authors have made substantial contributions to: (1) the conception and design of the study (MV, AK, RA), (2) acquisition of data (MV, LW, TM, LS, MD, SC, RA), (3) analysis and interpretation of data (MV, AK, RA), (4) drafting the article or revising it critically for important intellectual content (MV, AK, LW, TM, SC, MD, LS, RA), and (5) final approval of the version to be submitted (MV, AK, LW, TM, SC, MD, LS, RA).
Acknowledgments
The authors thank our clinical and research faculty and staff and the patients and their families for participating.
References
- 1.Pui C.H., Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat. Rev. Drug Discov. 2007;6:149–165. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 2.Krajinovic M., Labuda D., Sinnett D. Glutathione S-transferase P1 genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukaemia. Pharmacogenetics. 2002;12:655–658. doi: 10.1097/00008571-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Sepe D.M., McWilliams T., Chen J., Kershenbaum A., Zhao H., La M. Germline genetic variation and treatment response on CCG-1891. Pediatr. Blood Cancer. 2012;58:695–700. doi: 10.1002/pbc.23192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray L., Vujkovic M., McWilliams T., Cannon S., Devidas M., Stork L. TPMT and MTHFR genotype is not associated with altered risk of thioguanine-related sinusoidal obstruction syndrome in pediatric acute lymphoblastic leukemia: a report from the Children's Oncology Group. Pediatr. Blood Cancer. 2014;61:2086–2088. doi: 10.1002/pbc.25057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relling M.V., Hancock M.L., Rivera G.K., Sandlund J.T., Ribeiro R.C., Krynetski E.Y. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl. Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 6.Davies S.M., Borowitz M.J., Rosner G.L., Ritz K., Devidas M., Winick N. Pharmacogenetics of minimal residual disease response in children with B-precursor acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2008;111:2984–2990. doi: 10.1182/blood-2007-09-114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aplenc R., Thompson J., Han P., La M., Zhao H., Lange B. Methylenetetrahydrofolate reductase polymorphisms and therapy response in pediatric acute lymphoblastic leukemia. Cancer Res. 2005;65:2482–2487. doi: 10.1158/0008-5472.CAN-04-2606. [DOI] [PubMed] [Google Scholar]
- 8.Krajinovic M., Lemieux-Blanchard E., Chiasson S., Primeau M., Costea I., Moghrabi A. Role of polymorphisms in MTHFR and MTHFD1 genes in the outcome of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2004;4:66–72. doi: 10.1038/sj.tpj.6500224. [DOI] [PubMed] [Google Scholar]
- 9.de Jonge R., Hooijberg J.H., van Zelst B.D., Jansen G., van Zantwijk C.H., Kaspers G.J. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood. 2005;106:717–720. doi: 10.1182/blood-2004-12-4941. [DOI] [PubMed] [Google Scholar]
- 10.Stanulla M., Schrappe M., Brechlin A.M., Zimmermann M., Welte K. Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a case-control study. Blood. 2000;95:1222–1228. [PubMed] [Google Scholar]