SUMMARY
RORγt+ Th17 cells are important for mucosal defenses, but also contribute to autoimmune disease. They accumulate in the intestine in response to microbiota and produce IL-17 cytokines. Segmented filamentous bacteria (SFB) are Th17-inducing commensals that potentiate autoimmunity in mice. RORγt+ T cells were induced in mesenteric lymph nodes early after SFB colonization and distributed across different segments of the gastrointestinal tract. However, robust IL-17A production was restricted to the ileum, where SFB makes direct contact with the epithelium and induces serum amyloid A proteins 1 and 2 (SAA1/2), which promote local IL-17A expression in RORγt+ T cells. We identified an SFB-dependent role of type 3 innate lymphoid cells (ILC3), which secreted IL-22 that induced epithelial SAA production in a Stat3-dependent manner. This highlights the critical role of tissue microenvironment in activating effector functions of committed Th17 cells, which may have important implications for how these cells contribute to inflammatory disease.
INTRODUCTION
The vertebrate gastrointestinal (GI) tract is colonized by hundreds of distinct species of microorganisms that collectively maintain a mutualistic relationship with the host (Macpherson and Harris, 2004). This mutualism is critically dependent on a state of balanced immune activation, which fosters cohabitation between the host and microbiota, whilst providing optimal protection against opportunistic pathogens (Honda and Littman, 2012). It is now appreciated that the composition of the microbiome can contribute significantly to this immunological balance, in part through the capacity of individual bacterial or viral species to profoundly influence distinct arms of the immune response by themselves or in concert with other microbial species (Hooper et al., 2012; Virgin, 2014). For instance, Bacteroides fragilis and mixtures of various strains of Clostridia, which colonize the large intestine, have been shown to promote intestinal and systemic immune tolerance through regulatory T cells (Atarashi et al., 2013; Lathrop et al., 2011; Round et al., 2011); and segmented filamentous bacteria (SFB) induce antigen-specific T-helper-17 cells (Th17) cell differentiation in the small intestine (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009; Yang et al., 2014).
Th17 cells constitute a subset of activated CD4+ T cells that are characterized by the production of the cytokines IL-17A, IL-17F, and IL-22 (Korn et al., 2009). These cytokines act on a broad range of hematopoietic and non-hematopoietic cells to regulate central aspects of host immunity, including granulopoiesis, neutrophil recruitment, and the induction of antimicrobial peptides (Weaver et al., 2007). Th17 cells thus execute important functions in both epithelial homeostasis and host defense against various extracellular pathogens, such as Candida albicans and Pseudomonas aeruginosa (Stockinger and Veldhoen, 2007). Conversely, over-exuberant Th17 responses may promote auto-inflammatory diseases, such as Crohn’s disease, rheumatoid arthritis (RA), psoriasis, and multiple sclerosis (MS) (Furuzawa-Carballeda et al., 2007). While genetic polymorphisms significantly factor into the onset of these diseases, emerging evidence also highlights the influence that environmental factors, such as diet and microbial composition, can impose on such propensities. Accordingly, recent studies have illustrated the potential of SFB to exacerbate Th17-mediated disease in murine autoimmune models of both RA and MS (Lee et al., 2011; Wu et al., 2010), although the intermediate molecular steps connecting SFB to a distal immune response are ill defined.
SFB colonization of the small intestine promoted global transcriptional changes in host epithelia, including the induction of antimicrobial peptides and stress response genes, such as serum amyloid A (SAA1 and SAA2) (Ivanov et al., 2009). SAA is typically induced in response to infection and acute injury and can promote inflammation, in part through elicitation of proinflammatory cytokine production and recruitment of granulocytes, monocytes, and T lymphocytes (Uhlar and Whitehead, 1999). The effect of SAA on the immune response is context-driven (Cray et al., 2009; Eckhardt et al., 2010; Ivanov et al., 2009), much like that of Th17 cells. Insofar as SFB and Th17 cells are intertwined, the question of whether SAA impacts aspects of Th17 biology remains to be addressed.
Th17 cells along with several other innate-like cell lineages, including specific subsets of γδ T cells (γδ17) and type 3 innate lymphoid cells (ILC3), are regulated by the transcription factor RAR-related orphan receptor gamma (RORγt) (Chien et al., 2013; Ivanov et al., 2006; Spits and Di Santo, 2011). However, in contrast to the requirement for antigen recognition in the context of MHC to drive Th17 cell activation, γδ17 and ILC3 effector functions are elicited independently of antigen presentation. For example, the pro-inflammatory cytokine IL-23 triggers rapid IL-17 and IL-22 secretion by γδ17 cells and ILC3s, respectively, upon ligation of the highly-expressed IL-23 receptor (IL-23R). As γδ17 cells and ILC3s often reside in proximity to exposed mucosal surfaces, their activation typically precedes antigen-specific T cell differentiation and recruitment (Martin et al., 2009; Sutton et al., 2009). Whether this has bearing on the function of newly-recruited T cells is unclear.
We utilized models of acute SFB colonization to investigate the mechanism of Th17 cell induction in a spatiotemporal context. We found that, following early induction of SFB-specific RORγt+ Th17 cells in the mesenteric lymph nodes, there was distribution of such cells throughout the length of the gut, from duodenum to colon, but IL-17A expression was largely confined to the terminal ileum, the site of SFB attachment to epithelium (Ivanov et al., 2008). We have identified an SFB-triggered circuit in which ILC3 secretion of IL-22 is critical for local epithelial production of SAA1 and SAA2, which act directly on poised Th17 cells to amplify effector cytokine production. These findings suggest that tissue microenvironments contribute to the acquisition of effector functions by polarized activated effector and memory cells.
