Abstract
We are exposed to a growing number of chemicals in our environment, most of which have not been characterized in terms of their toxicological potential or mechanisms. Here, we employ a chemoproteomic platform to map the cysteine reactivity of environmental chemicals using reactivity-based probes to mine for hyper-reactive hotspots across the proteome. We show that environmental contaminants such as monomethylarsonous acid and widely used pesticides such as chlorothalonil and chloropicrin possess common reactivity with a distinct set of proteins. Many of these proteins are involved in key metabolic processes, suggesting that these targets may be particularly sensitive to environmental electrophiles. We show that the widely used fungicide chlorothalonil specifically inhibits several metabolic enzymes involved in fatty acid metabolism and energetics, leading to dysregulated lipid metabolism in mice. Our results underscore the utility of using reactivity-based chemoproteomic platforms to uncover novel mechanistic insights into the toxicity of environmental chemicals.
Introduction
Many complex human diseases cannot be explained by genetics alone and likely have environmental or gene-environment causes (Hemminki et al., 2006; Hunter, 2005). We are exposed to an ever-growing number of chemicals, many of which have been linked to adverse health effects, and most of which have not been characterized in terms of their toxicological potential or mechanisms (Bookman et al., 2011; Rappaport, 2011). It is therefore likely that there are more chemicals in our environment than previously anticipated that act as potential drivers of disease. These agents may go unnoticed due to adverse health effects that occur through yet unknown mechanisms.
While there have been many efforts made to characterize the toxicology of environmental chemicals such as traditional long-term toxicity assessments in animal models and human epidemiological studies, to high throughput screening and modern systems biology approaches to map gene and protein expression changes, these approaches are often correlative or provide insufficient detail on toxicological mechanisms (Bucher, 2013; Collins et al., 2008; Dix et al., 2007; Huang et al., 2011; Shukla et al., 2010). This is because gene and protein expression, epigenetic, or metabolite changes that manifest as cellular and systemic phenotypes arising from chemical exposure are indirect outcomes that result from direct chemical interactions with specific molecular targets. Deconvoluting these indirect effects to predict the direct molecular targets has been very challenging. We believe that understanding the direct chemical-protein interactions of environmental chemicals is critical in informing our understanding of downstream molecular events and linking them to pathological effects, providing a direct approach to identifying environmental drivers of disease.
Of particular concern among chemicals in our environment are reactive electrophiles that we are directly exposed to or that form through bioactivation (Rappaport et al., 2012). These environmental electrophiles have the potential to covalently react with nucleophilic amino acid hotspots within the proteome, leading to protein dysfunction and subsequent pathological effects (Rappaport et al., 2012). These “hyper-reactive” nucleophilic hotspots include cysteine, serine, and lysine residues that are often involved in important biological functions such as catalysis, regulation, post-translational modifications, redox balance, metal binding, and protein-protein interactions (Shannon and Weerapana, 2015). Though we are exposed to a large array of potentially reactive chemicals, we have little to no understanding regarding their interactions with hyper-reactive proteome hotspots and the resulting effects on protein function, downstream biochemistry, or ensuing pathophysiological consequences. While there have been many innovative approaches to test the mutagenicity of DNA-damaging reactive electrophiles (Araya and Fowler, 2011; Meier and Gartner, 2014), there has been a considerable lag in technologies that enable global assessment of chemical reactivity with the proteome. At this time, there is a major gap in our knowledge of how reactive chemicals elicit adverse health effects through chemical-protein interactions.
Historically, the toxicological mechanisms underlying many reactive electrophiles have been thought to occur through indiscriminate covalent modifications on proteins or DNA leading to mutations or non-specific toxicities. However, recent studies have revealed that even highly reactive endogenous electrophiles preferentially and selectively react with certain sites on specific protein targets, indicating that the interactions of electrophilic compounds with the proteome are driven not only by their reactivity, but also by their affinity to certain protein microenvironments (Codreanu et al., 2014; Wang et al., 2014; Weerapana et al., 2008; Weerapana et al., 2010). Here, we have employed a chemoproteomic platform that uses reactivity-based probes to comprehensively map the proteome-wide targets of reactive environmental chemicals in complex biological systems. We show that certain environmental chemicals show specific reactivity with a set of previously unrecognized protein targets, particularly enriched in enzymes that play key roles in metabolism. Taking the widely applied fungicide chlorothalonil (CTN) as a representative example, we use this reactivity-based chemoproteomic platforms to directly identify specific enzymes and transporters involved in fatty acid metabolism and energetics as in vivo targets of CTN in mice, leading to distinct downstream biochemical alterations in the lipidome.
Results and Discussion
Using Reactivity-Based Probes to Map Proteome-Wide Interactions of Environmental Chemicals
Among amino acid side-chains that may be adducted by reactive electrophiles, cysteines are particularly sensitive to modification due to their inherent nucleophilicity (Pace and Weerapana, 2013; Shannon and Weerapana, 2015; Weerapana et al., 2010). This nucleophilicity can be enhanced by local protein microenvironments which lower the inherent pKa of certain cysteines (Poole, 2015). These “hyper-reactive” cysteines have been previously shown to be important in diverse protein functions, including catalysis, redox balance, protein-protein interactions, metal binding, and functional regulation (Wang et al., 2014; Weerapana et al., 2010). Weerapana et al. described a powerful chemoproteomic platform that utilized a bioorthogonal cysteine-reactive probe, iodoacetamide-alkyne (IAyne), to globally profile these hyper-reactive cysteines across thousands of protein targets directly in complex proteomes (Weerapana et al., 2008; Weerapana et al., 2010). The authors showed that hyper-reactive cysteines identified by this method were highly enriched in sites important to protein function, indicating that this strategy could be used to assess proteome-wide hyper-reactive and functional sites (Wang et al., 2014; Weerapana et al., 2010).
