Abstract
Bioluminescent imaging is an emerging biomedical surveillance strategy that uses external cameras to detect in vivo light generated in small animal models of human physiology or in vitro light generated in tissue culture or tissue scaffold mimics of human anatomy. The most widely utilized of reporters is the firefly luciferase (luc) gene; however, it generates light only upon addition of a chemical substrate, thus only generating intermittent single time point data snapshots. To overcome this disadvantage, we have demonstrated substrate-independent bioluminescent imaging using an optimized bacterial bioluminescence (lux) system. The lux reporter produces bioluminescence autonomously using components found naturally within the cell, thereby allowing imaging to occur continuously and in real-time over the lifetime of the host. We have validated this technology in human cells with demonstrated chemical toxicological profiling against exotoxin exposures at signal strengths comparable to existing luc systems (~1.33 × 107 photons/second). As a proof-in-principle demonstration, we have engineered breast carcinoma cells to express bioluminescence for real-time screening of endocrine disrupting chemicals and validated detection of 17β-estradiol (EC50 = ~ 10 pM). These and other applications of this new reporter technology will be discussed as potential new pathways towards improved models of target chemical bioavailability, toxicology, efficacy, and human safety.
Keywords: Bioluminescent imaging, Luciferase, Lux, Toxicology, Bioavailability, Estrogen, Tissue scaffold, Bioreporter
1. INTRODUCTION
Bioluminescent imaging is an emerging biomedical surveillance strategy that uses external cameras to detect in vivo light generated in small animal models of human physiology or in vitro light generated in tissue culture or tissue scaffold mimics of human anatomy. Mammalian cells expressing reporter proteins that are capable of producing an optical signal are progressively becoming more widely employed by investigators from diverse backgrounds to interrogate a variety of cellular functions in cell culture and living animals [1]. These applications include, but are not limited to, tumorigenesis and cancer treatment [2, 3], gene expression and regulation [4, 5], cell trafficking [6, 7], viral and pathogenic infection [8], protein stability and function [9, 10], and protein-protein interactions [11, 12]. Currently, the mainstays of optical reporter proteins used for these applications are fluorescent proteins (GFP and its other color variants) and bioluminescent proteins (luciferase enzymes) isolated from insects and marine organisms. The optical signal generated by these proteins allows for visualization of the cellular events of interest, but each type of these reporters is associated with its own disadvantages. Fluorescent proteins, for instance, require an initial excitation for light emission. It is this excitation that can produce high levels of background fluorescence from endogenous biological structures in cultured cells and whole animal imaging, thus greatly reducing the sensitivity and resolution of this technique and interfering with data interpretation.
With little to no endogenous bioluminescent activity in mammalian cells and tissues, the use of luciferase proteins has the advantage over their fluorescent counterparts of near background-free detection, especially for in vivo whole animal imaging. To date, the main reporter proteins used for these applications have been the firefly luciferase (FLuc), the Renilla luciferase (RLuc), and the Gaussia luciferase (GLuc). These bioluminescent proteins, regardless of their native host organisms, function in a similar fashion. They catalyze the oxidation of a substrate (luciferin for FLuc and coelenterazine for RLuc and GLuc) in the presence of oxygen to produce light [13-15]. These substrates, however, cannot be synthesized in host cells, and therefore must be added exogenously. In cell culture based applications, the substrate addition is often associated with cell lysis, therefore only providing measurements on a single time point basis. For in vivo whole animal imaging, the required substrate is usually injected directly without animal sacrifice. Although this allows for repeated monitoring of a single subject, due to rapid substrate uptake and consumption [16, 17], detection using these luciferase proteins has relatively short temporal dynamics, and is thus handicapped for long term monitoring applications. Meanwhile, these substrates are relatively unstable and expensive, adding a financial burden to the utilization of these systems [18].
