Abstract
The nuclear pore complex mediates nucleocytoplasmic transport of macromolecules in eukaryotic cells. Transport through the pore is restricted by a hydrophobic selectivity filter comprising disordered phenylalanine-glycine-rich repeats of nuclear pore proteins. Exchange through the pore requires specialized transport receptors, called exportins and importins, that interact with cargo proteins in a RanGTP-dependent manner. These receptors are highly flexible superhelical structures composed of HEAT-repeat motifs that adopt various degrees of extension in crystal structures. Here, we performed molecular-dynamics simulations using crystal structures of Importin-β in its free form or in complex with nuclear localization signal peptides as the starting conformation. Our simulations predicted that initially compact structures would adopt extended conformations in hydrophilic buffers, while contracted conformations would dominate in more hydrophobic solutions, mimicking the environment of the nuclear pore. We confirmed this experimentally by Förster resonance energy transfer experiments using dual-fluorophore-labeled Importin-β. These observations explain seemingly contradictory crystal structures and suggest a possible mechanism for cargo protection during passage of the nuclear pore. Such hydrophobic switching may be a general principle for environmental control of protein function.
Introduction
Fundamental processes, such as transcription and translation, depend on molecules crossing the nuclear envelope in both directions. The sites of transition are the nuclear pore complexes (NPCs) (1), which permit the passive passage of metabolites and small proteins (2,3). The central channel of the NPC is a hydrophobic meshwork formed by nucleoporins (4), which contain intrinsically disordered regions of phenylalanine-glycine (FG)-rich repeats (5). Macromolecules larger than ∼40 kDa require specialized receptors as transport mediators to traverse the permeability barrier of the nuclear envelope efficiently (6–8), although exceptions are known for larger proteins with unusual surface hydrophobicity (9,10).
Most of the proteins that mediate nuclear transport belong to the homologous Importin-β/Karyopherin superfamily (11,12). Importin-β is a highly versatile molecule that is able to transport a variety of cargoes by binding them directly or through an adaptor protein (13–15). Importin-β:cargo complexes form in the cytosol and then cross the nuclear pore and enter the nucleus, where the complexes dissociate upon binding of the small GTPase Ran, in its GTP-bound form (16–18). The Importin-β:RanGTP complex shuttles back to the cytoplasm, where GTP hydrolysis is triggered and RanGDP is released, closing the cycle (19,20). Its unusual structural characteristics give Importin-β great flexibility, enabling it to undergo the rapid structural transitions involved in transport (21–26). Importin-β is an α-solenoid protein composed of 19 structurally conserved HEAT (Huntingtin, elongation factor 3, a subunit of protein phosphatase 2A, TOR (target of rapamycin 1)) repeats (27). Each HEAT repeat comprises two antiparallel α-helices connected by a loop (Fig. 1 A). The N-terminal A-helix is exposed at the outside, whereas the B-helix is located at the inner surface. This consecutive arrangement results in the overall superhelical shape of Importin-β (22,25,28–30). Solenoid proteins exhibit only very few contacts between residues distant in primary sequence, conveying a remarkable flexibility (25,31,32). The central HEAT repeats of Importin-β are more flexible compared with the remaining HEATs, resulting in an N-terminal and a C-terminal arch connected by hinge regions (16).
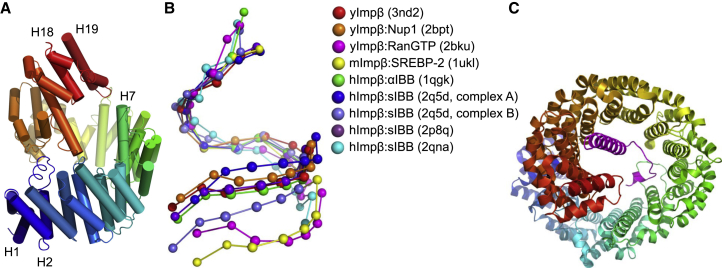
Figure 1.
Conformations of Importin-β. (A) Structure of Importin-β, shown in cartoon representation with α-helices represented by cylinders, colored by HEAT repeats from the N-terminus (blue) to the C-terminus (red). (B) Conformational variety of crystal structures of Importin-β (PDB IDs are given in the legend). The centers of mass of the HEAT repeats are shown. Importin-β from different organisms (yImpβ: yeast; mImpβ: mouse; hImpβ: human). (C) Importin-β bound to the sIBB domain (magenta). To see this figure in color, go online.
Both cargoes and RanGTP bind to the concave inner surface of Importin-β, although in different regions (24). The binding site for cargoes is located in the central HEAT repeats (28). The adaptor proteins Importin-α and Snurportin1, which bridge the interaction between Importin-β and cargoes, contain an α-helical Importin-β binding (IBB) domain that is enclosed by the C-terminal HEAT repeats (13,22,30,33–36). RanGTP attaches to the N-terminally located CRIME (for Crm1, Importin-β, etc.) domain (16,37,38), which is the most conserved sequence motif among nuclear transport receptors (11).
To mediate transport across the nuclear envelope, Importin-β has to interact with hydrophobic patches on FG nucleoporins of the NPC (5,39). The phenylalanine side chains can interact with the outer convex surface of Importin-β, where they insert into hydrophobic clefts between adjacent HEAT repeats (40–42), allowing the complexes to cross the nuclear envelope efficiently. The key factor for successful nuclear transport is the high hydrophobicity of the outer surface of nuclear transport receptors (9).
Structural studies on Importin-β have produced a disparate image of its conformational flexibility. Small-angle x-ray scattering (SAXS) studies and molecular-dynamics (MD) simulations suggested a rather extended conformation of free Importin-β in solution (31,32,43), whereas most crystal structures, with some conflicting exceptions, displayed more compact conformations (Fig. 1 B). No clear correlation has been observed between the binding mode of the interacting proteins and the conformation of the superhelix, and MD simulations uniformly predicted large differences in the flexibility of the different Importin-β complexes (32).