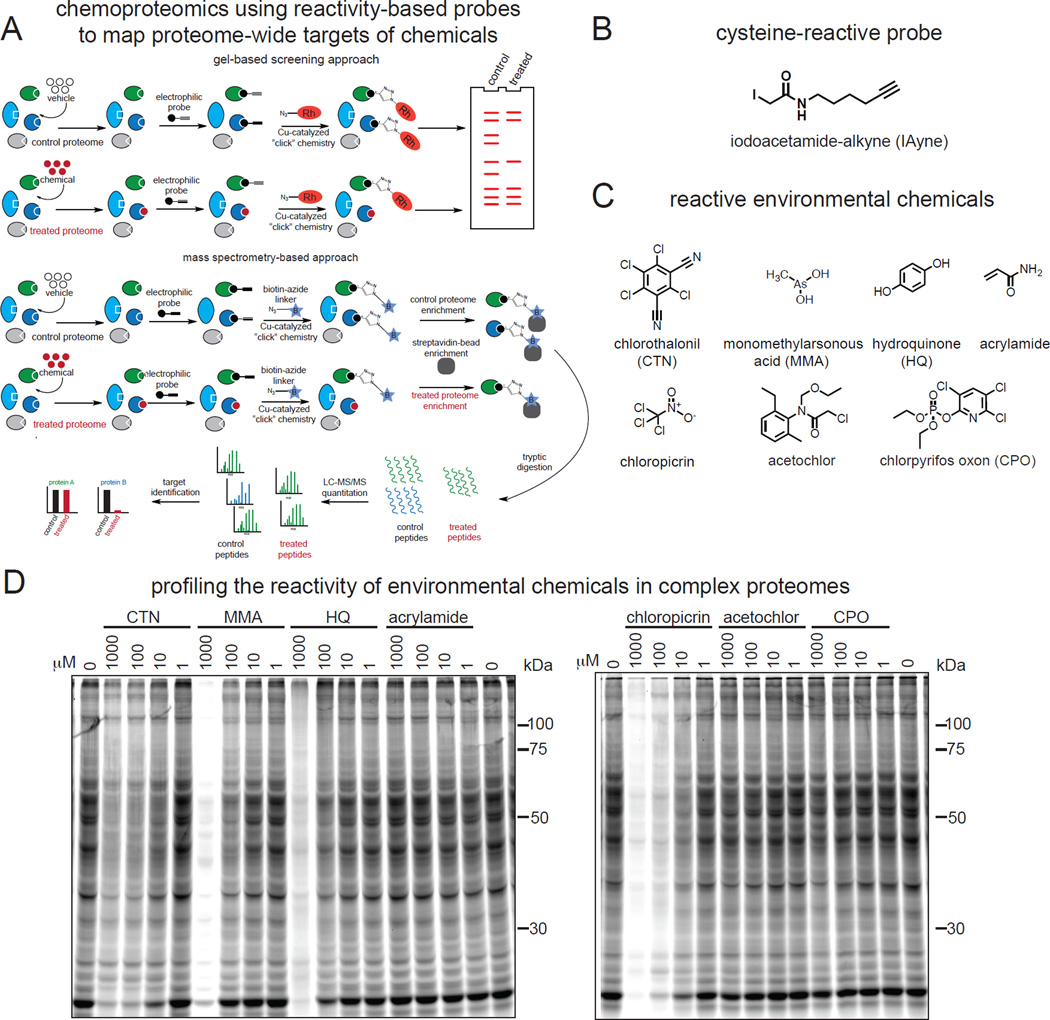
In this study, we set out to address the knowledge gap in reactive chemical toxicities and identify potentially novel mechanisms of action by mapping the proteome-wide interactions of a representative and diverse set of highly used reactive environmental electrophiles. We adapted the chemoproteomic strategy put forth by Weerapana et al. (Weerapana et al., 2010), in which we competed these chemicals against the binding of the cysteine-reactive IAyne probe to hyper-reactive sites in native tissue proteomes (Fig. 1A–1C). We have employed two complementary strategies to achieve this goal—a gel-based fluorescence assay for rapid chemical screening and a proteomic-based approach for detailed target identification. Briefly, we treated proteomes with a panel of representative reactive environmental chemicals to allow covalent binding to sensitive sites. Proteomes were then treated with the cysteine-reactive probe IAyne to competitively label available hyper-reactive cysteines, followed by appending an azide-linked rhodamine or biotin enrichment handle using copper-catalyzed azide-alkyne cycloaddition or “click chemistry,” for fluorescent or proteomic-based detection, respectively. Fluorescently tagged proteomes were separated by SDS-PAGE and biotin tagged proteomes were enriched and digested to tryptic peptides for liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic analyses. We then determined the reactivity of the environmental chemicals by either loss of fluorescent labeling or loss of MS signal.
Figure 1. Mapping proteome-wide interactions of reactive environmental chemicals using reactivity-based probes and chemoproteomic profiling.
(A–C) Here we mapped the direct chemical-protein interactions of reactive environmental chemicals, including metals (MMA), chemical breakdown products (HQ, acrylamide), and pesticides (CTN, acetochlor, chloropicrin, CPO) directly in complex proteomes. We employed a gel-based fluorescence assay for screening and a proteomic approach for target identification. Proteomes were pre-treated with either vehicle or environmental electrophiles, competitively labeled with the cysteine-reactive iodoacetamide-alkyne (IAyne) probe, then an azide-linked rhodamine or biotin enrichment handle was appended using “click chemistry” for fluorescent or proteomic detection, respectively. Fluorescently tagged proteomes were separated by SDS-PAGE and biotin tagged proteomes were enriched and digested to tryptic peptides for liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic analyses. Reactivity of environmental chemicals was determined by loss of fluorescent labeling or MS signal. (D) Shown are gel-based analyses of environmental chemical reactivity with cysteine-reactive sites on proteins in mouse liver proteomes. Proteomes were pre-treated in vitro with chemicals at the designated concentrations (30 min) and then competed with IAyne labeling (10 µM, 60 min), rhodamine-conjugated, and cysteine-reactivity visualized by SDS-PAGE and in-gel fluorescence. The gel-based analyses reveal the substantial cysteine reactivity of MMA, HQ, CTN, and chloropicrin. Shown are representative gels from n=3. Cysteine-reactivity in kidney is shown in Fig. S1.
We tested representative classes of widely used or commonly exposed reactive environmental chemicals, including metals, chemical breakdown products, and several pesticide classes for their concentration-dependent cysteine-reactivity in mouse liver and kidney proteomes against IAyne labeling of cysteine-reactive sites. Using the gel-based detection method, we observed dose-dependent reactivity of certain chemicals such as monomethylarsonous acid (MMA), a highly toxic bioactivated arsenical (Watanabe and Hirano, 2013); hydroquinone (HQ), a bioactivated metabolite of benzene (McHale et al., 2012); the widely used fungicide chlorothalonil (CTN) (Wilkinson and Killeen, 1996); and the broad-spectrum pesticide chloropicrin (Oriel et al., 2009) (Fig. 1D; Fig. S1). In contrast, chemicals such as the chloroacetanilide herbicide acetochlor, organophosphorus insecticide chlorpyrifos oxon (CPO), and the common industrial chemical and food contaminant acrylamide showed little to no observable reactivity at the tested concentrations (Fig. 1D; Fig. S1). These seemingly unreactive chemicals may still bind to protein targets but their reactivity may be masked in gel-based approaches or these chemicals may target other amino acid hotspots in the proteome. For example, our previous studies using activity-based protein profiling have shown that chlorpyrifos oxon (CPO) and triphenyl phosphate (TPP) react with the catalytic serines of several serine hydrolases (Medina-Cleghorn et al., 2014; Morris et al., 2014; Nomura et al., 2008; Nomura et al., 2005; Nomura et al., 2011). Alternatively, these chemicals may not form stable protein adducts, may be rapidly oxidized by detoxifying enzymes, or may react with other non-protein biological nucleophiles such as glutathione or nucleic acids.