The only substrate-free bioluminescent reporter system that has been developed to date is the bacterial bioluminescence (lux) system. This system is intrinsically different from other bioluminescent systems because of its ability to synthesize/recycle all required substrates endogenously within the host cells to produce light in a fully autonomous fashion. This is made possible because the lux system contains genes (luxCDEfrp) encoding proteins that are capable of providing the substrate required for light production. Specifically, the luxC, luxD, and luxE gene products are used for synthesizing and recycling a long chain fatty aldehyde substrate from endogenous compounds. For efficient light production in mammalian cells, a sixth gene, frp, encoding a flavin oxidoreductase is also included to facilitate efficient FMNH2 recycling. Co-expression of these genes together with the luciferase genes (luxAB) can produce sufficient substrate for autonomous bioluminescent expression and therefore obviates the step of exogenous substrate addition that is essential for the conventional Luc systems.
Due to this unique feature, the lux system is highly amenable towards continuous on-line biosensing. Indeed, it is extensively used as a prokaryotic bioreporter for environmental assessment [19, 20] and pathogenic infection [21-23]. The advantage offered by the lux system over other substrate-requiring luciferase systems has made it an attractive target for development into a mammalian reporter, but it is not until very recently that it has been adapted to express at a functional level for autonomous bioluminescent production in mammalian cells [24]. Here we present proof-of-principle demonstrations of the utilities of autonomous bioluminescent human cell lines for real-time detection of chemical bioavailability.
2. MATERIALS AND METHODS
2.1 Vector construction
The DNA fragment containing the luxCDABEfrp sequences was assembled de novo by GeneArt. During the synthesis process, the removal of intervening restriction and regulatory sequences was performed according to standard GeneArt synthesis protocols, and each of the lux genes was codon optimized to mimic human codon usage patterns for improved expression efficiency in human cells. Divergent viral-derived 2A elements were used to link adjacent lux genes in the polycistronic single vector. Originally identified in foot-and-mouth disease virus, 2A elements are in-frame linker regions that separate two genes driven off a single promoter. A highly conserved D(V/I)EXNPGP motif at the C-terminus of these peptides is essential for the function of 2A peptides, as the last G-P bond is “cleaved” during translation [25]. Because the 2A linker regions permit continuous translation of the mRNA to protein, the stop codons of all lux genes except for the gene most distal from the promoter were removed. These modifications allowed for the synthetic assembly of a single DNA construct consisting of luxC-2A linker-luxD-2A linker-luxA-2A linker-luxB-2A linker-luxE-2A linker-frp. The lux genes were organized in such a way to replicate their orders within the lux operon as found in their native bacterial host.
The synthetic fragment was then cloned into a mammalian expression vector containing a cytomegalovirus immediate early (CMV IE) promoter, resulting in vector pCMV-Lux for constitutive expression. To create a reporter construct for tetracycline response element (TRE)-regulated expression of the luxCDABEfrp genes, the TRE and its associated minimal CMV promoter were removed from the pTRE-Tight-BI vector (Clontech) to replace the CMV IE promoter, resulting in vector pTet-Lux. The schematic representation of the polycistronic lux mammalian expression vector is shown in Figure 1.
Figure 1.
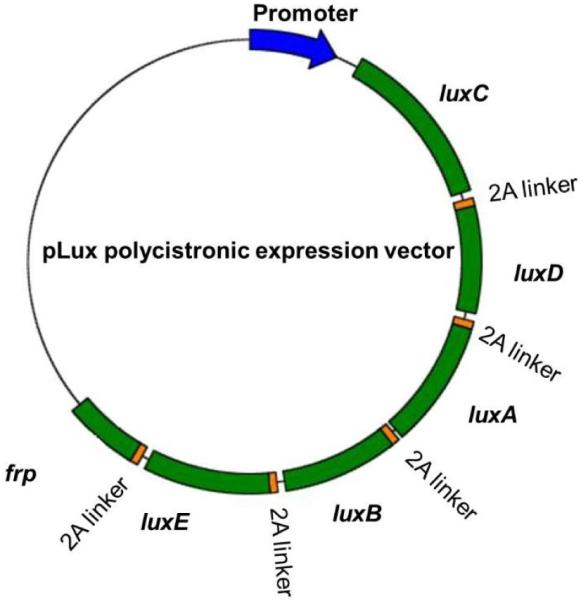
A schematic representation of the single promoter polycistronic vector for lux expression in human cell lines.