As a possible explanation for these discrepancies, we speculated that Importin-β displays environmentally induced plasticity. To test the possible impact of solvent polarity on the conformation of Importin-β, we carried out extended MD simulations of free Importin-β and of Importin-β in complex with the IBB domains of Importin-α (αIBB) or Snurportin1 (sIBB) in solvents of different polarity. In the simulations, free Importin-β and both IBB domain complexes adopted a significantly more compact structure in methanol than in aqueous solution. We subsequently confirmed this prediction by biophysical experiments, which showed an increased energy transfer between two fluorophores installed on Importin-β in more hydrophobic solvents. These conformational changes may play a role in the nuclear import cycle.
Materials and Methods
MD simulations
For the MD simulations, the crystal structures of the molecular complexes of Importin-β bound to the IBB domains of Importin-α (PDB ID: 1QGK) (20) and Snurportin1 (32) (PDB ID: 2P8Q) were used. The protonation states of titratable groups were determined using WHAT IF (44). For the simulations of free Importin-β, the IBB domain was removed from the crystal structures. The structures were placed in cubic boxes of sufficient size, such that the minimal distance between the protein and the box edges was at least 2 nm. Subsequently, the structures were solvated with either water or methanol, resulting in an average number of ∼76,800 water and 32,500 methanol molecules, respectively. Na+ and Cl− ions were added at a 0.15 M concentration.
All MD simulations were carried out with the GROMACS program package (45) (CVS version from 2007-07-20 and version 4.0) using the OPLS-AA force field (46) and the SPC water model (47) with 0.15 M Na+ and Cl− ions. These parameters were previously used for simulations of α-solenoid proteins and were shown to give reliable results (31,32,48,49).
For the simulations in methanol, a box with 1728 methanol molecules, preequilibrated with the OPLS force field at 298 K and 1 bar, was used (50). Radii of gyration were calculated with the GROMACS tool g_gyrate. SAXS curves were simulated using the FOXS program (51,52). All structural figures were created with Pymol (53). For details regarding the simulation parameters and the calculation of the statistical uncertainty, see Supporting Materials and Methods in the Supporting Material.
Protein expression and labeling
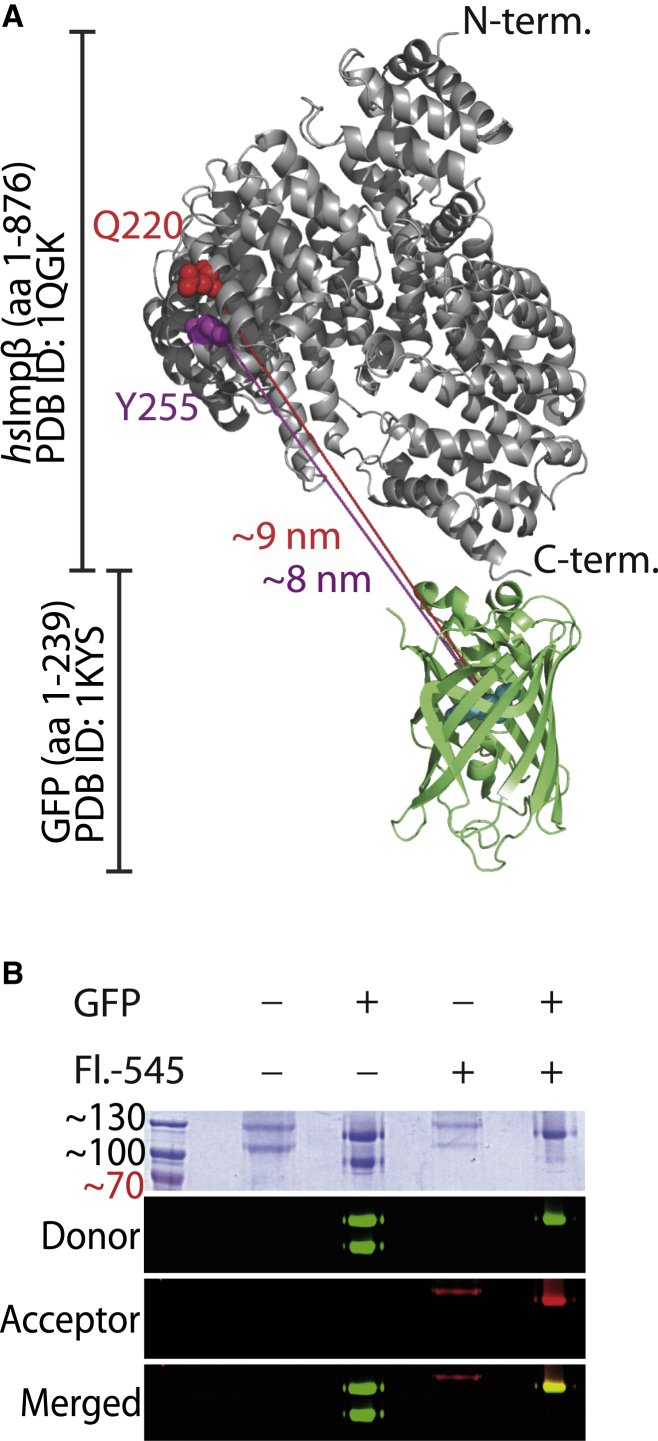
The construct hsImpβ-sfGFP-His6 (human Importin-β fused to superfolder green fluorescent protein (sfGFP) with a C-terminal hexa-histidine tag) was cloned into pCDFDuet-1 (Merck Millipore), and amber codons introduced by site-directed mutagenesis to create hsImpβQ220TAG-sfGFP-His6 and hsImpβY255TAG-sfGFP-His6. Proteins were expressed in E. coli BL21(DE3) transformed with plasmids to encode target proteins and MjCNPheRS/tRNACUA, and purified according to standard protocols (Supporting Materials and Methods). Purified proteins were labeled with dibenzocyclooctyne-conjugated (DBCO) dyes in ∼20-fold molar excess for 2 h in the dark, followed by native PAGE purification and electroelution. Native PAGE purification was necessary to remove remaining free fluorophore and a truncated form of hsImpβ-sfGFP (see Fig. 4 B, lower band in lane 2). The extent of labeling of hsImpβ-sfGFP Q220AzF-Fl-545 and hsImpβ-sfGFP Y255AzF-Fl-545 was qualitatively assessed by in-gel fluorescence, which showed a single dual-fluorophore-labeled band after the final purification step (Fig. S7). Samples were supplemented with glycerol (5–10%) as a cryoprotectant and snap-frozen in liquid nitrogen.