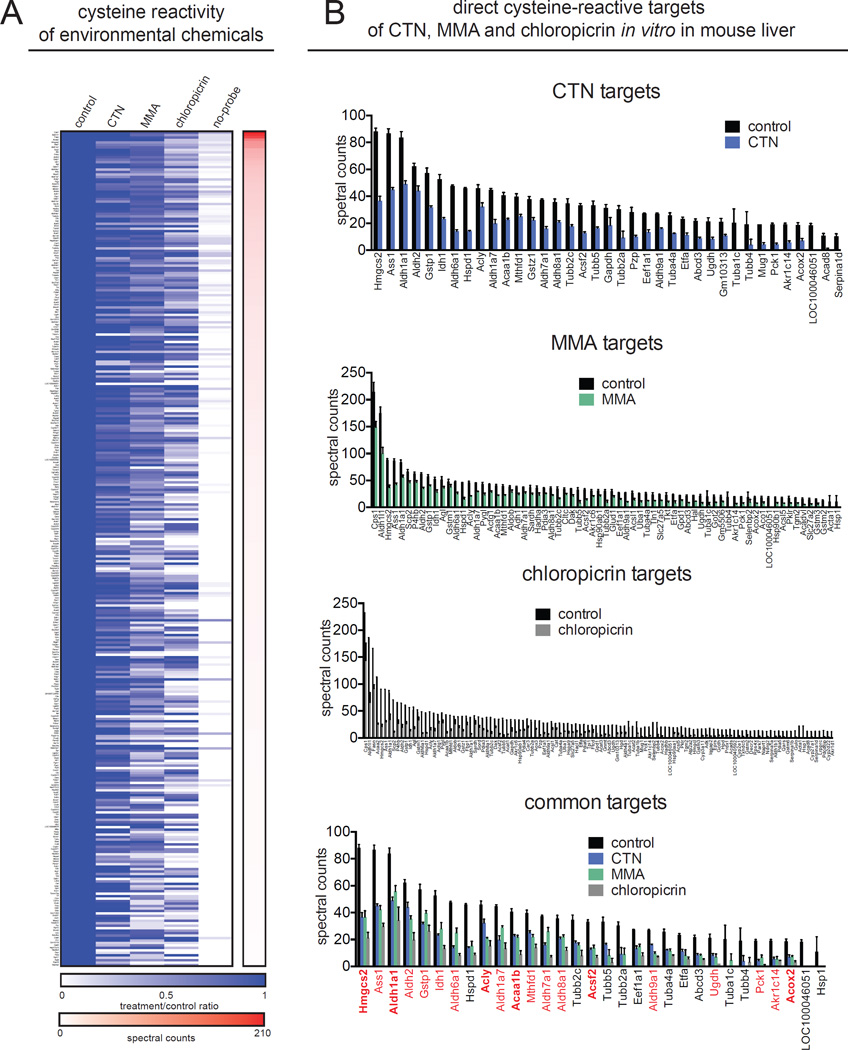
To identify the direct protein targets of the chemicals that showed reactivity by our gel-based detection methods, we first applied our LC-MS/MS-based competitive chemoproteomic analysis to MMA, CTN, and chloropicrin at 100 µM in vitro in mouse liver proteomes (Fig. 2A; Table S1). Based on literature and chemical properties, these chemicals were determined as likely to directly and covalently interact with protein targets in complex proteomes, as compared to potentially indirect mechanisms of HQ such as generation of oxidative stress. Out of 339 total targets enriched by IAyne labeling (>5 spectral counts, >2-fold above no-probe DMSO controls), we found that CTN, MMA, and chloropicrin interact with 37, 69, and 119 protein targets, respectively (Fig. 2B; Table S1). First, we note that by using this chemoproteomic approach, we were able to enrich cysteine-reactive protein targets across hundreds of proteins spanning diverse protein classes (Table S1). Second, while each reactive agent shows a distinct protein target profile, CTN, MMA, and chloropicrin collectively bind 31 protein targets in common, spanning many unrelated protein classes that include metabolic enzymes, cell structure proteins, chaperones, and translation-related proteins. Third, we also show that the targets of individual chemicals are not merely restricted to the most abundant IAyne enriched targets, but instead span the entire range of detected protein abundances, suggesting that these interactions result from a combination of affinity and reactivity towards these targets and not just through non-specific reactivity with the most abundant IAyne-enriched targets (Fig. 2A, 2B). Conversely, there are many high-abundance IAyne enriched proteins that were not bound by CTN, MMA, or chloropicrin. Most of the targets of CTN, MMA, and chloropicrin identified by our chemoproteomic approach have not been previously identified as targets of these agents (Fig. 2B; Table S1). Interestingly, 19 of the 31 shared targets are enzymes involved in key metabolic processes (highlighted in red in Fig. 2B), and six of these enzymes are involved in different aspects of lipid metabolism (in red and bold in Fig. 2B). Hmgcs2, Acaa1b, Acsf2, and Acox2 are all involved in energetic fatty acid oxidation; Acly is involved in producing acetyl-CoA for de novo lipogenesis, and Aldh1a1 catalyzes the oxidation of retinaldehyde to form the critical signaling molecule retinoic acid (Kanehisa and Goto, 2000). These agents binding covalently to reactive nucleophiles on their target enzymes may disrupt lipid metabolism in particular, representing a novel mechanism of toxicity not previously associated with these chemicals. Thus, we show that structurally dissimilar reactive environmental chemicals share certain specific and overlapping targets predominantly involved in metabolism. These protein targets may be particularly susceptible to disruption by environmental electrophiles, and exposure to multiple agents may synergistically affect protein function to exert significant toxicities.
Figure 2. Target identification of reactive environmental chemicals.
(A) Competitive chemoproteomic analysis for direct protein target identification of CTN, MMA, and chloropicrin (reactive chemicals from Fig. 1D) was performed using mouse liver proteome treated with vehicle or chemical (100 µM, 30 min) prior to IAyne labeling (10 µM, 30 min) or vehicle (no-probe control). Each biological replicate uses a liver homogenate from a different 6 week old male C57BL/6 mouse. Subsequent biotin conjugation, avidin-enrichment of targets, and analysis of trypsin-digested peptides by LC-MS/MS revealed enrichment of 339 total proteins by IAyne labeling spanning diverse protein classes (>5 spectral counts, >2-fold above no-probe DMSO controls). Data in (A) are expressed as heat maps showing ratios of treatment compared to control. IAyne-enriched targets are sorted by their spectral counts. (B) MMA, CTN, and chloropicrin significantly displace IAyne labeling of protein targets in vitro in mouse liver (p<0.05 compared to vehicle-treated controls using two-way ANOVA with Tukey’s multiple comparisons test). MMA, CTN, and chloropicrin possess 31 overlapping targets. Highlighted in red are metabolic enzyme targets and in bold are those enzymes that are involved in lipid metabolism. Data in (B) are presented as mean ± sem; n=3 per group. Raw proteomic data and statistical analyses are in Table S1. HQ targets are shown in Fig. S2.
In addition to CTN, MMA, and chloropicrin, we also profiled HQ and acrylamide reactivity at 100 µM in vitro in mouse liver proteome. We identified 12 protein targets of HQ, most of which do not overlap with those of CTN, MMA, and chloropicrin (Fig S2; Table S1). This lack of overlap may be due to the indirect mechanism of HQ in generating reactive oxygen stress that may be leading to cysteine oxidation (Kalf, 1987), compared with the potentially covalent reactive mechanisms of CTN, MMA, and CPR. Consistent with the gel-based approaches, we observed no direct targets of acrylamide (Table S1). It is possible that this apparent lack of HQ and acrylamide reactivity may be due to insufficient concentration or the need for bioactivation, or that these compounds are rapidly oxidized by detoxifying enzymes, may form transient or unstable protein adducts, or may react with other biological nucleophiles. We also note the possibility that we may be masking hyper-reactive cysteines present in living cells that may become oxidized during tissue proteome preparation.