2.2 Mammalian cells maintenance and transfection
Human breast carcinoma T-47D cells were regularly maintained in phenol-red free RPMI-1640 medium (Hyclone) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 0.01 mM non-essential amino acids (Life Technologies), 1X antibiotic-antimyotic (Life Technologies) and 0.01 mM sodium pyruvate (Life Technologies). For estrogen treatment assays, assay medium was modified from growth medium by substituting 10% charcoal/dextran-treated FBS (CD-FBS) (Hyclone). Neomycin (CalBiochem) at the concentration of 500 µg/ml was used for selection and maintenance of stably transfected cells.
HEK293 Tet-On cells expressing the tetracycline-regulated transactivator were obtained from Clontech and maintained in Dulbecco's Modified Eagle Medium (DMEM) (Hyclone) supplemented with 100 μg Neomycin/ml, 10% FBS, 0.01 mM non-essential amino acids, 1X antibiotic-antimyotic and 0.01 mM sodium pyruvate. For doxycycline induction assays, assay medium was modified from growth medium by addition of 10% Tet System Approved FBS (Clontech).
All transfections were carried out in 6-well tissue culture plates (Corning). The day prior to transfection, cells were plated at a density of 5 × 105 cells/well (for HEK293 Tet-On cells) or 7.5 × 105 cells/well (for T-47D cells). Vector DNA was introduced into the cells using Lipofectamine 2000 transfection reagent (Life Technologies) following the manufacturer’s instructions. Transfected cells were either assayed twenty-four hours post transfection (section 2.3), or subjected to stable transfectant selection (section 2.4).
2.3 Bioluminescent measurement of Tet-Lux reporter cells in response to doxycycline
Doxycycline-induced bioluminescent expression of pTet-Lux was examined in HEK293 Tet-On cells in transient expression assays. Transfections were carried out in assay medium (DMEM containing 10% Tet System Approved FBS) as described previously. Twenty-four h post transfection, cells were washed with sterile PBS and counted using the Scepter 2.0 handheld automated cell counter (Millipore). Approximately 1 × 106 cells were plated in each well in 1 ml assay medium in a black 24-well plate. Doxycycline was added at final concentrations of 0 (control) or 100 ng/ml in replicate wells. Bioluminescent measurements were performed immediately after doxycycline induction using the IVIS Lumina imaging system (PerkinElmer) using a 10 min integration time every 30 min for 19 h.
2.4 Selection of stable bioluminescent T-47D/Lux cells
T-47D cells transfected with pCMV-Lux were subjected to stable transfectant selection. Twenty-four hours post-transfection, transfected cells were harvested and diluted into new six-well plates in growth medium without antibiotic selection. Starting the next day, selection of stably transfected clones was performed by refreshing with selective medium containing 500 µg Neomycin/ml every 2 - 3 days until all untransfected cells had died and stably transfected cells had formed visible colonies. The colonies were then removed by trypsinization and expanded into individual lines in growth medium supplemented with Neomycin at a 500 µg/ml concentration.
To screen for autonomous bioluminescence following transfection with pCMV-Lux, each isolated cell line was grown in individual 25 cm2 tissue culture flasks and harvested for imaging upon reaching ~80% confluence. Cells were collected in 1 ml of RPMI-1640 growth medium and plated in each well of a black 24-well plate. Bioluminescent measurements were carried out using the IVIS Lumina Imaging System with a 10 min integration time every 30 min for 24 h. The cell line displaying the highest bioluminescent production was designated T-47D/Lux and used in further analysis.