Figure 4.
Dual-fluorophore labeling of hsImportin-β. (A) Cartoon model of hsImportin-β showing the positions of AzF incorporation and their approximate distances to the sfGFP fluorophore. The figure was created using Pymol v0.99 and PDB entries 1QGK and 1KYS. (B) Importin proteins with or without genetically fused sfGFP-tag were purified from E. coli and labeled with DBCO-Fl.-545 where indicated. Fluorescence was analyzed using a Typhoon phosphoimager. To see this figure in color, go online.
Förster resonance energy transfer measurements
Fluorescence scans were performed on a Fluoromax-4 (Horiba Scientific) with a 10 × 2 mm fluorescence cuvette (104002F-10-40; Hellma Analytics), with the following settings: temperature = 20°C, band width = 3/3 nm, resolution = 1 nm, and integration time = 0.1 s. The emission spectra were normalized to maximum = 1 and minimum = 0. The relative Förster resonance energy transfer (FRET) efficiency (see Fig. 5, B and C) was calculated as EFRET = IA/(IA + ID), where ID and IA are the fluorescence intensities at donor emission maximum (508 nm) and acceptor emission maximum (575 nm), respectively, from a single emission scan after excitation of the donor (470 nm).
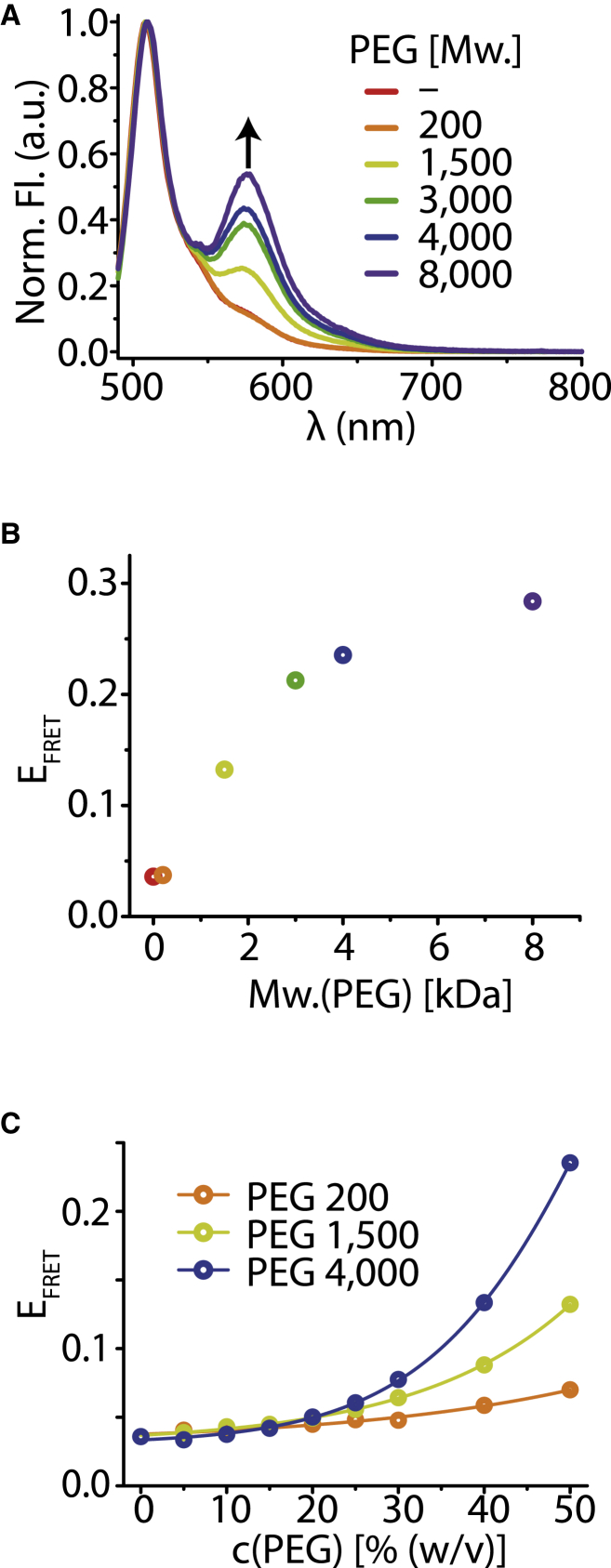
Figure 5.
hsImpβ-sfGFP Q220AzF-Fl-545 shows increased FRET in the presence of PEG. (A) Addition of 50% (w/v) PEG of various molecular weights leads to an increase in fluorescence emission at 575 nm. (B) The relative FRET efficiency of hsImpβ-sfGFP Q220AzF-Fl-545 calculated from the emission spectra in (A). (C) Relative FRET efficiency increases with PEG concentration. To see this figure in color, go online.
Results
Previous structural studies have focused on the influence of cargo binding on the conformation of Importin-β. However, it is possible that the observed structural discrepancies are the result of the different solvents used in crystallization. These solvents differed substantially in polarity, mainly due to the amount of polyethylene glycol (PEG) present in the solution (Table S2) (22,30,33,36). This behavior may be relevant in vivo since Importin-β experiences a more hydrophobic environment in the channel of the NPC.
Structure and dynamics of Importin-β in aqueous solution
To test this hypothesis, we chose to use x-ray crystal structures of Importin-β bound to the sIBB or αIBB domain and of free Importin-β, after in silico removal of the IBB domain, as representative examples to carry out unbiased all-atom MD simulations in explicit solvent (for an overview of the simulations, see Table S1).