RESULTS
Selective IL-17A induction in RORγt+ T cells in ileum of SFB-colonized mice
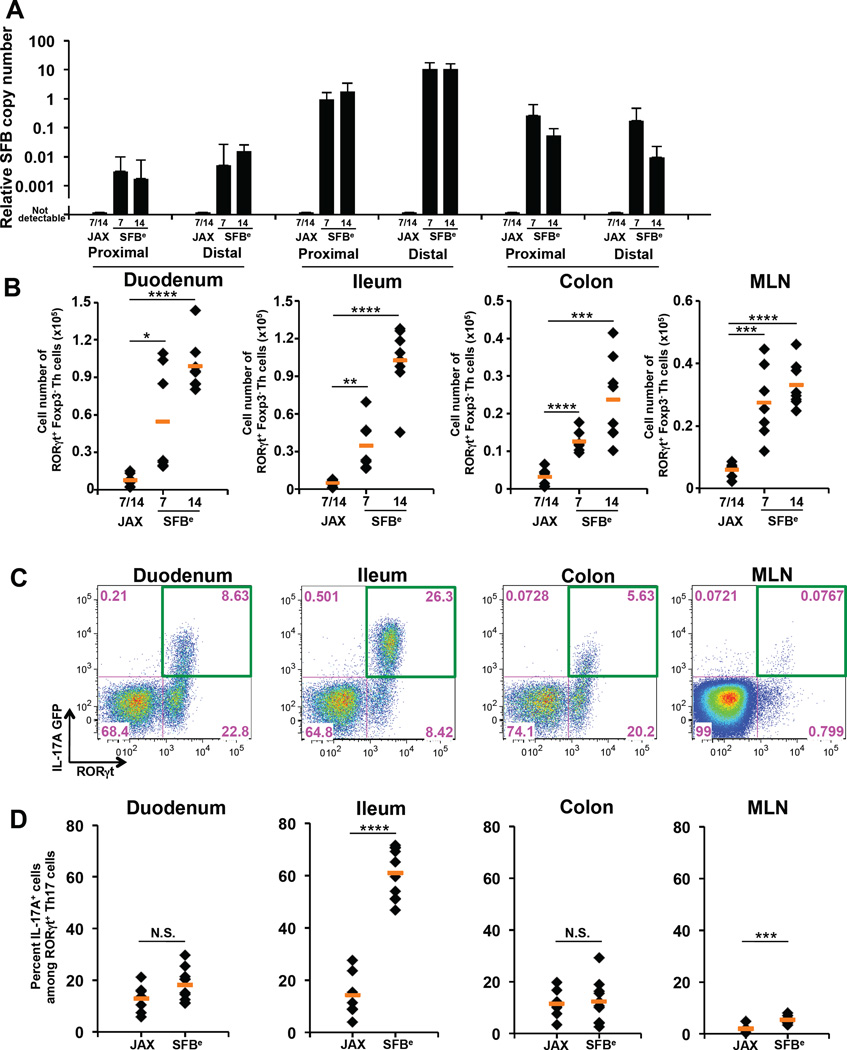
SFB colonization results in a striking increase in both the number and proportion of Th17 cells among total CD4+ T cells within the small intestine lamina propria (SILP) (Ivanov et al., 2009). To explore the mechanism of Th17 cell induction, we orally introduced fecal contents from Il-23rgfp/gfp Rag2−/− mice, which have about 100-fold enriched fecal SFB (SFBe) compared to WT animals (Yang et al., 2014), into SFB-free (JAX) mice and subsequently traced the kinetics of Th17 cell differentiation in different compartments of the GI tract on days 7 and 14. As expected, SFB colonization was most abundant in the terminal ileum (Figure 1A and S1A). After 5 days of SFBe gavage, we observed a marked increase in proportion and number of CD4+ T cells expressing RORγt, which defines the Th17 cell program, in the mesenteric lymph nodes (MLN) that drain the intestine (Figure S1B). Adoptively transferred CFSE-labeled SFB-specific T cells from 7B8 T cell receptor transgenic mice (Yang et al., 2014) proliferated and up-regulated RORγt expression in the MLN by day 4 after SFB colonization (Figure S1C). Expansion of these cells was then observed throughout the SILP by 7 or 14 days post SFB gavage (Figure 1B, S1C). Using fluorescently-labeled MHC class II tetramers with a bound immunodominant SFB antigen, we found SFB-specific RORγt+ cells throughout the gut; however, they were enriched in the ileum, possibly reflecting additional expansion in response to local antigen (Figure S1D).
Figure 1. Site of SFB colonization correlates with maximal IL-17A induction in RORγt+ T cells.
(A) Quantitative PCR (qPCR) analysis of SFB genomic 16S in duodenum, ileum and colon (proximal and distal segments of each) from SFB-colonized (SFBe; gavaged with SFB-rich fecal contents) or SFB-free (JAX; gavaged with SFB-free fecal contents) mice at 7 and 14 days post gavage. Copy number of SFB 16S was normalized by copy number of mouse genomic Il-23r gene. The experiment was repeated more than 3 times with similar results combined. Data are represented as mean with SD.
(B) Number of RORγt+ CD4+ T cells in intestinal lamina propria (LP) or MLN of SFB-colonized (SFBe; n=7) or SFB-free (JAX; n=7) mice on 7 and 14 days post gavage. CD4+ T cells were isolated from the indicated tissues, and Foxp3 RORγt+ CD4+ T cells were quantified by FACS.
(C) IL-17A (GFP+) production in Foxp3 RORγt+ CD4+ T cells from intestinal LPs and MLN in IL-17A reporter (Il17agfp/gfp) mice at 14 days post SFB colonization. IL-17A–producing Th17 cells were monitored by FACS after staining with anti-GFP antibody without re-stimulation.
(D) The proportion of IL-17A producing Th17 cells among Foxp3− RORγt+ CD4+ T cells following SFB colonization. Each dot represents a single mouse, and the orange bar indicates average of relative gene expression of each group. Data were from at least two independent experiments (JAX; n=7. SFBe; n=9).
N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S1.
To track expression of IL-17A, we colonized Il17agfp/gfp reporter mice with SFBe. Strikingly, co-staining of RORγt and GFP indicated that the majority of RORγt+ cells expressed IL-17A mRNA only in the terminal ileum, but not in other regions of the GI tract (Figures 1C, D). Induction of IL-17A in poised Th17 cells thus appears to occur primarily at the site of SFB colonization.