2.5 Correlating population size with bioluminescence in T-47D/Lux cells
Actively growing T-47D/Lux cells were trypsinized and harvested from 75 cm2 tissue culture flasks and counted using a hemacytometer. Groups of either 2.5 × 105, 1 × 105, 5 × 104, 2.5 × 104, 1 × 104, 5 × 103, or 2.5 × 103 cells were plated in triplicate in 1 ml of phenol red-free RPMI-1640 complete growth medium in each well of a clear-bottom, tissue culture-treated, black 24-well plate. A negative control for monitoring background was performed in triplicate wells containing 1 ml of medium without cells. Bioluminescence was determined in the IVIS Lumina imaging system using a 10 min integration time every 30 min for 16 h. Statistical analysis was performed using Student’s t tests with a significant p value of 0.05. To establish the relationship between population size and bioluminescent signal, the total flux values in photons/second (p/s) were correlated to cell numbers using the Pearson’s linear regression model.
2.6 Bioluminescent T-47D/Lux estrogen assay
The bioluminescent T-47D estrogen screening assay was carried out in clear-bottom, tissue culture treated, black 24-well plates. Approximately 1 × 104 T-47D/Lux cells were seeded into each well in 1 ml growth medium and were allowed to attach for 24 h. Before addition of the estrogen 17β-estradial (E2), medium was refreshed with 1 ml CD-FBS containing assay medium. E2 (using HPLC grade ethanol as the solvent) was added at final concentrations of 0 pM (control), 0.1 pM, 1 pM, 10 pM, 100 pM, 1 nM, 10 nM, or 100 nM in triplicate wells. Solvent concentration remained constant at 0.1% (v/v) across all wells. Bioluminescent measurements were obtained using the IVIS Lumina imaging system with a 10 min integration time every 24 h for 6 days. Cells were incubated at 37°C in a 5% CO2 environment between measurements.
3. RESULTS AND DISCUSSION
3.1 The use of lux for continuous, real-time imaging of chemical bioavailability
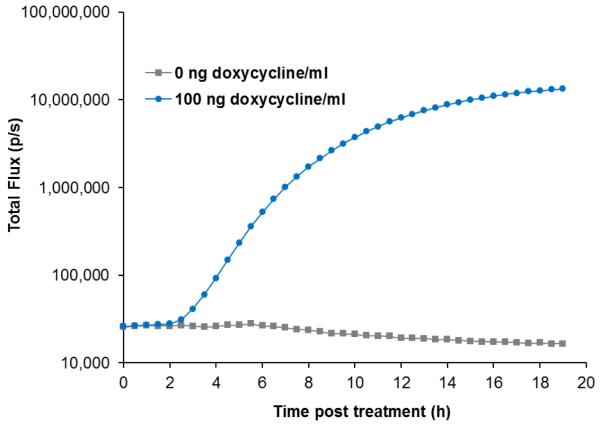
The functionality of the lux cassette as a fully autonomous, real-time reporter for target-regulated gene expression was investigated using a well-characterized Tet-On mammalian expression system. HEK293 Tet-On cells constitutively expressing the Tet-On transactivator were transfected with a tetracycline response element regulated lux reporter construct (pTet0-Lux) and the dynamics of doxycycline-induced bioluminescent response was determined over a 19 h imaging course (Figure 2). Without doxycycline induction, the Tet-lux reporter cells displayed a background luminescence of ~ 2.57 × 104 p/s, which decreased gradually over time, possibly due to compromised cellular metabolism caused by nutrient depletion from the medium and the inability to regulate the CO2 level and humidity in the imaging chamber. Exposure to 100 ng doxycycline/ml induced bioluminescent production that could be differentiated from un-induced control as early as 2.5 h post treatment (p < 0.05). From this point on, light production continued to increase rapidly with increasing treatment time, reaching a total flux of ~ 1.33 × 107 p/s after 19 h of exposure, which was approximately 800-fold change in light production compared to un-exposed control. Under this expression strategy, the maximal light production induced by doxycycline was greater than that from equal numbers of cells expressing identical genes under the control of a constitutive CMV IE promoter (data not shown), which was in agreement with previously published studies [26].