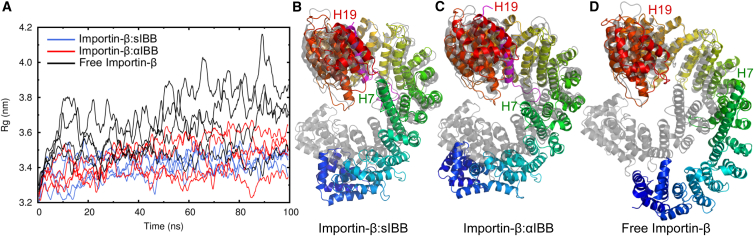
In all simulations using Importin-β:sIBB (PDB ID: 2P8Q) as the starting point (Fig. 1 C), Importin-β underwent rapid structural changes (Fig. 2 A, blue lines). Within 100 ns, the complex opened noticeably, expanding the radius of gyration (Rg) from 3.17 to 3.45 ± 0.01 nm (calculated from the last 20 ns of the trajectories) (Table 1; for the estimation of errors, see Supporting Materials and Methods). Therefore, the closed conformation of the Importin-β:sIBB complex is not stable during MD simulations in aqueous solution. Instead, Importin-β:sIBB exhibits a large conformational plasticity, especially in the N-terminal arch. However, all representative structures showed a larger opening of the superhelix than the crystal structure (see Fig. S1). Similar results were obtained with αIBB-bound Importin-β (PDB ID: 2QGK) (22) (Fig. 2 A, red lines).
Figure 2.
Importin-β in aqueous solution. (A) Running average of the Rg of Importin-β complexed with the sIBB domain (blue) or the αIBB domain (red) compared with free Importin-β (black). (B–D) Snapshots after 100 ns of MD simulation of the sIBB complex (B), αIBB complex (C), and free Importin-β (D) compared with the corresponding closed crystal structures in gray. For clarity, the structures were aligned at the less variable HEAT repeats 11–19. To see this figure in color, go online.
Table 1.
Radius of gyration of Importin-β
| Importin-β in Complex with | Rg (nm) Crystal Structure | Rg (nm) Simulation in Water | Rg (nm) Simulation in Methanol |
|---|---|---|---|
| sIBB | 3.17 | 3.45 (0.01) | 3.32 (0.01) |
| αIBB | 3.16 | 3.48 (0.06) | 3.37 (0.01) |
| — | 3.17/3.16a | 3.71 (0.08) | 3.50 (0.03) |
The Rg of Importin-β in water was averaged over all independent simulations. Only the last 20 ns of each simulation were included in the calculation. The estimated statistical uncertainty is given in parentheses (see Supporting Materials and Methods).
The conformational changes of Importin-β in aqueous solution are reflected in the backbone root mean-square deviation (RMSD) with respect to the closed crystal structure (see Fig. S2). In each frame of the trajectory, all backbone atoms of the complete scaffold of Importin-β were aligned to the closed crystal structure. Subsequently, the RMSD was calculated separately for each HEAT repeat of the aligned structures. For the sIBB complex (Fig. S2 A), the inner groove of Importin-β (HEAT repeats H8–H12) was the most rigid, and the N- and C-termini showed higher flexibility. This pattern of flexibility is in agreement with previous studies that analyzed the flexibility of Importin-β and found that it was mainly determined by movements around two major hinges at H4-5 and H14-15 (32). For the αIBB complex, the pattern was slightly different (Fig. S2 B). In three out of four simulations the pattern was similar to that of the sIBB complex, whereas in one simulation the N-terminus was drastically distorted.
The predominant flexibility of the N-terminus of Importin-β also becomes evident from a comparison of snapshots from the trajectories with the closed crystal structure (Fig. 2, B and C). In all simulations, the IBB domain remained bound to Importin-β. Whereas the conformation of the α-helical part of the IBB domain was largely unchanged (with RMSD values of 0.1–0.3 nm, compared with the crystal structure), the N-terminal loop stretched slightly to adapt to the more open conformation of Importin-β, reaching RMSD values of up to 0.5 nm.
These MD simulations indicate that in water, at physiological salt concentration, the Importin-β:IBB domain complexes adopt markedly more extended conformations compared with the crystal structures. In the simulations, the C-terminal arch was closed and tightly wrapped around the cargo, whereas the N-terminus was quite flexible. Experimental evidence for such an open conformation of Importin-β in solution comes from SAXS studies on the ligand-free state (25). On the basis of these data, it was proposed that free yeast Importin-β in solution has an extended Rg of 3.9 nm.
To check our MD simulations against the SAXS experiments, and to understand the influence of cargo binding on the conformation and flexibility of Importin-β, we carried out simulations of human Importin-β from which the bound IBB domain had been removed (Fig. 2 A, black lines). In our simulations, free Importin-β clearly showed a more pronounced opening and a larger flexibility than the IBB domain complexes (Fig. 2 D): during 100 ns of simulation, Rg values of up to 4.2 nm were reached, with an average Rg of 3.71 ± 0.08 nm (Fig. 2 D; Table 1). These conformational changes occurred along the entire superhelix of Importin-β, with the largest fluctuations being observed within the N- and C-terminal HEAT repeats (Fig. S2 C). We calculated theoretical SAXS curves from snapshots from the last 50 ns of all simulations. The resulting averaged scattering profile correctly reproduced the features observed in SAXS studies of yeast Importin-β (KAP95), including the characteristic shoulder at q ≈ 1 nm−1 (Fig. S3) (25,43).
To summarize, in all of the MD simulations in aqueous solution with a physiological ion concentration, all closed crystal conformations of Importin-β turned out to be unstable and quickly adopted a more relaxed and open conformation. Even though the IBB domain complexes were found to be more compact than free Importin-β, a noticeable opening of the Importin-β superhelix was also seen in both of the Importin-β:IBB domain complexes under study.
Sensitivity of Importin-β to solvent polarity
The observation that the closed conformations of free Importin-β and the Importin-β:IBB domain complexes opened up during MD simulations in water led us to ask whether the abundance of compact crystal structures may be due to the crystallization conditions. The good agreement between the simulations and SAXS experiments supports this idea. In fact, most crystals of Importin-β were obtained using PEG, which facilitates protein crystallization by lowering the polarity of the medium, as a precipitant (Table S2). The crystals of the open conformation of the Importin-β:sIBB complex, in contrast, were obtained in the absence of PEG (30).