Serum amyloid A proteins modulate Th17 cell cytokine production
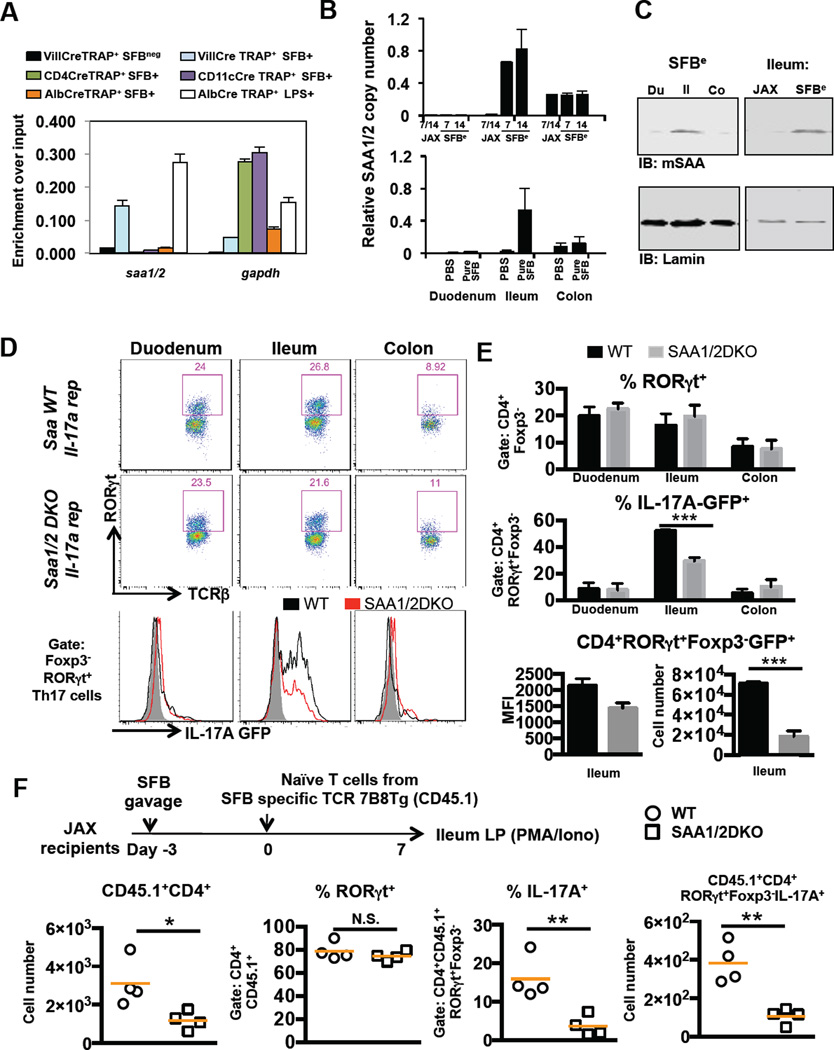
Recent studies have shown that the tissue microenvironment confers distinct transcriptional and functional properties to otherwise similar lymphoid and myeloid cells (Burzyn et al., 2013; Cipolletta et al., 2012; Lavin et al., 2014; Lewis et al., 2011). Our observation of the spatial restriction of IL-17A–producing Th17 cells raised the possibility that SFB induces changes in the microenvironment of the underlying lamina propria, thus contributing to the differentiation state of poised Th17 cells. Consistent with this possibility, germ-free (GF) mice monocolonized with SFB up-regulated expression of several secreted molecules in the terminal ileum, among which SAA1 and SAA2 (referred to as SAA1/2, as they are highly homologous) were the most strongly induced (Ivanov et al., 2009). Because SFB attaches to the epithelium, we focused on secreted molecules that changed expression within ileal epithelial cells for their potential influence on the functional status of adjacent Th17 cells. To identify epithelium-specific translated RNAs, we employed in vivo translating ribosome affinity purification (TRAP), which was developed to identify cell type-specific transcripts in tissues containing heterogeneous cell populations (Heiman et al., 2014). TRAP Villin-Cre, TRAP CD11c–Cre, TRAP CD4-Cre, and TRAP Albumin-Cre mice expressing a conditionally induced GFP ribosome L10 fusion protein from the rpl10 locus in epithelial cells, dendritic cells, T cells, and hepatocytes, respectively, were colonized with SFB for two weeks or left untreated, after which GFP pull-down of tissue lysates and qRT-PCR using transcript-specific primers were performed to identify translating mRNAs in specific cell types. Using primers that amplify both SAA1 and SAA2 mRNAs, we found that both were highly transcribed and translated in epithelial cells from SFB+ mice (Figure 2A), consistent with previous reports of SAA protein expression in small and large intestinal epithelial cells (Derebe et al., 2014; Eckhardt et al., 2010). We next assessed the spatiotemporal induction of SAA1/2 mRNAs in the epithelial fraction of the intestine upon acute colonization with SFB (Figure 2B). Strikingly, SAA1/2 mRNA induction was confined to the terminal ileum and was modulated in synchrony with SFB colonization (Figure 1A and S1A). Differential SAA expression was confirmed by immunoblotting of the EDTA-epithelial cell-rich fractions from different regions of the GI tract of animals with or without SFB colonization (Figure 2C). Notably, this localization corresponds to the site of SFB attachment and the location of IL-17A–producing Th17 cells.
Figure 2. Role of epithelial SAA1/2 in ileal Th17 cell production of IL-17A.
(A) Translating ribosome affinity purification (TRAP) assay for identification of the cell type expressing SAA1/2. Four individual lines of TRAP mice, conditionally expressing a GFP tagged ribosome L10 fusion protein in intestinal epithelial (Villin-Cre), T (CD4-Cre), myeloid (CD11c–Cre), or liver (Albumin-Cre) cells were generated. After two weeks of SFB colonization, ribosomes in small intestinal lysates collected from each line were enriched by GFP pull-down. Actively translating SAA1/2 mRNAs were quantified by qRT-PCR. Black and white bar indicates TRAP signal from SFB-free animals and light blue, dark blue, purple, and orange bars indicate TRAP signal from SFB positive animals. White bar indicates TRAP signal from tissue harvested from JAX animals at 6h following intraperitoneal administration of LPS.
(B) Quantification of SAA1/2 mRNA in intestinal epithelial cells of duodenum, ileum and colon from SFBe (Day7; n=8, Day 14; n=3), pure SFB (n=4), or JAX (n=3), or PBS-gavaged (n=8) mice. Data were normalized using Gapdh. Similar experiments were performed three times.
(C) Representative western blots of lysates from the EDTA-epithelial-rich fraction of different regions of the GI tract in animals with or without SFB colonization.