Figure 2.
Continuous real-time monitoring of the bioluminescent response to doxycycline treatment of the HEK293 Tet-lux reporter cells.
3.2 Long term continuous monitoring of bioluminescent production
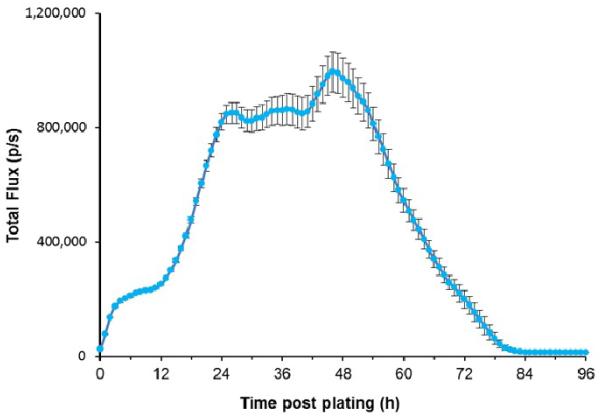
Continuous imaging of approximately 1 × 106 stable T-47D/Lux cells was attempted for 96 h to monitor the cellular response to ambient environment inside the imaging chamber, which was different from the well-controlled incubator environment. As shown in Figure 3, bioluminescent output increased in the first 6 h, and then remained relatively stable between 6 h and 12 h with an average hourly total flux increase of 6.62 (± 0.10) × 103 p/s. A rapid increase of light production was observed between 12 h to 26 h with an average increase in total flux of 4.28 (± 0.26) × 104 p/s per hour, followed by another relatively stable phase between 26 h and 40 h. Following this stable phase was a short but relatively rapid rise in bioluminescent expression between 40 h and 48 h, increasing from 8.50 (± 0.59) × 105 p/s to 9.75 (± 0.67) × 105 p/s. After peaking at approximately 48 h post plating, light production rapidly decrease indicating comprised cellular health and cell death. No significant bioluminescence was observed after ~ 84 h of imaging.
Figure 3.
Ninety-six hour continuous monitoring of T-47D/Lux cells incubated in the IVIS Lumina imaging chamber.
Because of the strong and constitutive activity of the CMV IE promoter used to drive bioluminescent expression, it is possible to relate light production dynamics to either a change in cell numbers or cellular health and metabolism. Since the doubling time of T-47D cells is greater than 20 hours, the rapid increase in bioluminescence in the first few hours observed was not likely caused by cell proliferation. It is speculated that switching from a carefully controlled 5% CO2 incubator environment to the atmospheric condition in the imaging chamber causes a rapid increase in the medium pH value, which may affect the intracellular redox potential and NADPH availability for the bioluminescent reaction. The inability to control the CO2 level, humidity and air temperature in the imaging chamber in combination with the continuing consumption of nutrients present in the medium represents a unique situation where cellular metabolism and health is challenged by unspecified factors whose effects on living systems cannot be easily detected using the conventional reporter systems. The substrate-free nature of the lux system eliminates the requirements of cell destruction and substrate addition, thus permitting cellular growth and metabolic response to be visualized continuously in real-time.
3.3 Using bioluminescence as an indicator for population size
For these cells to be useful as a reporter, the bioluminescent output must be detectable over a dynamic population range. To determine minimum detectable cell numbers, T-47D/Lux cells at concentrations ranging from 2.5 × 103 to 2.5 × 105 were plated in triplicate in equal volumes of media over a constant surface area for signal detection (Figure 4A). Using a 10 min integration time, approximately 2.5 × 103 cells could be significantly differentiated from medium background (p < 0.01). It was also revealed that the bioluminescent flux correlated tightly to the number of cells present in a population in a linear fashion (R2 > 0.99) (Figure 4B), suggesting that the bioluminescent output could be used to as an indicator of population size.
Figure 4.