To test whether less polar solvents can indeed induce closed structures of Importin-β, we carried out MD simulations of cargo-free and cargo-bound Importin-β in methanol as a hydrophobic mimic of both the PEG solution and the interior of the nuclear pore. The simulations were started from three different conformations of the sIBB complex (Fig. S4, A and B) or the αIBB complex (Fig. S4, C and D) using crystal structures or, as controls, open conformations extracted from previous simulations in water. Simulations of free Importin-β were started from the closed structure and from an open conformation with an Rg of 3.9 nm (Fig. S4 E). Contrary to what was observed in aqueous solution, the closed crystal structure of the Importin-β:sIBB complex opened only slightly in methanol (Fig. 3 A, black line). Strikingly, all simulations of open conformations showed a marked decrease of the Rg of the complex, reaching average values between 3.32 ± 0.01 nm (Importin-β:sIBB) and 3.50 ± 0.03 nm (free Importin-β), thus indicating a pronounced shift in the conformational equilibrium (Table 1). In all simulations starting from an open conformation with little distortion of individual HEAT repeats, the N-terminus closed spontaneously within 10–20 ns (Fig. 3 A, cyan lines). This conformational change brought the termini of Importin-β into contact, forming a closed ring, which then remained stable during the rest of the simulation (Fig. 3 B). The final conformation was even more compact than in the crystal structure (Fig. S4 F).
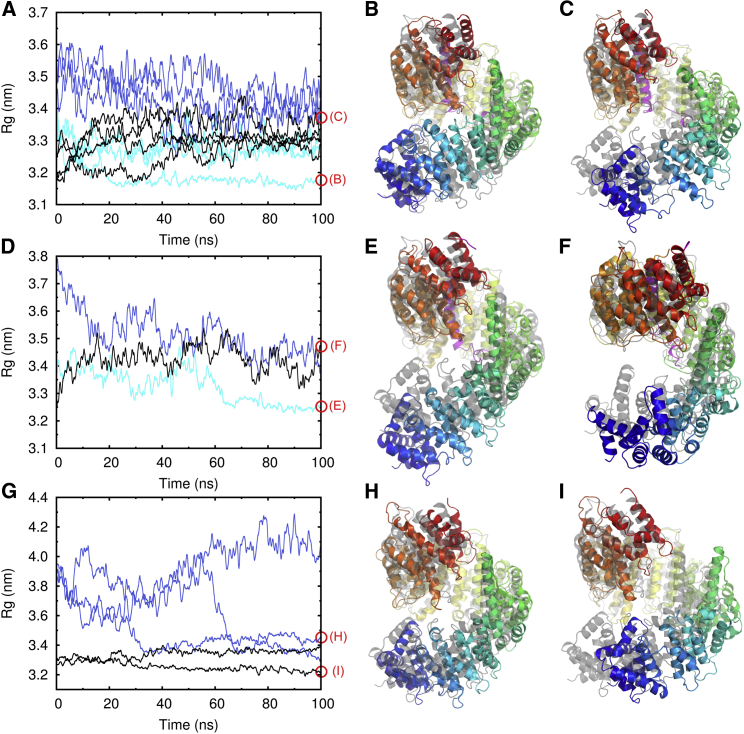
Figure 3.
Importin-β in methanol. (A) Running average of the Rg of the Importin-β:sIBB complex. Simulations started from the closed conformation (black) and from open conformations with little deformation of individual HEAT repeats (cyan) and stronger deformation (blue). (B and C) Snapshots after 100 ns of simulations compared with the crystal structures (in gray). (D) Rg of the Importin-β:αIBB complex. (E and F) Snapshots after 100 ns of simulation. (G) Running average of the Rg of free Importin-β. (H and I) Snapshots after 100 ns of simulation time. Red circles indicate where the displayed snapshots were taken. To see this figure in color, go online.
In cases where the starting structure displayed more local distortions of individual HEAT repeats, the conformational rearrangement was slower (Fig. 3 A, blue lines). Within 100 ns, a noticeable decrease in Rg was observed, but a conformation with stable contacts between the N- and C-terminal regions was not reached (Fig. 3 C). However, an overall compaction and a transient ring closure of Importin-β were also seen in these cases.
The Importin-β:αIBB and sIBB complexes exhibited similar Rg values in methanol, with final values reaching 3.25–3.5 nm (Fig. 3 D). However, the corresponding structures showed a larger conformational variety (Fig. 3, E and F). The contacts between the N- and C-terminal HEAT repeats were less stable, as reflected by the larger RMSD (0.5–0.8 nm) compared with the crystal structure (Fig. S4 G).
For uncomplexed Importin-β, the less polar solvent methanol also stabilized a compact structure of the superhelix. Even after removal of the IBB domain, the structure remained rather stable (Fig. 3 G, black lines, and H). Two of the three simulations of Importin-β that started from a completely open conformation, with no sequence distal contacts between HEAT repeats, completed closure within 40 and 70 ns (Fig. 3 D, blue lines). Although contacts between the N- and C-termini were formed, a certain conformational heterogeneity within the ensemble of closed structures remained (Figs. 3, H and I, and S4 H). The deviations from the crystal structure are due to a lack of stabilization of the C-terminus and the central arch of Importin-β by the IBB domain.
The simulations show that despite their conformational differences, free Importin-β and both of the IBB domain complexes adopt a markedly more compact structure in methanol than in aqueous solution. This reaction to the polarity of the environment is therefore an intrinsic property of Importin-β. The large rearrangements, which lead to closure of the Importin-β superhelix, take place very rapidly, on a timescale of ∼100 ns.
A major conformational change of a large and highly flexible system such as Importin-β is a stochastic process that requires the crossing of numerous energetic barriers. The observed differences between the trajectories corresponding to independent simulations of the same system are thus not surprising. Nevertheless, from the simulations alone we cannot exclude the possibility, however remote, that the observed trends could be artifacts stemming from insufficient convergence of the simulations. We therefore addressed the significance of the results from the simulations by conducting experiments.