CTN shows selective reactivity with protein targets in vivo in mouse kidney
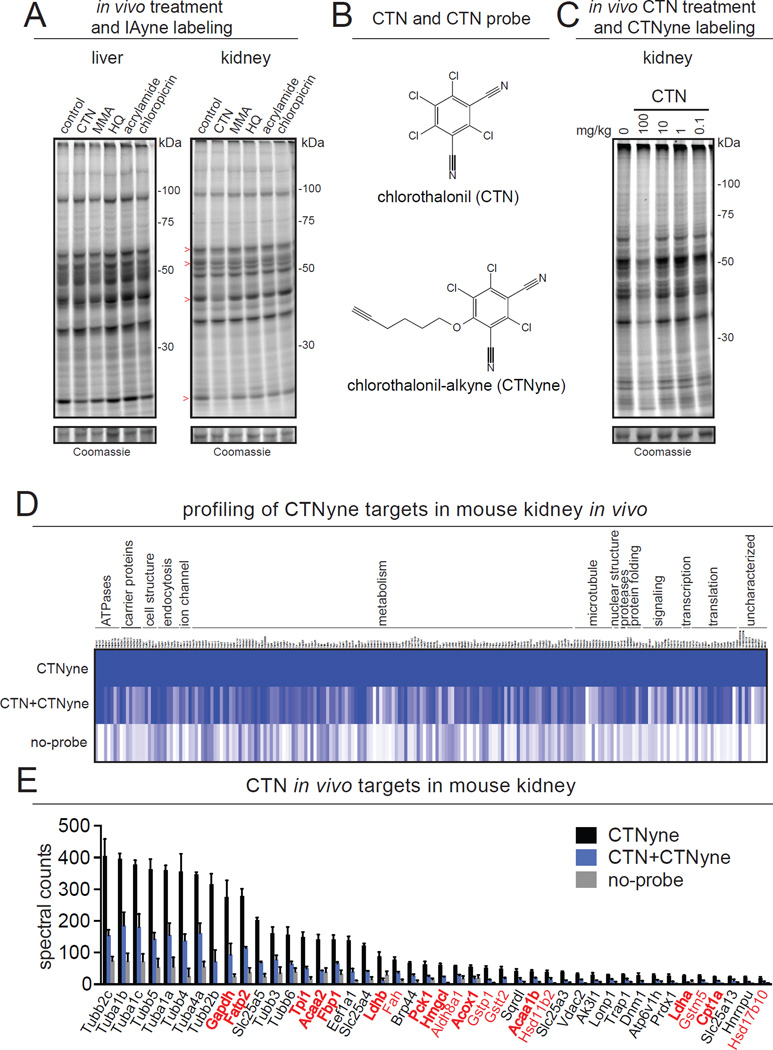
Building on our in vitro profiling, we next wanted to validate our chemoproteomic strategy using an in vivo model of exposure in mice. We treated mice with the maximally tolerated doses of CTN, MMA, HQ, acrylamide, and chloropicrin at 100, 1, 50, 100, and 4 mg/kg ip, respectively, and subsequently labeled liver and kidney proteomes ex vivo with IAyne to detect in vivo cysteine reactivity (Fig. 3A). While we did not observe reactivity using gel-based methods in liver, CTN showed considerable in vivo reactivity in mouse kidney (Fig. 3A). We thus selected CTN for further study. CTN is one of the most widely used pesticides, with 9 million pounds applied per year in US agriculture alone (Grube et al., 2011). Despite its heavy usage, CTN has also been shown to cause renal toxicity and kidney carcinogenesis in multiple animal models, but the underlying toxicological mechanisms remain poorly understood (Wilkinson and Killeen, 1996). Thus, our finding that CTN showed selective reactivity in mouse kidney compared to liver was interesting (Fig. 3A).
Figure 3. CTN binds metabolic enzymes in vivo involved in energetics and fatty acid metabolism.
(A) Reactivity of CTN, MMA, HQ, acrylamide, and chloropicrin was assessed by in vivo treatment in mice (at maximum tolerated dose of 100, 1, 50, 100, and 4 mg/kg, ip 2 h), and subsequent labeling of liver and kidney proteomes with IAyne (10 µM, 30 min) and visualization of cysteine-reactive targets by SDS-PAGE and in-gel fluorescence. CTN reactivity in vivo is displayed through displacement of IAyne binding ex vivo, indicated by “>” signs. (B) We developed a tailored bioorthogonal probe, CTN-alkyne (CTNyne) to comprehensively identify mouse in vivo CTN targets in the relevant organ, kidney. (C) CTNyne labels multiple protein targets in mouse kidney (10 µM, 30 min), and is displaced by in vivo CTN treatment (100 mg/kg, ip, 2h). (D, E) In vivo CTN-specific protein targets were identified by labeling in vivo vehicle- and CTN-treated mouse kidney proteomes with CTNyne (50 µM, 30 min), followed by biotin conjugation, avidin enrichment, and tryptic peptide analysis by LC-MS/MS. (D) 216 protein targets were significantly enriched by the CTNyne probe compared to no-probe control. Data are presented as a heat map of total CTNyne-enriched targets normalized to the average spectral counts of CTNyne control for each protein. (E) 44 proteins were identified as CTN targets that were >20 spectral counts in abundance and significantly (p<0.05 by two-tailed t-test) competed greater than 2-fold by in vivo CTN treatment. 19 of these targets are metabolic enzymes (red emphasis), 12 of which are involved in glycolytic, gluconeogenic, or fatty acid metabolic pathways (bold red emphasis). Data in (A, C) are representative gels from n=3–4 mice/group. Data in (D, E) are from n=3–4 mice/group and raw proteomic data is in Table S2. Data in (E) are presented as mean ± sem; n=3 per group. In vivo liver targets of CTN are shown in Fig. S3.
Encouraged by these results, we conducted a comprehensive assessment of CTN reactivity in vivo in mouse liver and kidney by developing a tailored bioorthogonal probe, CTNyne (Fig. 3B; Fig. S3). The CTNyne probe is a structural analog of CTN that contains a bioorthogonal alkyne handle for click chemistry conjugation after labeling in proteomes. We competitively labeled liver and kidney proteomes from mice treated in vivo with vehicle or CTN with the CTNyne probe and appended a rhodamine reporter for in-gel fluorescence analysis. We confirmed that CTNyne labels multiple targets in kidney, and that many of these targets were competed by in vivo CTN-treatment, demonstrating the increased specificity for CTN targets using the CTNyne probe (Fig. 3C). While CTNyne labeled many protein targets in liver, most of these targets were not displaced by in vivo CTN treatment, recapitulating our previous observation with IAyne labeling (Fig. S3).
CTN affects metabolic enzymes involved in energetics and fatty acid metabolism
Based on the findings from our in-gel fluorescence analysis, we next sought to identify the in vivo targets of CTN in mouse kidney using our CTNyne probe and gain mechanistic insight into CTN toxicity. We used the kidney proteomes of mice exposed in vivo to vehicle or CTN for an ex vivo competitive labeling by our CTNyne probe, followed by LC-MS/MS-based chemoproteomic analysis. Through this approach, we identified 216 protein targets that were significantly enriched by the CTNyne probe compared to no-probe controls (Fig. 3D; Table S2). From these enriched proteins, we identified 44 targets that were significantly competed by in vivo CTN treatment more than 2-fold and were detected at >20 spectral counts in abundance (Fig. 3E; Table S2). In contrast to our findings in vivo in mouse kidney, we identified only 5 in vivo targets in mouse liver (Fig S3; Table S2).
Of the 44 in vivo kidney targets of CTN that we identified, we were intrigued to find that a substantial proportion of these targets (19 proteins) were enzymes involved in the metabolism of small-molecule metabolites (red emphasis, Fig. 3E). The majority of these enzymes (12 targets) are involved in energetics, participating in glycolytic, gluconeogenic, or fatty acid metabolic pathways (bolded red emphasis in Fig. 3E). These enzymes include the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase (Gapdh), triosephosphate isomerase 1 (Tpi1), and lactate dehydrogenases (Ldha, Ldhb), and the gluconeogenic enzymes fructose-1,6-bisphosphatase 1 (Fbp1) and pyruvate carboxykinase 1 (Pck1). Enzyme targets involved in fatty acid transport or oxidation include fatty acid transport protein 2 (Fatp2), 3-ketoacyl-CoA thiolase 1b and 2 (Acaa1b and Acaa2), peroxisomal acyl-CoA oxidase 1 (Acox1), and carnitine palmitoyltransferase 1a (Cpt1a), as well as the ketone body-generating enzyme 3-hydroxymethyl-3-methylglutaryl-CoA lyase (Hmgcl). Collectively, our results indicated that CTN might alter renal energetics and fatty acid metabolism.