(D–F) Role of SAAs in intestinal LP Th17 cell differentiation following SFB colonization. (D,E) IL-17A reporter littermates in WT (black, n=5) or Saa1/2 double knock-out (DKO) (grey, n=5) backgrounds were colonized with SFB for 7 days. RORγt+ cells among CD4+ LP T cells and their IL-17A producing capacity were measured by FACS. MFI and cell numbers of CD4+RORγt+Foxp3GFP+ Th17 cells were quantified. (F) Naïve CD4+ T cells from SFB-specific TCR transgenic (7B8) mice were adoptively transferred into SFB-colonized WT (circles, n=4) or Saa1/2 DKO (squares, n=4) littermates (as indicated in diagram). Total number of donor cells recovered from ileum (far left), percentage of RORγt-expressing donor-derived cells (center left), percentage of IL-17A–expressing cells upon ex vivo re-stimulation by PMA/Ionomycin (center right), and their total number (far right) were monitored by FACS at 7 days post transfer. N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. Data are represented as mean with SD. See also Figure S2.
We next asked whether SAA1/2 contribute to functional differentiation of Th17 cells in vivo. Wild-type (WT) or SAA1 and SAA2 (DKO) mutant mice (Eckhardt et al., 2010) were crossed with Il17agfp/gfp reporter mice in an SFB-free background. Seven days after SFBe gavage, WT and DKO littermates were colonized at similar levels (Figure S2A). On day 7, at the peak of the Th17 response, we observed in WT and DKO animals comparable proportions of Foxp3neg RORγt+ CD4+ T cells in different segments of the GI tract. Within the terminal ileum, however, production of IL-17A–GFP in the poised Th17 cells was significantly reduced in DKO mice, both proportionally and on a per-cell basis (Figure 2 D–E). In addition, IL-17A and IL-17F mRNAs in the SILP of DKO animals were significantly reduced relative to WT littermates (Figure S2A). Unlike IL-17A and IL-17F, the chemokine receptor CCR6, a marker of Th17 cells, was expressed in similar amounts in wildtype and SAA1/2DKO animals (Figure S2B), consistent with SAA1/2 contributing to a specific subset of genes in the Th17 cell program.
To exclude a possible role for SAA1/2 in T cell development or repertoire bias, we colonized WT and DKO hosts with SFBe feces and adoptively transferred into them naïve CD4+ T cells from 7B8 transgenic mice (Yang et al., 2014). One week post-transfer, we noted that comparable proportions of donor-derived cells became RORγt+ within the terminal ileum of both DKO and WT animals. However, production of IL-17A by the RORγt+ 7B8 cells upon PMA/Ionomycin restimulation was markedly attenuated in DKO recipients (Figure 2F). Similar results were observed in WT and DKO recipients of T cells from 7B8 transgenic mice bred onto the IL17A–IRES-GFP reporter background (Figure S2C). Although almost all SFB-specific donor-derived T cells found in the ileum of SFB-colonized mice expressed RORγt regardless of the host genotype, we observed a reduction in total number of these cells recovered in SAA1/2 DKO animals (Figure 2F). This suggests that, in addition to a role in promoting Th17 cytokine production, SAA may promote local Th17 cell proliferation and/or retention.
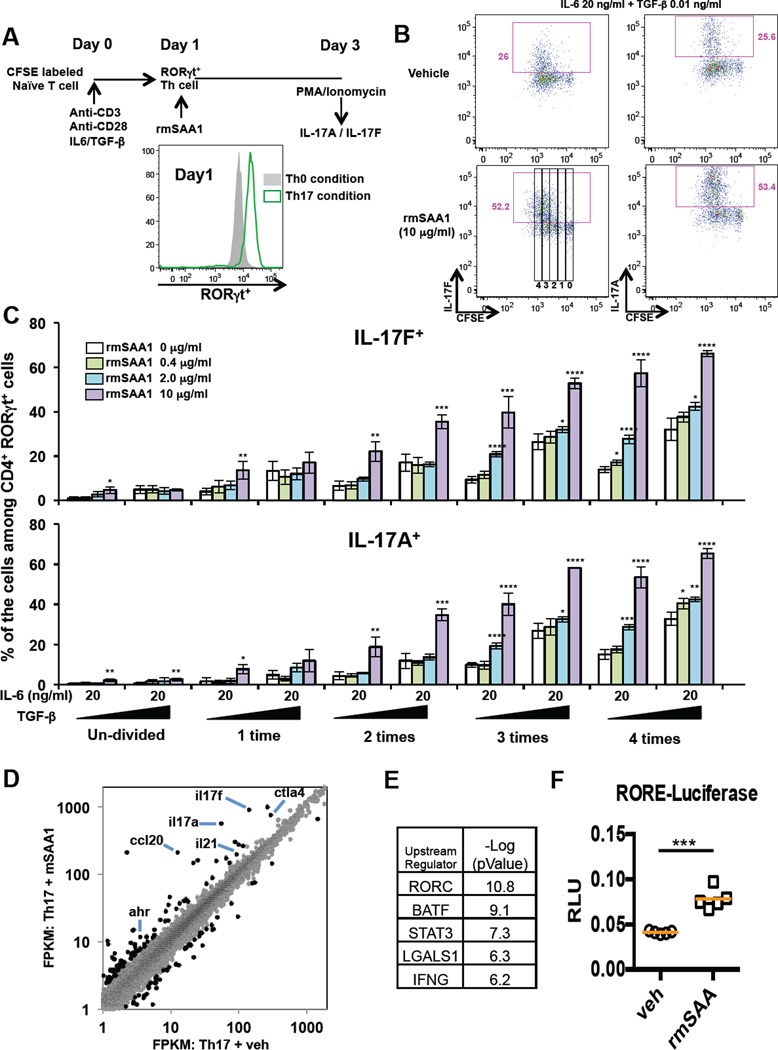
Next, we treated murine naïve CD4+ T cells cultured in Th17 polarizing conditions with recombinant mouse SAA1 (rmSAA1). SAA1 significantly enhanced IL-17A and IL17-F production in RORγt+ CD4+ T cells cultured in suboptimal Th17 polarizing conditions (Figures 3A–C and S3A). Importantly, this effect was reversed when anti-mSAA blocking antibody was introduced (Figure S3A). In contrast, CCR6 expression was not changed by rmSAA1 treatment (Figure S3B). Based on our in vivo observation, we also asked whether mSAA1 exerts a direct effect on cell proliferation in addition to Th17 signature cytokine production. Labeling of naïve CD4+ T cells with CFSE and intracellular Ki67 staining showed enhanced proliferation of RORγt+ Th17 cells upon treatment with rmSAA1 (Figures 3A–C and S3C), but the higher proportions of cells expressing IL-17A and IL-17F were observed independently of the extent of proliferation (Figure 3C). Furthermore, in vitro differentiation of naïve CD4+ T cells toward Th1, Th2, and iTregs was not significantly affected in the presence of recombinant mouse SAA1 (Figure S3D). Similarly, rhSAA augmented IL-17A production during human Th17 cell priming (Figure S3E). Importantly, this effect was reversed when SAA blocking antibody was introduced into culture. These results suggest that SAA acts directly on both mouse and human Th17 cells to enhance their differentiation and effector function.