Increased energy transfer in dual-fluorophore-labeled Importin-β in hydrophobic solvents
We aimed to test the above theoretical observations biochemically by assessing the conformational plasticity of Importin-β in response to differences in the hydrophobicity of the environment. To this end, we produced dual-fluorophore-labeled Importin-β to observe shape changes by measuring the FRET efficiencies. Site-specific coupling of fluorophores to Importin-β is impeded by the presence of more than 20 cysteine residues in the natural sequence, precluding the use of thiol chemistry. Therefore, we genetically fused sfGFP (54) as the donor fluorophore to the C-terminal end of hsImportin-β (hsImpβ-sfGFP). The fluorescence properties of sfGFP are unaffected in the fusion construct (Fig. S5). To introduce the acceptor fluorophore, we used genetic-code expansion to install amino acids with side chains containing bioorthogonal reactive functional groups at structurally permissive sites. These amino acids are activated by evolved aminoacyl-tRNA synthetases and incorporated in response to amber (UAG) codons by corresponding cognate tRNAs. We chose the surface-exposed residues Gln-220 and Tyr-255 for incorporation of p-azido-l-phenylalanine (AzF) using the evolved MjCNPheRS/tRNACUA pair (55) (Fig. 4 A). Full-length protein was produced only when AzF was supplied with the growth medium (Fig. S6), confirming the successful incorporation of the amino acid in response to the amber codon. We used the azide group to site-specifically conjugate the fluorophore DBCO-Fl-545 in a bioorthogonal strain promoted azide-alkyne coupling (SPAAC) (Figs. 4 B and S7). In fluorescence emission scans (λex. = 470 nm) of hsImpβ-sfGFP Q220AzF-Fl-545, we observed an additional intensity around 575 nm, which depended on the presence of the acceptor fluorophore and therefore may indicate the presence of intramolecular FRET (Fig. S8). Treating the labeled protein with Proteinase K removed this additional intensity at ∼575 nm (Fig. S9 A). When the unlabeled hsImpβ-sfGFP was supplemented with free DBCO-Fl-545 dye and treated with Proteinase K, no such change at ∼575 nm was observed (Fig. S9 B). Hence, we conclude that a small amount of FRET occurs between sfGFP and Fl-545 in hsImpβ-sfGFP Q220AzF-Fl-545, which may be used to report on changes in the protein’s conformation in response to the surrounding medium. As a functional test for correct folding, we incubated permeabilized HeLa cells with labeled hsImpβ-sfGFP proteins. As shown in Fig. S10, the protein was detected in the nucleus and also at the nuclear envelope, indicating physiological interactions with components of the nuclear transport machinery.
First, we investigated the FRET efficiency in the presence of increasing concentrations of methanol, similar to the conditions used in the simulation experiments (Fig. S11). Indeed, we observed an increase in FRET with increasing methanol concentration, indicating that hsImportin-β contracts in hydrophobic environments. Next, we tested the impact of the addition of PEG of various molecular weights on the FRET efficiency in hsImpβ-sfGFP Q220AzF-Fl-545 (Fig. 5 A). PEG creates a more crowded and hydrophobic environment, an effect that escalates with increasing chain length. At 50% (w/v) PEG 200 the fluorescence intensity at 575 nm of hsImpβ-sfGFP Q220AzF-Fl-545 rose slightly, whereas higher-molecular-weight PEG at the same concentration showed increasingly stronger effects. Titrating the concentration of PEG 200, PEG 1500, and PEG 4000 over the range of 0–50% (w/v) revealed that relative FRET efficiency increased with PEG molecular weight and concentration (Figs. 5, B and C, and S12). The increase in acceptor fluorescence is indeed a result of enhanced energy transfer because a mixture of hsImpβ-sfGFP and DBCO-Fl-545 did not react to the addition of PEG in this manner (Fig. S13). This effect was not a simple result of molecular crowding, since Ficoll-70 (molecular weight 70,000), which induces crowding but is not hydrophobic, did not show a similar effect on the fluorescence properties of hsImpβ-sfGFP Q220AzF-Fl-545 (Fig. S14). This compaction was reversible, as expected for Importin-β exiting the NPC, as demonstrated by stepwise dilution of hsImpβ-sfGFP Q220AzF-Fl-545 from 50% PEG solutions (Fig. S15). We obtained similar results using an alternative spectroscopic method when we measured the interaction of the fluorophores by donor fluorescence lifetime instead of energy transfer. Again, increasing concentrations and molecular weights of PEG reduced the donor fluorophore lifetime to a similar degree as observed by FRET (Fig. S16).
These experiments confirm our simulation results showing that Importin-β indeed adopts a more compact conformation in hydrophobic environments.
Discussion
The high intrinsic flexibility of Importin-β has received considerable attention, and it is generally accepted that it is of paramount importance for the fast binding and release of a wide variety of cargoes (21–26). However, previous studies have been centered on the function of conformational changes in cargo binding, and little attention has been paid to their possible role in crossing of the nuclear pore. Recent findings, as well as our simulations, suggest that due to its unique and complex hydrophobic core, Importin-β in solution fluctuates in equilibrium between different conformational states, comprising completely open as well as quite compact conformations of the superhelix (25,31,33). Cargo binding shifts the equilibrium toward more closed conformations by selecting rather strained conformers of Importin-β from this large pool of different conformations, with the corresponding entropy changes being crucial for the overall thermodynamics of the system. Similar observations were made for the exportin Crm1, which cooperatively adopts more compact conformations upon binding RanGTP and cargo (56). However, available structural models of Importin-β do not fully support this perception. Whereas models built from SAXS experiments show free Importin-β in rather open conformations, x-ray crystallography has yielded predominantly closed, compact structures of both free and cargo-bound Importin-β.
In our MD simulations, we observed that binding of the IBB domain of the adaptor proteins Importin-α and Snurportin1 to the inner surface of Importin-β indeed stabilized more compact conformations of the superhelix, in line with the current model. However, in our simulations, the IBB domain stabilized only the C-terminal arch of Importin-β (HEAT repeats 12–19), which is in direct contact with the helical part of the IBB domain. Other domains of the molecule underwent large conformational changes, adopting an open conformation. This flexibility may have functional implications, e.g., for RanGTP binding and unbinding.