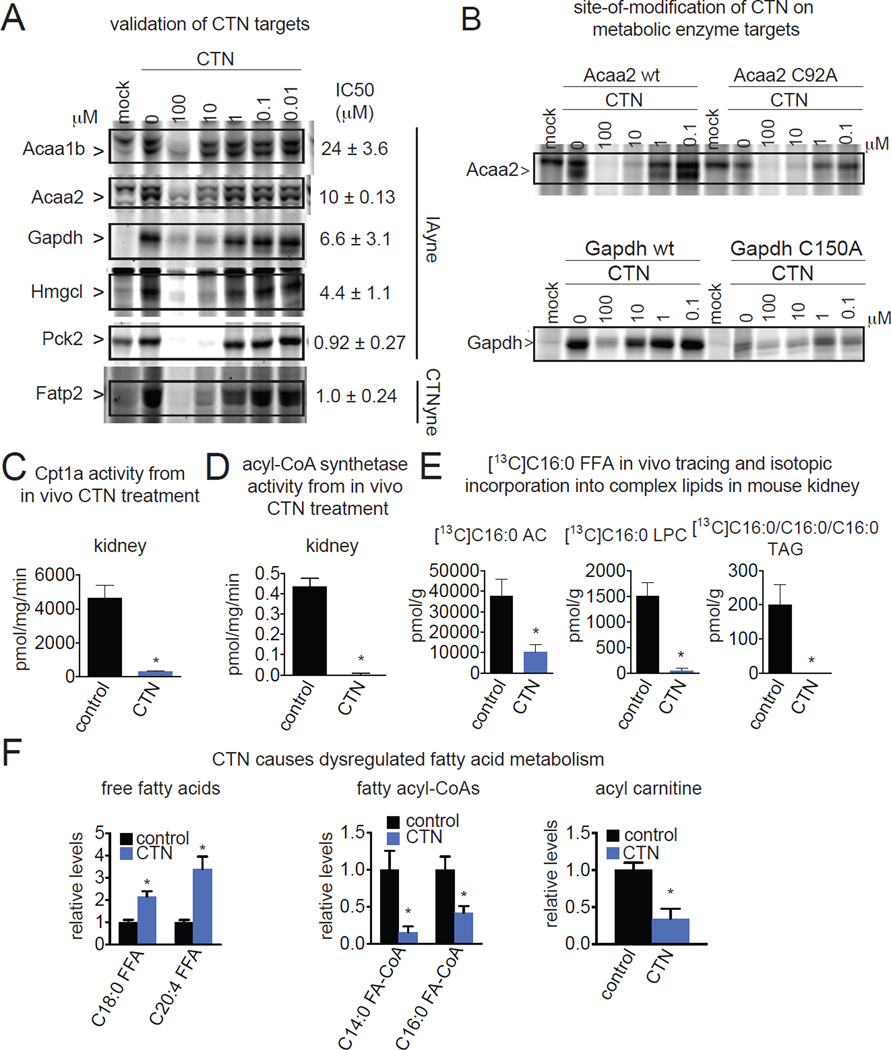
To validate these metabolic enzymes as true CTN-specific hits, we recombinantly overexpressed representative targets from each pathway in HEK293T cells and subsequently performed in vitro competition studies with CTN against IAyne or CTNyne labeling. Indeed, we show that Acaa1b, Acaa2, Gapdh, Hmgcl, and Pck1 were labeled by IAyne and this labeling was displaced in a concentration-dependent manner by CTN, indicating that CTN specifically bound to one or more hyper-reactive cysteine on these proteins (Fig. 4A). While we found that CTN displaced binding of CTNyne to Fatp2 (Fig. 4A), Fatp2 was not labeled by IAyne (data not shown), suggesting that CTN may bind to another hyper-reactive residue on this protein besides cysteine. We calculated 50 % inhibitory concentration (IC50) values of 0.9–24 µM for CTN binding to Acaa1b, Acaa2, Gapdh, Hmgcl, Pck2, and Fatp2 (Fig. 4A). We note that the IC50s are a rough estimate of relative reactivity, since the reaction is covalent and there is no equilibrium. Furthermore, we demonstrate that CTN is binding to catalytic cysteines on key energetic enzymes through blunted or eliminated IAyne labeling upon mutatgenesis of Acaa2 and Gapdh catalytic cysteines to C92A and C150A, respectively (Fig. 4B; Fig. S4).
Figure 4. CTN inhibits the activity of several metabolic enzymes involved in energetics and fatty acid metabolism leading to dysregulated lipid metabolism.
(A) Representative metabolic enzyme targets of CTN in vivo in mouse kidney were validated by recombinant target overexpression in HEK293T and COS7 cells. Lysates from mock- or target-overexpressing cells were preincubated with vehicle or 0.01–100 µM CTN (30 min), followed by labeling with IAyne or CTNyne (10 µM, 30 min). 50 % inhibitory concentration (IC50) values are reported. (B) CTN binds the catalytic cysteine on Acaa2 (C92) and Gapdh (C150), as confirmed by CTN-sensitive IAyne labeling of Acaa2 and Gapdh is substantially lower in Acaa2 C92A and Gapdh C150A-overexpressing compared to wild-type Acaa2 and Gapdh-overexpressing HEK293T lysates. Expression controls for wild-type and mutant enzymes are in Fig. S4, ascertained by qPCR analysis of RNA harvested alongside these experiments showing equivalent expression. (C, D) Carnitine palmitoyltransferase and fatty acyl-CoA synthetase activity are significantly inhibited in in vivo CTN-treated (100 mg/kg ip, 2 h) mouse kidney proteomes compared to vehicle-treated controls. Carnitine palmitoyltransferase activity was assessed ex vivo using C16:0 free fatty acid (FFA) and trimethyl-d9-L-carnitine measuring C16:0-d9-carnitine formation by SRM-based LC-MS/MS. Acyl-CoA synthetase activity was assessed ex vivo using [U-13C]C16:0 FFA and CoA, measuring [U-13C]C16:0-CoA formation by SRM-based LC-MS/MS. (E) Isotopic incorporation of [U-13C]C16:0 FFA into complex lipids in vivo in kidney was measured ex vivo by SRM-based LC-MS/MS from mice treated with vehicle or CTN (100 mg/kg, ip 1 h), followed by [U-13C]C16:0 acid (100 mg/kg ip, 1 h). (F) Consistent with inhibition of CTN target fatty acid transport and oxidation enzymes, in vivo CTN treatment (100 mg/kg ip, 2h) leads to elevated FFA levels, decreased C14:0 and C16:0 fatty acyl-CoA (FA-CoA), and decreased C12:0 acyl carnitine (AC) levels in mouse kidney, compared to vehicle-treated controls. Data in (A, B) are representative gels from n=3/group. Data in (C–F) are presented as mean ± sem; n=3–5 mice/group. Significance is presented as *p<0.05 compared to vehicle-treated controls using two-tailed tests. Data in (F) show fatty acid-derived metabolites with significantly (p<0.05) altered abundances in two independent experiments. Raw metabolomic data is in Table S3 and a more detailed overview of the metabolomics data is shown in Fig. S4.