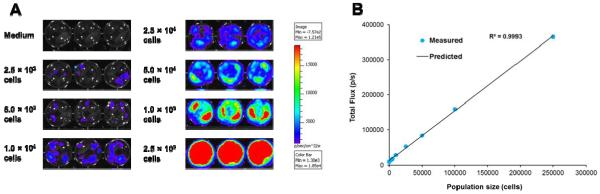
(A) Pseudocolor images of bioluminescent production from different numbers of T-47D/Lux cells. (B) Correlation between population size and bioluminescent output.
3.4 Bioluminescent estrogen assay
Human breast cancer T-47D cells are estrogen sensitive, meaning that their proliferative behavior is regulated by estrogen [27]. In this study, the constitutively bioluminescent T-47D/Lux cells were examined for their potential application in an estrogen screening assay. To determine if the bioluminescent dynamics could be used to denote estrogen-stimulated cell proliferation, equal numbers of cells were exposed to varying concentrations of E2. Due to the inability to maintain long term cellular health in the imaging chamber (Figure 3), continuous imaging was not attempted. Instead, cells were incubated at optimal growth condition (5% CO2 at 37°C) with bioluminescence being measured every 24 h. Because cells remained intact and attached to the growing surface during measurements, it was possible to track the bioluminescent dynamics of the same population throughout the course of exposure (Figure 5A). While exposure to 0.1 pM E2 was not capable of increasing bioluminescence significantly compared to unexposed control throughout the 6-day exposure period, a significant change in bioluminescent production (p < 0.05) was observed 3 days after exposure to concentrations of E2 as low as 1 pM. It was also shown that E2 stimulated distinctive bioluminescent dynamics in a time- and dose-dependent manner. Treatment with 1 pM E2 resulted in a significant change in bioluminescence after 3 days of exposure, but the signal was no longer distinguishable from vehicle control after 4 days. Higher concentrations of E2 were able to elicit light production after 3 days of exposure and were significantly distinguishable from the control for the rest of the exposure period. Based on the bioluminescent measurement after 4 days of exposure, it was determined that 17β-estradiol concentrations to induce maximal and half-maximal bioluminescence in this assay were approximately 0.1 nM and 10 pM (Figure 5B), which were comparable to those measured using the traditional E-SCREEN method (EC50 = 7 - 12 pM using MCF-7 cells) [28].
Figure 5.
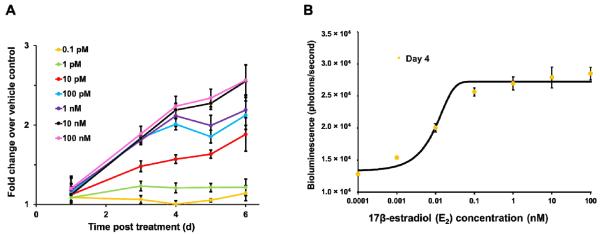
(A) Repeated imaging of the same population of bioluminescent T-47D/Lux cells throughout the course of estrogen exposure. (B) Dose-dependent response of T-47D/Lux cells after 4 days of exposure to various concentrations of E2.
The traditional E-SCREEN assay was developed 27 years ago for measuring the physiological signature of the action of estrogen (i.e., induction of cell proliferation) and is still widely used for identification of chemicals with potential estrogenic activities [29]. This assay compares the cell yield following 5-day to 6-day incubation in the presence or absence of estrogen. The original method of quantifying cell numbers involved direct nuclei counting, but a higher throughput and less laborious colorimetric MTT assay has been widely adopted as the endpoint measurement of viable cell counts. Regardless of the method used, cell lysis is required prior to data acquisition, which therefore provides only single time point snapshots. On the other hand, using the autonomous bioluminescence emitted from T-47D/Lux cells as the endpoint of measurement, the same cell population can be monitored repeatedly throughout the course of exposure; therefore, cell proliferation can be captured progressively at any time point of interest. Results show that the bioluminescent signal emitted from these modified T-47D cells responds to estrogen in a dose-dependent manner, exemplified by a typical sigmoidal curve that is similarly obtained using other endpoint measurements (Figure 5B) [29]. The minor discrepancies between our results and those reported in other studies are most likely due to logistical factors such as differences between cell line clones and culture conditions [30]. However, in general, using bioluminescent output as an indicator for cell proliferation, estrogen can be detected in the picomolar range.