Figure 3. SAA1 promotes IL-17A production in polarized Th17 cells in vitro.
(A) Schematic representation of the experimental protocol. CFSE-labeled naïve CD4+ T cells from WT mice were activated under non-polarizing (Th0: Anti-CD3 and Anti-CD28) or Th17 polarization condition (Th17: Anti-CD3, Anti-CD28, IL-6 and TGF-β). At 24h, RORγt+ T cells were further cultured with recombinant mouse SAA1 (rmSAA1) under Th17 polarization conditions for 48h. (B) IL-17A and IL-17F production by in vitro-polarized mouse Th17 cells (CD4+ RORγt+) at 48h after rmSAA1 (10 µg/ml) addition. Cells were re-stimulated with PMA/Ionomycin for 5h, and cytokine production and CFSE dilution were monitored by FACS analysis. (C) Representative data from polarized cells (48h) (4 experiments total). The frequencies of IL-17A- and IL-17F–producing cells during each cell division are displayed using the gates represented in (B). Data are shown as mean with SD. (D) RNAseq studies from two biological replicates comparing in vitro Th17 cells cultured in 20 ng/ml IL-6 and 0.1 ng/ml TGF-β with or without 2 µg/ml rmSAA1 for 48h. Dots represent FPKMs of individual mRNA transcripts in the respective cell type (grey). 195 genes that were significantly different between vehicle and SAA treatment (p-value<0.05, fold change greater than 2) are highlighted in black. (E) Ingenuity Pathway Analysis for upstream regulators of the 195 SAA-affected genes in Th17 cells. (F) A ROR-responsive element (RORE) firefly luciferase reporter was used to monitor RORγt transcriptional activity in Th17 cells. The RORE-luciferase reporter construct was introduced by nucleofection into Th17 cells polarized for 48h and cultured with or without rmSAA1 (2 µg/ml), and luciferase activity was measured 24h later. Data were normalized by renilla reporter activity. Results from four technical repeats in each condition were averaged. Results are representative of three independent experiments. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S3.
To further study how SAA influences the transcriptional program of Th17 cells, we performed RNAseq with control and rmSAA1-treated murine Th17 cells. We identified a specific subset of the Th17 gene expression program that was specifically affected by rmSAA1 treatment (Figures 3D and S3F). Unbiased Ingenuity Pathway Analysis predicted RORγt, the key regulator of the Th17 differentiation program, as the top upstream regulatory factor sensitive to SAA perturbation in vitro (Figure 3E). ROR-element driven luciferase assays in Th17 cells confirmed that RORγt transcriptional activity was potentiated during rmSAA1 stimulation in vitro (Figure 3F).
SFB-induced SAA1/2 requires IL-22 expression and epithelial Stat3 signaling
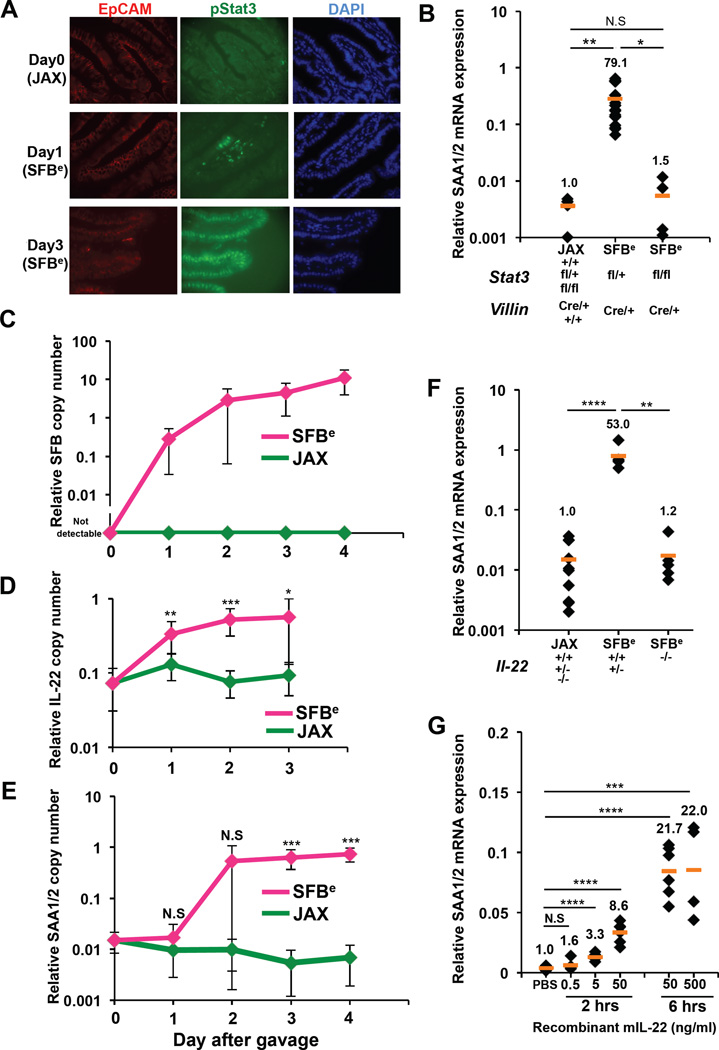
We next sought to understand the upstream signaling pathway that regulates ileal SAA induction in response to SFB colonization. SFB-free C57BL/6 mice were co-housed with SFB-positive mice (Taconic) for two weeks before epithelial-specific translating RNAs were assessed. RNASeq profiling of polysome-associated mRNAs pulled-down from SFB-colonized TRAP Villin-Cre mice revealed a significant enrichment of actively translated genes that are known to be regulated by the transcription factor Stat3 (Figure S4A). To corroborate this analysis, we used immunofluorescence to probe for Stat3 activity (pStat3) in small intestinal tissue sections of mice acutely colonized with SFBe feces. We detected pStat3 within 24h of SFB colonization (Figure 4A). Strikingly, SFBe fecal-induced upregulation of SAA1/2 was abrogated almost entirely in mice with intestinal epithelial cell-specific inactivation of Stat3 (Figure 4B). Stat3 activation is elicited by multiple cytokines, including IL-10 and members of the IL-20 subfamily, of which IL-22 is the best characterized (Rutz et al., 2014). Interestingly, we detected a significant increase in IL-22 mRNA within the terminal ileum shortly after colonization with SFBe, and this preceded the induction of epithelial SAA1/2 expression by one day (Figure 4C–E). This result suggested that IL-22 activated Stat3, which then induced SAA1/2 expression. Indeed, treatment with a neutralizing anti-IL-22 antibody prevented SAA1/2 induction following SFB colonization (Figure S4B). Consistent with this result, SAA1/2 mRNA induction by SFB was almost completely abrogated in IL-22 deficient mice (Figure 4F). In addition, IL-17A production among RORγt+ Th17 cells in ileum was partially blocked in animals receiving IL-22 blocking antibodies, similar to what was observed in SAA1/2 DKO mice (Figure S4C).