Though we were unsuccessful at recombinantly overexpressing Cpt1a, we confirmed that the endogenous kidney carnitine palmitoyltransferase activity from in vivo CTN-treated mice was significantly inhibited compared to vehicle-treated controls, as measured by an activity assay using trimethyl-d9-L-carnitine and C16:0 free fatty acid (C16:0 FFA) substrates measuring C16:0-trimethyl-d9-carnitine product formation by LC-MS/MS (Fig. 4C). We also wanted to determine whether CTN inhibits the activity of Fatp2, an enzyme with both peroxisomal fatty acyl-CoA synthetase activity and cytosolic fatty acid transport activity (Falcon et al., 2010). We indeed show that acyl-CoA synthetase activity was completely inhibited in the kidneys of mice treated in vivo with CTN compared to vehicle-treated controls, as measured by an activity assay using [U-13C]C16:0 FFA) substrate and measuring [U-13C]C16:0 fatty acyl-CoA (FA-CoA) product formation by single-reaction monitoring (SRM)-based LC-MS/MS (Fig. 4D). Next, to determine whether fatty acid transport might be impaired in vivo due to inhibition of Fatp2, we performed an in vivo isotopic fatty acid tracing study and showed that ip administered isotopically labeled [U-13C]C16:0 FFA incorporation into representative complex lipids in mouse kidney was significantly lower in CTN-treated mice, including isotopically labeled species of acyl carnitine (AC), lysophosphatidylcholine (LPC), and triacylglycerols (TAGs) (Fig. 4E). While we cannot directly attribute these activities to Fatp2, these data taken together suggest that fatty acid transport and acyl-CoA synthetase activities attributable to Fatp2 are inhibited in vivo by CTN in the kidney.
We next wanted to assess the downstream biochemical effects that may arise from inhibition of the metabolic enzyme targets of CTN, especially those involved in fatty acid metabolism. We performed SRM-based LC-MS/MS metabolomic analyses on kidneys from mice treated in vivo with CTN to identify alterations in the lipidome conferred by CTN treatment. Consistent with our results suggesting that CTN disrupts fatty acid transport both into cells through Fatp2 inhibition and into mitochondria through Cpt1a inhibition, and causes impairments in fatty acid oxidation, we observe expected increases in FFA levels and depletions in the levels of FA-CoAs (Fatp2 and β-oxidation products) and acyl carnitines (AC, Cpt1a product) in mouse kidney from in vivo CTN treatment (Fig. 5E; Fig. S4; Table S3). CTN has previously been shown to react directly with glutathione causing glutathione depletion and toxicity (Wilkinson and Killeen, 1996). However, we show that, at the doses used here, both liver and kidney glutathione levels were unchanged upon CTN administration (Fig. S4).
Collectively, we show that the major fungicide CTN inhibits metabolic enzymes in kidney that are involved in energetic metabolism, leading to downstream biochemical alterations in glycolysis and fatty acid metabolism.
Conclusions
Here, we have used a chemoproteomic platform employing reactivity-based chemical probes to map the proteome-wide interactions of environmental chemicals and uncover novel mechanisms through which these chemicals may cause adverse health effects. We show here that individual environmental electrophiles engage specific protein targets with privileged reactivity, and do not non-specifically target all proteins or merely react with the most abundant targets. This indicates that these agents possess affinities to highly reactive nucleophiles on specific proteins, suggesting that the mechanisms of toxicity may be through interactions with these selected proteins to cause distinct downstream adverse biochemical events. By competing these chemicals against a cysteine-reactive probe, we show that seemingly dissimilar reactive environmental agents including the fungicide CTN, the arsenical MMA, and the insecticide chloropicrin possess both distinct and overlapping targets in the proteome, suggesting that these protein targets may be particularly susceptible to reactive electrophiles. It will be of future interest to examine whether prolonged or combined exposure to these and other reactive chemicals causes sustained and cumulative adduction of these common targets, leading to adverse biochemical and pathophysiological effects.
We note that the other reactive environmental chemicals we screened that did not show cysteine reactivity by our gel-based fluorescence method may still react with protein targets. These chemicals may potentially bind to hyper-reactive cysteines on particular targets that are simply poorly resolved by SDS-PAGE, may bind at even higher concentrations, or may bind to other hyper-reactive amino acid side-chains. For example, while the flame retardant TPP and the bioactivated insecticide metabolite CPO do not show cysteine reactivity, we have previously shown that these chemicals covalently bind to and inhibit catalytic serines of several serine hydrolases (Medina-Cleghorn et al., 2014; Morris et al., 2014; Nomura et al., 2008; Nomura and Casida, 2011). Nonetheless, the enzyme targets that we identify here represent multiple, potentially novel mechanisms of toxicity that may even be in common across broad classes of environmental electrophiles. We also note that we may still be missing some direct protein targets of the chemicals that we showcase in this study. The cysteine-reactive probe may be labeling multiple sites on each protein, and thus we may be obscuring chemical-protein interactions when we quantify enriched proteins based on total peptides for each protein. Additionally, certain chemicals may also require bioactivation in specific tissues in vivo before they may bind highly reactive nucleophilic residues. It will be of future interest to complement our study with a more comprehensive quantitative proteomics approach for identifying sites of probe modification and sites of chemical displacement of probe binding.
We also demonstrate the ability to apply the chemoproteomic strategies described in this study to assess in vivo reactivity of environmental chemicals, which enables assessment of target organs that may be particularly susceptible to toxicity from environmental agents. Using the fungicide CTN as an example, we showed that CTN exhibits particular reactivity with protein targets in vivo in mouse kidney compared to liver. Through the compound-specific CTNyne probe, we identified 44 direct protein targets of CTN in vivo in mouse kidney. We found that this target list was enriched in metabolic enzymes involved in glycolytic and fatty acid metabolism and that the activities of these enzymes were seemingly inhibited. Through subsequent metabolomic profiling, we observed a dysregulation of lipid metabolism, consistent with our target list. The inhibition of glycolytic and gluconeogenic metabolism, fatty acid transport, and fatty acid oxidation enzymes, potentially leading to altered levels of key metabolites and perturbed metabolic networks represents a novel mechanism of toxicity for CTN.
While the doses of CTN used in this study to identify direct in vivo targets were high, the apparent IC50 values of CTN against the targets that we have validated here are within an order of magnitude of the concentration of CTN that has been observed in human serum. Barr et al. previously reported up to 25 ng/g of CTN found in human maternal and cord sera, which equates to ~100 nM (Barr et al., 2010). Acaa2, Acaa1b, Gapdh, Fatp2, and Pck1 showed 9.4 ± 1.2 %, 3.4 ± 3.4 %, 20 ± 8.4 %, 13 ± 6.7 %, and 6.8 ± 6.8 % reduction in IAyne labeling compared to vehicle-treated controls. Acaa2 and Gapdh were significantly inhibited (p<0.05 compared to vehicle-treated controls using a two-tailed t-test) at 100 nM.
Indeed, studies in animal models have linked CTN to selective kidney toxicity and carcinogenesis, but the mechanisms have remained unclear (Wilkinson and Killeen, 1996). Here, we suggest that the kidney-specific toxicity of CTN may be due to the inhibition of energetic and fatty acid metabolism enzymes, which may impair cell function, cause cellular necrosis, and subsequent kidney dysfunction. When considered in the context of chronic exposure these toxicities may develop into pathophysiologies such as inflammation, fibrosis, and cancer. Because CTN presumably acts through covalent modification of these targets, cumulative exposure may cause an accumulation of protein adducts on these targets leading to their eventual inhibition.
While we have identified novel potential toxicological mechanisms of CTN, we still do not understand the basis for the observed selective reactivity of CTN in kidney compared to liver. Previous studies have postulated that CTN may cause kidney-specific toxicity through glutathione conjugation and further bioactivation by β-lyase, an enzyme highly expressed in renal proximal tubules (Wilkinson and Killeen, 1996). Thus, while we show here that CTN itself directly interacts with the observed in vivo CTN targets, CTN may also undergo further kidney-specific bioactivation to inhibit the observed targets. It will be of future interest to investigate the role of CTN metabolites in inhibiting the CTN targets, and to perform long-term and low-dose chronic treatment studies to determine whether CTN may cause renal toxicity or carcinogenesis through inhibiting these targets.