Strikingly, in all simulations with a more hydrophobic solvent, such as methanol, the structure of Importin-β favored closed conformations. Rapid closure was seen in all simulations of Importin-β, whether or not it was bound to an IBB domain. Further, the superhelix adopted a conformation that largely resembled the closed conformations seen in most Importin-β crystal structures. This unusually strong influence of environment polarity on Importin-β structure is corroborated by a clear correlation between the crystallization conditions and the observed Importin-β conformation. In fact, most crystals containing closed conformations were obtained in solutions of high PEG concentrations with a markedly decreased polarity, whereas the crystal structure obtained from a PEG-free solution showed a much more open conformation (Table S2). Similar conformational plasticity of Importin-β in response to changes in the polarity of the solvent (by addition of different types of alcohols) was observed in a recent study (57).
Overall, our biochemical and theoretical studies suggest that the closed structures of cargo-bound Importin-β do not represent the predominant conformation in the cytoplasm, but rather conformations of Importin-β in a less polar environment.
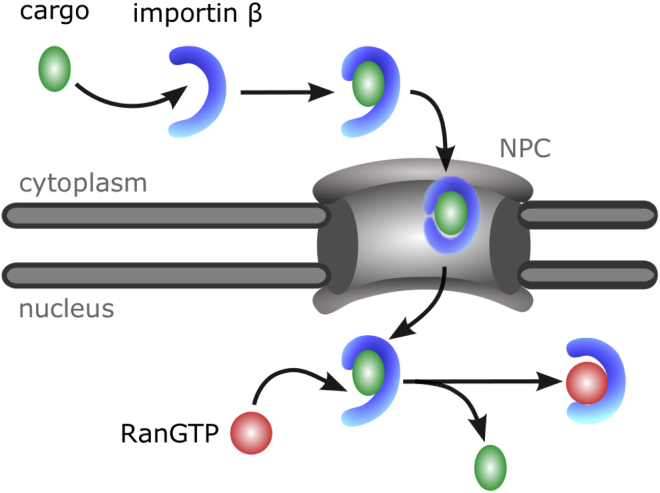
This finding suggests an unexpected functional role of the environment during nucleo-cytoplasmic transport, where Importin-β is subject to changes in the surrounding medium. In the cytoplasm or nucleoplasm, it is exposed to aqueous solvent, whereas inside the central channel of the NPC, the high concentration of hydrophobic FG repeat peptides reduces the polarity of the solvent (4) in a manner similar to that observed for PEG. We therefore assume that Importin-β also adopts a more compact conformation upon entering the nuclear pore. It is tempting to speculate that the transitions between open and closed conformations correspond to the varying requirements for the stability of Importin-β complexes with cargo molecules during different stages of the transport process (Fig. 6). In the cytoplasm, Importin-β binds cargoes at its concave inner surface. An open conformation of the superhelix would make the binding sites more accessible and facilitate cargo recognition. During the passage through the nuclear pore, the inside of the superhelix needs to be protected against interactions with other molecules present in the channel to prevent dissociation of the cargo-transporter complex. Closed conformations would prevent the complex from dissociating during passage. After it is imported into the nucleus, the IBB domain is displaced from Importin-β by RanGTP, which would be facilitated by an opening of the complex in the polar environment of the nucleoplasm. Such environment-sensitive conformational plasticity may in fact be quite a general mechanism to modulate or control protein function in response to environmental changes. Such changes may be due to molecular crowding, solute concentration changes, or molecular transitions between different intracellular compartments.
Figure 6.
Proposed model for Importin-β-dependent transport. In the aqueous solution of the cytoplasm, Importin-β adopts predominantly open conformations, rendering its inner surface more accessible to ligand binding (upper part). When cargo-bound Importin-β enters the NPC, contact with the hydrophobic permeability barrier leads to closure of the Importin-β superhelix, thus preventing complex dissociation during passage (central part). After the complex exits from the pore, the N-terminus opens up again, rendering Importin-β accessible to RanGTP binding (lower part). To see this figure in color, go online.
Author Contributions
K.H. produced recombinant proteins. K.H. and Q.V. performed FRET experiments. I.B. carried out fluorescence microscopy experiments. N.D. performed MD simulations. I.G. contributed analytic tools. K.H., N.D., H.G., and H.N. wrote the manuscript. All authors designed experiments, analyzed data, and commented on the manuscript.
Acknowledgments
We thank Clement Blanchet, Daniel Wohlwend, Thomas Monecke, and Ulrich Zachariae for helpful discussions, and Thomas Monecke for carefully reading the manuscript.
This work was supported by grants from the Human Frontiers Science Program (grant RGP53/2004), the Deutsche Forschungsgemeinschaft (GR 2079/4-1 to H.G., Ke660/14-1 to R.K., and SFB860 to J.E., R.F., and H.N.), the Emmy-Noether Programme of the Deutsche Forschungsgemeinschaft, the Free Floater Programme of the University of Göttingen (to H.N.), and the Cluster of Excellence and DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain (to J.E., H.G., and H.N.).
Editor: Scott Feller.
Footnotes
Kangkan Halder and Nicole Dölker contributed equally to this work.
Helmut Grubmüller and Heinz Neumann contributed equally to this work.
Nicole Dölker’s present address is Structural Computational Biology Group, Spanish National Cancer Research Centre (CNIO), Madrid, Spain.
Supporting Materials and Methods, 16 figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00596-2.
Contributor Information
Helmut Grubmüller, Email: hgrubmu@gwdg.de.
Heinz Neumann, Email: hneumann@uni-goettingen.de.
Supporting Citations
References (58–60) appear in the Supporting Material.