4. CONCLUSIONS
Bioluminescent imaging is a noninvasive biomedical surveillance strategy that applies living cells carrying light emitting reporter genes as biological flashlights to profile disease states, therapeutic/vaccine efficacy, and drug toxicity. Current reporter gene technology, however, only allows imaging to occur along an intermittent timeline of single time point data snapshots. As the most recent addition to the bioluminescent reporter protein family for mammalian expression, the lux system is the only reporter that permits fully autonomous signal production without the requirement of exogenous excitation or substrate addition, making it an ideal candidate for continuous, real-time, data intensive imaging of cellular events in cell culture and living animals. In this study, we have validated this technology in human cells with demonstrated chemical toxicological profiling against antibiotic and exotoxin exposures at photon emission strengths comparable to existing firefly luciferase reporter systems. We have also engineered breast cancer cells to express the lux system for the real-time screening of chemicals displaying endocrine disrupting activity and validated detection of 17β-estradiol at low picomolar concentrations and EC50 values comparable to traditional cell-destructive, single time point methods. These and other applications of this novel bioimaging approach enables more accurate prediction of human relevant drug/vaccine safety and efficacy and lessens the burden of in vivo animal applications. Therefore, this new reporter technology can be developed as potential new pathways towards improved models of target chemical bioavailability, toxicology, efficacy, and human safety.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Science Foundation Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET) under award number CBET-0853780, the National Institutes of Health, National Institute of Environmental Health Sciences (NIEHS) SBIR program, award number 1R43ES022567-01, the National Cancer Institute, Cancer Imaging Program, award number CA127745-01, and the Army Defense University Research Instrumentation Program.
REFERENCES
- [1].Baker M. The whole picture. Nature. 2010;463(7283):977–980. doi: 10.1038/463977a. [DOI] [PubMed] [Google Scholar]
- [2].Rehemtulla A, Stegman LD, Cardozo SJ, et al. Rapid and quantitative assessment of cancer treatment response using In Vivo bioluminescence imaging. Neoplasia. 2000;2(6):491–495. doi: 10.1038/sj.neo.7900121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Venisnik KM, Olafsen T, Loening AM, et al. Bifunctional antibody-Renilla luciferase fusion protein for in vivo optical detection of tumors. Protein Engineering Design & Selection. 2006;19(10):453–460. doi: 10.1093/protein/gzl030. [DOI] [PubMed] [Google Scholar]
- [4].Korpal M, Yan J, Lu X, et al. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nature Medicine. 2009;15(8):960–U169. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- [5].Rettig GR, McAnuff M, Liu DJ, et al. Quantitative bioluminescence imaging of transgene expression in vivo. Analytical Biochemistry. 2006;355(1):90–94. doi: 10.1016/j.ab.2006.04.026. [DOI] [PubMed] [Google Scholar]
- [6].Sheikh AY, Lin SA, Cao F, et al. Molecular Imaging of bone marrow mononuclear cell homing and engraftment in ischemic myocardium. Stem Cells. 2007;25(10):2677–2684. doi: 10.1634/stemcells.2007-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van der Bogt KEA, Sheikh AY, Schrepfer S, et al. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation. 2008;118(14):S121–U166. doi: 10.1161/CIRCULATIONAHA.107.759480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Andreu N, Zelmer A, Wiles S. Noninvasive biophotonic imaging for studies of infectious disease. Fems Microbiology Reviews. 2011;35(2):360–394. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang LC, Fu QX, Dong YF, et al. Bioluminescence imaging of Hepatitis C virus NS3/4A serine protease activity in cells and living animals. Antiviral Research. 2010;87(1):50–56. doi: 10.1016/j.antiviral.2010.04.010. [DOI] [PubMed] [Google Scholar]