Many of the metabolic enzyme targets inhibited by CTN and other reactive agents used this study involve active-site cysteines that directly contribute to the catalytic mechanisms. For example, Gapdh uses the active site cysteine to form a hemithioacetal intermediate and many of the fatty acid oxidation enzymes (e.g. Acaa1b and Acaa2) form thioester intermediates using their active site cysteines (Mathieu et al., 1997; Poirier et al., 2006; Seidler, 2013). The enrichment of metabolic enzymes amongst the observed targets may suggests that enzymes which use cysteines in their mechanism are particularly susceptible to reactive environmental electrophiles. In this study, we have demonstrated that chemoproteomic profiling using reactivity-based probes enables the mapping of direct proteome-wide interactions of reactive environmental chemicals, which provides mechanistic detail for downstream toxicities. We also show that metabolomic profiling can be coupled with chemoproteomic-based target identification towards demonstrating the biochemical outcomes associated with interactions of these chemicals with specific protein targets.
Looking forward, we anticipate that metabolomics approaches will enable identification of robust biomarkers of exposure and of toxicity for CTN and other environmental chemical agents. Furthermore, we foresee using chemoproteomic platforms to screen emerging industrial, environmental, or pharmaceutical chemicals against a suite of reactivity-based probes for other hyper-reactive amino acids will reveal potential off-targets and toxicological liabilities early on in the development pipeline. The advance knowledge of specific reactivities and detailed mechanistic insights revealed by this approach can be used to direct efforts towards optimizing the chemical structures and ensuring the selectivity and safety of future chemicals.
Methods
Mice
Male C57BL/6 mice (6–8 weeks old) were acutely (2 hours) exposed by intraperitoneal (ip) injection to 100 mg/kg CTN (TOCRIS) in a vehicle of 42:2.5:1:1 saline/ DMSO/ emulphor/ ethanol (10 µl/g mouse). For isotopic tracing studies, mice were first exposed to 100 mg/kg CTN by ip injection for 1 h, then exposed to 100 mg/kg 12C16:0 FFA (Sigma) or U-13C16:0 FFA (Cambridge Isotopes) dissolved in undiluted PEG40 castor oil (Spectrum) by ip injection for 1 hour. Following exposure, mice were sacrificed by cervical dislocation and livers and kidneys were immediately removed and flash frozen in liquid nitrogen. Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of California, Berkeley.
Processing of Mouse Brain, Liver, and Kidney Proteomes
Tissues were homogenized in Mg2+/Ca2+-free 1x phosphate-buffered saline (PBS, Corning), followed by a 1000 × g centrifugation of the homogenate. The resulting supernatant was collected and used for subsequent assays. Protein concentrations were determined by Bradford assay (Bio-Rad).
Cell Culture, Recombinant Overexpression, and Site-Directed Mutagenesis
HEK293T and COS7 cells were cultured in DMEM media containing 10% FBS and maintained at 37°C with 5% CO2. Recombinant cDNA constructs containing enzymes of interest in the SPORT6 vector were purchased from Thermo Open Biosystems and transiently transfected into HEK293T (Acaa1b, Acaa2, Fatp2, Gapdh) cells using Fugene HD (Promega) or into COS7 cells (Hmgcl, Pck1) using Lipofectamine 2000 (Life Technologies). Site-Directed Mutagenesis was performed using a QuikChange II XL kit (Agilent Technologies) and mutant plasmids were transiently transfected into HEK293T cells. Further details can be found in Supplemental Methods.
Electrophile Competition and Click Chemistry Procedures to Append Analytical Handles to IAyne- or CTNyne-Treated Mouse Proteomes
Proteome samples diluted in PBS (10, 25, or 50 µg in 50 µl PBS for gel-based analysis; 0.5 mg liver homogenate in 0.5 ml PBS or 1 mg kidney homogenate in 0.5 ml PBS) were subjected to vehicle or electrophile treatment for 30 min at 37°C. Then, IAyne (1 µM or 10 µM, gel-based assays, 10 µM MS-based assays, CHESS Gmbh) or CTNyne (10 µM, gel-based assays, 50 µM, MS-based assays) labeling was performed for 30 min (gel-based analysis) or 1 hour (MS-based analysis) at room temperature. Copper-catalyzed azide-alkyne cycloaddition “click chemistry” was performed using previously described methods (Nomura et al., 2010; Weerapana et al., 2010). Fluorescent and proteomic detection of IAyne or CTNyne labeled bands were performed as previously reported (Nomura et al., 2010; Weerapana et al., 2010). Further details can be found in Supplemental Methods.
Metabolomic profiling
Metabolomic profiling was performed using previously reported procedures (Benjamin et al., 2013). Additional details are reported in Supplemental Methods.
Activity Assays
Carnitine palmitoyltransferase activity was assayed by incubating kidney lysate from in vivo exposed mice at 10 µg in 400 µl PBS with 100 µM C16:0 FA-CoA lithium (Sigma-Aldrich), 100 µM trimethyl-d9-L-Carnitine (Cambridge Isotopes) for 5 min at 37°C. Reactions were quenched by addition of 1.2 ml 2:1 chloroform:methanol with dodecylglycerol (10 nmol, internal standard) and mixed by vortexing. The organic layer was collected and dried under a stream of N2 then resuspended in 120 µl chloroform for SRM based LC-MS/MS analysis targeting palmitoyl-d9-trimethyl-carnitine and analyzed as previously described.
Acyl-CoA synthetase activity was assayed as using a modified protocol based on Falcon et. al (Falcon et al., 2010). Briefly, 5 mg kidney lysate from mice treated in vivo with CTN or vehicle were incubated in 200 µl Tris-HCl (pH 8.5) buffer containing 10 mM ATP (Sigma), 10 mM MgCl2 (Sigma), 200 µM Coenzyme A (Sigma) and 10 µM U-13C16:0 FFA (Cambridge Isotopes) was added for 30 min at 37°C. Reactions were quenched with 750 µl Dole’s Reagent (20:5:1 2-propanol: n-heptane: 2M H2SO4), vortexed, and dried under a stream of N2. The product was then resuspended in a buffer of 4 ml 2:1:1 chloroform:methanol:PBS + 0.1% formic acid, containing 10 nmol pentadecanoic acid standard. The product was mixed by vortexing and the organic layer was collected, dried under a stream of N2, resuspended in 120 µl chloroform and analyzed by SRM based LC-MS/MS analysis targeting U-13C16:0 FA-CoA and analyzed as previously described.
Statistical analyses
For proteomic experiments comparing proteome-wide interactions of several environmental chemicals, a 2-way ANOVA with Tukey’s multiple comparisons test was used for statistical analysis. For proteomic and metabolomics analyses of in vivo CTN targets comparing vehicle versus CTN-treatment in liver or kidney, we used a two-tailed t-test coupled with a >2-fold difference to identify significant (p<0.05) CTN targets or CTN-induced changes in metabolite levels was used (Bourgon et al., 2010; McCarthy and Smyth, 2009).
Supplementary Material
Significance.