Figure 4. Requirement for IL-22 activation of epithelial Stat3 in SFB-dependent induction of SAA1/2.
(A) Immunohistochemistry analysis of phospho-Stat3 (green) in terminal ileum from SFB-colonized (SFBe) or SFB free (JAX) mice on day 1 or day 3 following gavage. EpCAM (red) and DAPI (blue) staining was used to identify the intestinal epithelial layer. The experiment was repeated 2 times with similar results.
(B) qRT-PCR analysis of SAA1/2 expression in the epithelial fraction of terminal ileum from Villin-Cre mice sufficient or deficient for epithelial Stat3 at 4 days after SFB colonization. Data were normalized using Gapdh.
(C) Kinetics of SFB colonization post oral gavage of SFBe or JAX fecal contents. SFB genomic 16S in terminal ileum was quantified by qPCR analysis. Copy number of the SFB genomic16S was normalized to host Il-23r genomic DNA (JAX; n=4. SFBe; n=7). (D,E) Kinetics of IL-22 (D) and SAA1/2 (E) mRNA expression post SFBe gavage. (D) Level of IL-22 mRNA from isolated ileal lamina propria cells was assessed (JAX; n=7. SFBe; n=8 in each time points). (E) Level of SAA1/2 mRNA from epithelial fraction in ileum (JAX; n=4. SFBe; n=7). Graphs (C–E) show accumulated data from two or three independent experiments as mean with SD. Day 0 samples were from animals not gavaged with additional fecal samples.
(F) SAA1/2 mRNA expression, determined by qRT-PCR, in epithelial fraction of terminal ileum from IL-22 sufficient or deficient mice at 4 days after SFB gavage (JAX gavaged: n=10, SFBe gavaged IL-22+/+ and +/− : n=5, SFBe gavaged IL-22−/− : n=5).
(G) SAA1/2 mRNA expression in small intestinal epithelial cell organoid cultures following rIL-22 treatment.
All qRT-PCR experiments were repeated two times with similar results, and data were normalized to GAPDH mRNA. Each dot represents a single mouse, and the orange bar indicates average relative gene expression of each group. N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S4.
To evaluate whether IL-22 can directly induce epithelial SAA1/2 expression, we generated small intestinal epithelial (SIE) organoids as previously described (Sato 2009) and exposed them to recombinant murine IL-22 (rIL-22). The addition of rIL-22 to SIE organoid cultures induced SAA1/2 mRNA in a dose-dependent fashion within two hours (Figure 4G). Coupled with the in vivo results, these findings indicate that SFB colonization initiates an IL-22-mediated Stat3 signaling cascade in epithelial cells, resulting in localized SAA1/2 expression.
SFB stimulates IL-23R-dependent IL-22 production by activated ILC3
In the intestine, IL-22 is produced by RORγt-expressing lymphoid cells that include ILC3, Th17, and γδ17 cells (Sawa et al., 2011). To investigate the source of IL-22 that directs epithelial SAA production following SFB colonization, we compared RNA expression profiles from small intestine Th17 cells and ILC3 in mice stably colonized with SFB. Based on normalized DESeq reads, IL-22 expression was ten-fold higher in ILC3s than in Th17 cells (Figure S5A). Examination of these data sets also revealed a surface antigen, killer cell lectin-like receptor subfamily B member 1B (Klrb1b), which was uniquely expressed in ILC3. We confirmed this finding at the level of protein by flow cytometry (Figure S5B), and subsequently incorporated Klrb1b into the fluorescent cell-labeling panel to dispense with the necessity of a reporter in studying the dynamic behavior of ILC3 upon SFBe colonization.
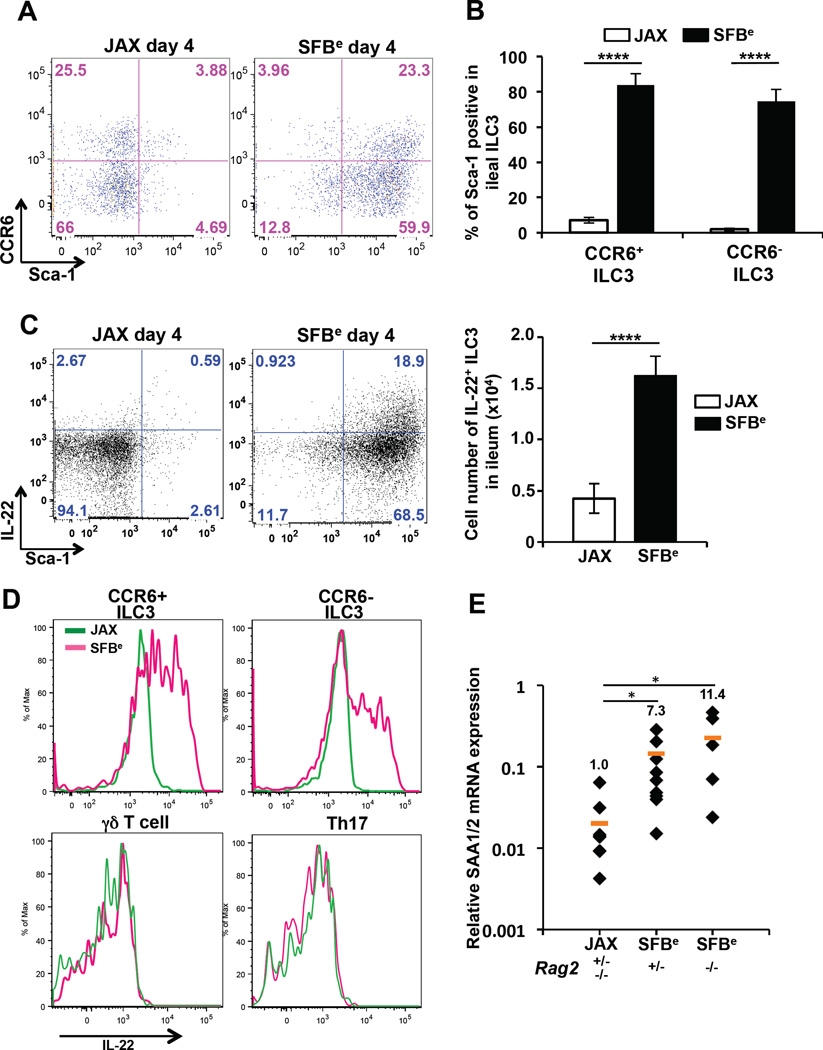