Supporting Material
References
- 1.Lim R.Y.H., Fahrenkrog B. The nuclear pore complex up close. Curr. Opin. Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Keminer O., Peters R. Permeability of single nuclear pores. Biophys. J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J., Goryaynov A., Yang W. Self-regulated viscous channel in the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2012;109:7326–7331. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramer A., Liashkovich I., Shahin V. Atomic force microscopy visualises a hydrophobic meshwork in the central channel of the nuclear pore. Pflugers Arch. 2008;456:155–162. doi: 10.1007/s00424-007-0396-y. [DOI] [PubMed] [Google Scholar]
- 5.Wälde S., Kehlenbach R.H. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Chook Y.M., Süel K.E. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook A.G., Conti E. Nuclear export complexes in the frame. Curr. Opin. Struct. Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Xu D., Farmer A., Chook Y.M. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr. Opin. Struct. Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naim B., Zbaida D., Reich Z. Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 2009;28:2697–2705. doi: 10.1038/emboj.2009.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R., Brattain M.G. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581:3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görlich D., Dabrowski M., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radu A., Blobel G., Moore M.S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber J., Cronshagen U., Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jäkel S., Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmeri D., Malim M.H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S.J., Matsuura Y., Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 17.Sun C., Fu G., Musser S.M. Choreography of importin-α/CAS complex assembly and disassembly at nuclear pores. Proc. Natl. Acad. Sci. USA. 2013;110:E1584–E1593. doi: 10.1073/pnas.1220610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C., Yang W., Musser S.M. Single-molecule measurements of importin alpha/cargo complex dissociation at the nuclear pore. Proc. Natl. Acad. Sci. USA. 2008;105:8613–8618. doi: 10.1073/pnas.0710867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorokin A.V., Kim E.R., Ovchinnikov L.P. Nucleocytoplasmic transport of proteins. Biochemistry Mosc. 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 20.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 21.Cingolani G., Lashuel H.A., Müller C.W. Nuclear import factors importin alpha and importin beta undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- 22.Cingolani G., Petosa C., Müller C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 23.Conti E., Müller C.W., Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Cook A., Bono F., Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 25.Forwood J.K., Lange A., Zachariae U., Marfori M., Preast C., Grubmüller H., Stewart M., Corbett A.H., Kobe B. Quantitative structural analysis of importin-beta flexibility: paradigm for solenoid protein structures. Structure. 2010;18:1171–1183. doi: 10.1016/j.str.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Lee S.J., Imamoto N., Tsukihara T. The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport. J. Mol. Biol. 2000;302:251–264. doi: 10.1006/jmbi.2000.4055. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B., Gleichmann T., Horne J., Jennings I.G., Scotney P.D., Teh T. Turn up the HEAT. Structure. 1999;7:R91–R97. doi: 10.1016/s0969-2126(99)80060-4. [DOI] [PubMed] [Google Scholar]
- 28.Lee S.J., Sekimoto T., Yoneda Y. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 29.Liu S.M., Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Wohlwend D., Strasser A., Ficner R. Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J. Mol. Biol. 2007;374:1129–1138. doi: 10.1016/j.jmb.2007.09.065. [DOI] [PubMed] [Google Scholar]
- 31.Kappel C., Zachariae U., Grubmüller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zachariae U., Grubmüller H. Importin-beta: structural and dynamic determinants of a molecular spring. Structure. 2008;16:906–915. doi: 10.1016/j.str.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Bhardwaj A., Cingolani G. Conformational selection in the recognition of the snurportin importin beta binding domain by importin beta. Biochemistry. 2010;49:5042–5047. doi: 10.1021/bi100292y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Görlich D., Henklein P., Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 35.Huber J., Dickmanns A., Lührmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitrousis G., Olia A.S., Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 37.Forwood J.K., Lonhienne T.G., Kobe B. Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics. J. Mol. Biol. 2008;383:772–782. doi: 10.1016/j.jmb.2008.07.090. [DOI] [PubMed] [Google Scholar]
- 38.Vetter I.R., Arndt A., Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 39.Otsuka S., Iwasaka S., Yoshimura S.H. Individual binding pockets of importin-beta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc. Natl. Acad. Sci. USA. 2008;105:16101–16106. doi: 10.1073/pnas.0802647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayliss R., Kent H.M., Stewart M. Crystallization and initial X-ray diffraction characterization of complexes of FxFG nucleoporin repeats with nuclear transport factors. J. Struct. Biol. 2000;131:240–247. doi: 10.1006/jsbi.2000.4297. [DOI] [PubMed] [Google Scholar]
- 41.Bayliss R., Littlewood T., Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 42.Isgro T.A., Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-beta. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Fukuhara N., Fernandez E., Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 2004;279:2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- 44.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 45.Van Der Spoel D., Lindahl E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Kaminski G.A., Friesner R.A., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 47.Berendsen H.J.C., Postma J.P.M., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 48.Dölker N., Blanchet C.E., Dickmanns A. Structural determinants and mechanism of mammalian CRM1 allostery. Structure. 2013;21:1350–1360. doi: 10.1016/j.str.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Zachariae U., Grubmüller H. A highly strained nuclear conformation of the exportin Cse1p revealed by molecular dynamics simulations. Structure. 2006;14:1469–1478. doi: 10.1016/j.str.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Böckmann R.A., Grubmüller H. Multistep binding of divalent cations to phospholipid bilayers: a molecular dynamics study. Angew. Chem. Int. Engl. 2004;43:1021–1024. doi: 10.1002/anie.200352784. [DOI] [PubMed] [Google Scholar]
- 51.Schneidman-Duhovny D., Hammel M., Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneidman-Duhovny D., Hammel M., Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLano W.L. DeLano Scientific; Palo Alto, CA: 2008. The PyMOL Molecular Graphics System. [Google Scholar]
- 54.Pédelacq J.D., Cabantous S., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 55.Young D.D., Young T.S., Schultz P.G. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry. 2011;50:1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monecke T., Haselbach D., Ficner R. Structural basis for cooperativity of CRM1 export complex formation. Proc. Natl. Acad. Sci. USA. 2013;110:960–965. doi: 10.1073/pnas.1215214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshimura S.H., Kumeta M., Takeyasu K. Structural mechanism of nuclear transport mediated by importin β and flexible amphiphilic proteins. Structure. 2014;22:1699–1710. doi: 10.1016/j.str.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 59.Charneau P., Mirambeau G., Clavel F. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 60.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.