We are exposed to a growing number of chemicals in our environment, most of which have not been characterized in terms of their toxicological potential or mechanisms. Research into understanding how chemicals may produce adverse effects has primarily focused on DNA-mutating or reversible endocrine disrupting mechanisms, and has largely ignored potential alternate toxicological mechanisms arising from chemical interactions with the >20,000 protein targets in the human proteome. Particularly concerning among chemicals in our environment are reactive electrophiles that have the potential to covalently react with amino acid hotspots and lead to protein dysfunction and pathological effects. Here, we have used a reactivity-based chemoproteomic platform to map the cysteine reactivity of environmental chemicals and show that contaminants such as monomethylarsonous acid and widely used pesticides such as chlorothalonil and chloropicrin possess common reactivity with a distinct set of sensitive proteins. Many of these proteins are involved in key metabolic processes, suggesting that these targets may be particularly sensitive to environmental electrophiles. We also show that the widely used fungicide chlorothalonil specifically inhibits several metabolic enzymes involved in fatty acid metabolism and energetics, leading to dysregulated lipid metabolism in mice. Our results highlight the utility of using reactivity-based chemoproteomic platforms to uncover novel mechanistic insights into the toxicity of environmental chemicals, and provide a potential framework for testing emerging chemicals.
Highlights.
We used a chemoproteomic platform to map the reactivity of environmental chemicals.
Several proteins involved in metabolism are targets of environmental electrophiles.
A major fungicide inhibits fatty acid metabolism leading to altered lipid profiles.
Acknowledgements
We thank Dr. Kathy Collins of the University of California, Berkeley for use of their Typhoon flatbed scanner. We thank the UC Davis Proteomics Core for MS-based proteomics detection. This work was supported by the Searle Scholar Award, the Center for Environmental Research on Toxics, the National Institutes of Health (P42ES004705; R01CA172667), the American Cancer Society Research Scholar Award (RSG-14-242-01-TBE), and BASF-CARA Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends in biotechnology. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr DB, Ananth CV, Yan XY, Lashley S, Smulian JC, Ledoux TA, Hore P, Robson MG. Pesticide concentrations in maternal and umbilical cord sera and their relation to birth outcomes in a population of pregnant women and newborns in New Jersey. Sci Total Environ. 2010;408:790–795. doi: 10.1016/j.scitotenv.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookman EB, McAllister K, Gillanders E, Wanke K, Balshaw D, Rutter J, Reedy J, Shaughnessy D, Agurs-Collins T, Paltoo D, et al. Gene-environment interplay in common complex diseases: forging an integrative model-recommendations from an NIH workshop. Genetic epidemiology. 2011;35:217–225. doi: 10.1002/gepi.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher JR. Regulatory Forum Opinion Piece*: Tox21 and Toxicologic Pathology. Toxicol Pathol. 2013;41:125–127. doi: 10.1177/0192623312450632. [DOI] [PubMed] [Google Scholar]
- Codreanu SG, Ullery JC, Zhu J, Tallman KA, Beavers WN, Porter NA, Marnett LJ, Zhang B, Liebler DC. Alkylation damage by lipid electrophiles targets functional protein systems. Molecular & cellular proteomics : MCP. 2014;13:849–859. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicological sciences : an official journal of the Society of Toxicology. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- Falcon A, Doege H, Fluitt A, Tsang B, Watson N, Kay MA, Stahl A. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am J Physiol-Endoc M. 2010;299:E384–E393. doi: 10.1152/ajpendo.00226.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube A, Donaldson D, Kiely T, Wu L. Pesticide Industry Sales and Usage: 2006 and 2007 Market Estimates. Environmental Protection Agency; 2011. [Google Scholar]
- Hemminki K, Bermejo JL, Forsti A. Opinion - The balance between heritable and environmental aetiology of human disease. Nat Rev Genet. 2006;7:958–965. doi: 10.1038/nrg2009. [DOI] [PubMed] [Google Scholar]
- Huang R, Xia M, Cho MH, Sakamuru S, Shinn P, Houck KA, Dix DJ, Judson RS, Witt KL, Kavlock RJ, et al. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environmental health perspectives. 2011;119:1142–1148. doi: 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- Kalf GF. Recent advances in the metabolism and toxicity of benzene. Critical reviews in toxicology. 1987;18:141–159. doi: 10.3109/10408448709089859. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Modis Y, Zeelen JP, Engel CK, Abagyan RA, Ahlberg A, Rasmussen B, Lamzin VS, Kunau WH, Wierenga RK. The 1.8 A crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: implications for substrate binding and reaction mechanism. Journal of molecular biology. 1997;273:714–728. doi: 10.1006/jmbi.1997.1331. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Zhang LP, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis. 2012;33:240–252. doi: 10.1093/carcin/bgr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cleghorn D, Heslin A, Morris PJ, Mulvihill MM, Nomura DK. Multidimensional profiling platforms reveal metabolic dysregulation caused by organophosphorus pesticides. ACS chemical biology. 2014;9:423–432. doi: 10.1021/cb400796c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Gartner A. Having a direct look: Analysis of DNA damage and repair mechanisms by next generation sequencing. Exp Cell Res. 2014;329:35–41. doi: 10.1016/j.yexcr.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PJ, Medina-Cleghorn D, Heslin A, King SM, Orr J, Mulvihill MM, Krauss RM, Nomura DK. Organophosphorus flame retardants inhibit specific liver carboxylesterases and cause serum hypertriglyceridemia. ACS chemical biology. 2014;9:1097–1103. doi: 10.1021/cb500014r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nature chemical biology. 2008;4:373–378. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Casida JE. Activity-based protein profiling of organophosphorus and thiocarbamate pesticides reveals multiple serine hydrolase targets in mouse brain. Journal of agricultural and food chemistry. 2011;59:2808–2815. doi: 10.1021/jf101747r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Leung D, Chiang KP, Quistad GB, Cravatt BF, Casida JE. A brain detoxifying enzyme for organophosphorus nerve poisons. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6195–6200. doi: 10.1073/pnas.0501915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chemistry & biology. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriel M, Edmiston S, Beauvais S, Barry T, O'Malley M. Illnesses Associated with Chloropicrin Use in California Agriculture, 1992–2003. Rev Environ Contam T. 2009;200:1–31. doi: 10.1007/978-1-4419-0028-9_1. [DOI] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS chemical biology. 2013;8:283–296. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. Peroxisomal beta-oxidation--a metabolic pathway with multiple functions. Biochimica et biophysica acta. 2006;1763:1413–1426. doi: 10.1016/j.bbamcr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Poole LB. The basics of thiols and cysteines in redox biology and chemistry. Free radical biology & medicine. 2015;80C:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM. Implications of the exposome for exposure science. Journal of exposure science & environmental epidemiology. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: characterizing exposures to reactive electrophiles. Toxicology letters. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler NW. Basic biology of GAPDH. Advances in experimental medicine and biology. 2013;985:1–36. doi: 10.1007/978-94-007-4716-6_1. [DOI] [PubMed] [Google Scholar]
- Shannon DA, Weerapana E. Covalent protein modification: the current landscape of residue-specific electrophiles. Current opinion in chemical biology. 2015;24:18–26. doi: 10.1016/j.cbpa.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Shukla SJ, Huang R, Austin CP, Xia M. The future of toxicity testing: a focus on in vitro methods using a quantitative high-throughput screening platform. Drug discovery today. 2010;15:997–1007. doi: 10.1016/j.drudis.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nature methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hirano S. Metabolism of arsenic and its toxicological relevance. Archives of toxicology. 2013;87:969–979. doi: 10.1007/s00204-012-0904-5. [DOI] [PubMed] [Google Scholar]
- Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nature chemical biology. 2008;4:405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CF, Killeen JC. A mechanistic interpretation of the oncogenicity of chlorothalonil in rodents and an assessment of human relevance. Regulatory toxicology and pharmacology : RTP. 1996;24:69–84. doi: 10.1006/rtph.1996.0065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.