At day four of SFBe colonization, up to 80% of ILC3s expressed the activation marker stem cell antigen-1 (Sca-1), which amounted to a 10-fold increase compared to the proportion of Sca-1+ ILC3s in mice gavaged with SFB-negative feces (Figure 5A–B). Unlike CCR6− ILC3s, CCR6+ ILC3s typically populate the isolated lymphoid follicles and cryptopatches (Lugering et al., 2010). Notably, both CCR6− and CCR6+ ILC3s expressed high level of Sca-1 on their surface in response to SFB colonization. Although ILC3s increased in size in response to SFB colonization, their absolute numbers remained unchanged (Figure S5C–E). Intracellular cytokine staining also revealed a striking increase in both the frequency and total number of ILC3s producing IL-22 (Figure 5C). We recapitulated these findings using the fecal contents from SFB mono-associated mice, supporting the notion that SFB alone is sufficient to direct the localized ILC3 response (Figure S5F). Importantly, at this early time point, other RORγt-dependent lineages, including γδ17 and (non-SFB-specific) Th17 cells, did not produce IL-22 (Figure 5D). Furthermore, epithelial cells from Rag2 mutant mice, harboring ILC3s but lacking B and T cells, also expressed SAA1/2 in response to SFB colonization (Figure 5E).
Figure 5. SFB colonization induces IL-22 production by ILC3s.
(A) Cell surface expression of Sca-1 and CCR6 on ileal SILP ILC3s from mice at 4 days after oral-gavage with JAX or SFBe fecal contents.
(B) Frequency of Sca-1-positive CCR6+ or CCR6− ILC3s in the terminal ileum from JAX- or SFBe-gavaged mice at 4 days after gavage. Data are represented as mean with SD (JAX gavaged: n=5, SFBe gavaged: n=6).
(C) IL-22 production in ileal SILP ILC3. Ileal LP cells from JAX- or SFBe-gavaged mice were isolated at 4 days after gavage and cultured in vitro with GolgiStop (without PMA/Ionomycin re-stimulation) for 4h. IL-22-producing ILC3s were analyzed by FACS, and cell numbers were calculated (right panel). Bar graph shows accumulated data as mean with SD (JAX gavaged: n=5, SFBe gavaged: n=6).
(D) IL-22 production in each indicated cell type isolated from the ileal LP of SFB-free (JAX; green line) and SFB-colonized (SFBe; magenta line) mice, analyzed at 4 days after gavage. (A–D) The experiment was repeated at least three times with similar results.
(E) SAA1/2 expression, analyzed by qRT-PCR, in the epithelial fraction from terminal ileum of Rag2-sufficient or -deficient mice, at 3 days after SFB (SFBe) or control (JAX) gavage. Data are from two independent experiments. Each symbol represents a single mouse, and the orange bar indicates average of relative gene expression in each group. Experiments were repeated two times with similar results and data were normalized by GAPDH mRNA and combined. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001.
See also Figure S5.
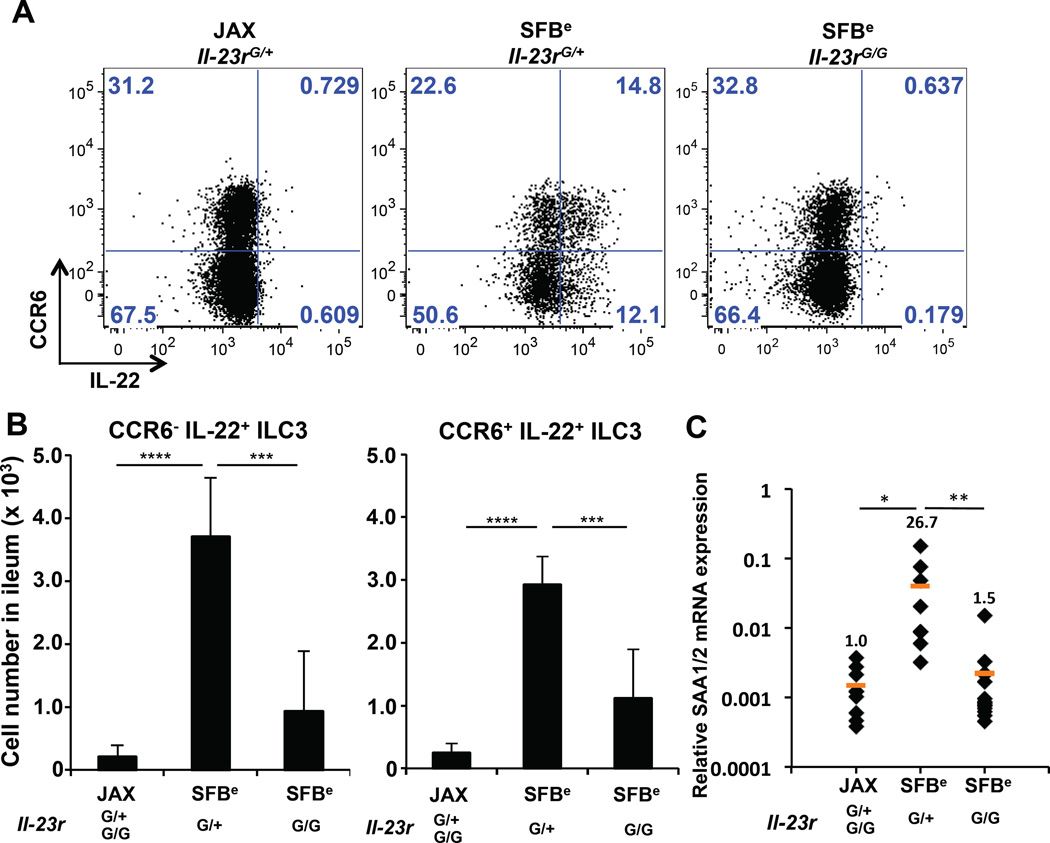
ILC3s residing in the colon secrete IL-22 in response to IL-23 (Eken et al., 2014). To determine whether this mechanism also governs terminal ileal production of IL-22 by ILC3s upon SFB colonization, we colonized IL-23R–sufficient and -deficient mice with SFBe feces and examined the spontaneous release of IL-22 by ILC3s. Both the frequency and absolute number of IL-22-secreting CCR6+ and CCR6− ILC3s declined sharply in IL-23R–deficient animals (Figure 6A–B). Consistent with this, induction of epithelial SAA1/2 mRNA was markedly blunted in IL-23R–deficient animals (Figure 6C). Altogether, these findings indicate that IL-23 signaling initiates IL-22-mediated crosstalk between ILC3s and small intestine epithelial cells, resulting in spatially-restricted SAA production, which contributes to local augmentation of Th17 effector functions (Figure S6).