- [10].Laxman B, Hall DE, Bhojani MS, et al. Noninvasive real-time imaging of apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):16551–16555. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pichler A, Prior JL, Luker GD, et al. Generation of a highly inducible Gal4 -> Fluc universal reporter mouse for in vivo bioluminescence imaging. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(41):15932–15937. doi: 10.1073/pnas.0801075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Verhaegen M, Christopoulos TK. Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Analytical Chemistry. 2002;74(17):4378–4385. doi: 10.1021/ac025742k. [DOI] [PubMed] [Google Scholar]
- [14].Fraga H. Firefly luminescence: A historical perspective and recent developments. Photochemical & Photobiological Sciences. 2008;7(2):146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- [15].Lorenz WW, McCann RO, Longiaru M, et al. Isolation and expression of a cDNA-encoding Renilla reniformis luciferase. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(10):4438–4442. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(1):377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Inoue Y, Kiryu S, Izawa K, et al. Comparison of subcutaneous and intraperitoneal injection of D-luciferin for in vivo bioluminescence imaging. European Journal of Nuclear Medicine and Molecular Imaging. 2009;36(5):771–779. doi: 10.1007/s00259-008-1022-8. [DOI] [PubMed] [Google Scholar]
- [18].Close DM, Hahn RE, Patterson SS, et al. Comparison of human optimized bacterial luciferase, firefly luciferase, and green fluorescent protein for continuous imaging of cell culture and animal models. Journal of Biomedical Optics. 2011;16(4):e12441. doi: 10.1117/1.3564910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Singh JS, Abhilash PC, Singh HB, et al. Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene. 2011;480(1-2):1–9. doi: 10.1016/j.gene.2011.03.001. [DOI] [PubMed] [Google Scholar]
- [20].Shin HJ. Genetically engineered microbial biosensors for in situ monitoring of environmental pollution. Applied Microbiology and Biotechnology. 2011;89(4):867–877. doi: 10.1007/s00253-010-2990-8. [DOI] [PubMed] [Google Scholar]
- [21].Francis KP, Joh D, Bellinger-Kawahara C, et al. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infection and Immunity. 2000;68(6):3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trcek J, Berschl K, Trulzsch K. In vivo analysis of Yersinia enterocolitica infection using luxCDABE. Fems Microbiology Letters. 2010;307(2):201–206. doi: 10.1111/j.1574-6968.2010.01983.x. [DOI] [PubMed] [Google Scholar]
- [23].Contag CH, Contag PR, Mullins JI, et al. Photonic detection of bacterial pathogens in living hosts. Molecular Microbiology. 1995;18(4):593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- [24].Close DM, Patterson SS, Ripp SA, et al. Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. Plos One. 2010;5(8):e12441. doi: 10.1371/journal.pone.0012441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szymczak AL, Vignali DAA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opinion on Biological Therapy. 2005;5(5):627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- [26].Qin JY, Zhang L, Clift KL, et al. Systematic Comparison of Constitutive Promoters and the Doxycycline-Inducible Promoter. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Soto AM, Murai JT, Siiteri PK, et al. Control of cell proliferation - evidence for negative control on estrogen-sensitive T-47D human breast cancer cells. Cancer Research. 1986;46(5):2271–2275. [PubMed] [Google Scholar]
- [28].Soto AM, Maffini MV, Schaeberle CM, et al. Strengths and weaknesses of in vitro assays for estrogenic and androgenic activity. Best Practice & Research Clinical Endocrinology & Metabolism. 2006;20(1):15–33. doi: 10.1016/j.beem.2005.09.001. [DOI] [PubMed] [Google Scholar]
- [29].Soto AM, Sonnenschein C, Chung KL, et al. The E-SCREEN assay as a tool to identify estrogens - an update on estrogenic environmental pollutants. Environmental Health Perspectives. 1995;103:113–122. doi: 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zacharewski T. In vitro bioassays for assessing estrogenic substances. Environmental Science & Technology. 1997;31(3):613–623. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.