Figure 6. IL-23R requirement for SFB-induced expression of IL-22 and SAA1/2 in ILC3 and epithelial cells, respectively.
(A) IL-22 production in CCR6+ and CCR6− ileal LP ILC3s from Il-23r–sufficient or -deficient mice. (B) Cell number of IL-22-producing CCR6− (left) or CCR6+ (right) ileal LP ILC3s in JAX- or SFBe-gavaged mice, at 4 days after gavage. Bar graphs show accumulated data from two independent experiments with a total of 10 SFB-free (JAX) and 5 SFB-colonized (SFBe) mice of each genotype, shown as mean with SD. (C) SAA1/2 expression, analyzed by qRT-PCR, in the epithelial fraction from terminal ileum of indicated mice (JAX gavaged; n= 9, SFBe gavaged Il-23rG/+; n=8, SFBe gavaged Il-23rG/G; n=14).This experiment was repeated twice with similar results. Each dot represents a single mouse, and the orange bar indicates average relative gene expression of each group. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001.
DISCUSSION
In this report, we describe a two-step process for the functional differentiation of intestinal Th17 cells following colonization of mice with SFB. The first step is the priming and polarization of antigen-specific CD4+ T cells in the tissue-draining lymph nodes, resulting in a poised state marked by the expression of RORγt, and the second is the activation of a cytokine gene expression program in a tissue microenvironment in which epithelial cell-derived factors SAA1 and SAA2 act on poised cells. Temporally, we observed specific SFB colonization of the ileum by 24h post-SFB gavage. Within 5 days, SFB antigen-presentation occurred in the MLN, where a significant number of RORγt+ Th17 cells were first observed. By day 7, RORγt+ Th17 cells accumulated in the lamina propria of the intestine. During the early phase of this commensal-induced host response, IL-22 and SAA1/2 mRNA were acutely up-regulated in the ileum within 3 days. Our studies revealed that activation of epithelial SAAs involved an SFB-initiated signaling pathway requiring IL-23 and ILC3-derived IL-22 (Figure S6). A similar IL-22-dependent pathway was recently described in SFB induction of Fut2, an enzyme that catalyzes fucosylation of glycans, thus providing nutritional substrates for commensals and contributing to barrier protection (Bry et al., 1996; Goto et al., 2014; Pickard et al., 2014).
Although we identified a requirement for the ILC3-dependent pathway during the early induction of epithelial SAA1/2, we cannot rule out that SFB attachment to the epithelial cells, which is accompanied by reorganization of the cortical actin network, contributes to or is required for SAA induction. Indeed, Honda and colleagues (accompanying paper) showed that rat SFB failed to attach to ileal epithelium and to induce SAA1/2 in mice, yet induced ILC3 IL-22 production. Thus, multiple signals may be required for epithelial cell induction of SAA1/2 or, alternatively, a single IL-22 signal may be sufficient, but its delivery to epithelial receptors may be compromised in mice colonized with rat SFB.
SAAs in Homeostasis and Autoimmune Diseases
SFB is considered a commensal microbe, as there is no evidence that it induces pathology despite its adhesion to the epithelial surface and its induction of host adaptive and innate immune responses. By virtue of its activation of epithelial Stat3, SFB may reinforce the terminal ileal epithelial firewall, which harbors the greatest bacterial load in the small intestine (Sekirov et al., 2010). In the large intestine, Stat3 was found to mediate protection from dextran sodium sulfate (DSS)-induced colon injury in an IL-22 dependent manner. In this model, SAA induction was similarly dependent on Stat3 and required for dampening local inflammation (Eckhardt et al., 2010; Neufert et al., 2010). Recently, Hooper and colleagues showed that SAAs bind with strong affinity to vitamin A derivatives (Martineau et al., 2015), which promote mucosal homeostasis (Hall et al., 2011). Therefore, in the context of SFB colonization, the elevation of SAA1/2 and Th17 cells may be considered a homeostatic mechanism that contributes to barrier integrity in the small intestine.
SAAs are significantly up-regulated in the joints and serum of rheumatoid arthritis patients (O'Hara et al., 2000) and are correlated with disease progression (Chambers et al., 1983). In mouse models, Th17 cells are also known to contribute to multiple autoimmune diseases (Genovese et al., 2010; Huh et al., 2011; Leppkes et al., 2009), and whether SAAs or differences in its binding of cofactors mediate these processes remain to be elucidated. As microbiota-specific helper T cells, including SFB-specific Th17 cells, circulate widely following their polarization in the gut-associated lymphoid tissues (Hand et al., 2012; Yang et al., 2014), it is possible that they contribute to autoimmune disease systemically through induction of their effector functions at sites where inflammatory mediators such as SAAs are elevated.
Another possibility is that under normal circumstances SAAs regulate the resolution of inflammatory responses. Th17 cells often produce IFNγ in addition to IL-17A at sites of inflammation (Ivanov et al., 2006), and this is thought to contribute to pathogenicity, but they can also give rise to IL-10-producing cells that may be important during the resolution of inflammation (Gagliani et al., 2015). Therefore, it will be important to study in detail the regulation of SAA expression in tissues during the course of autoimmune progression and whether their manipulation affects Th17-mediated pathology. Importantly, since we did not observe a complete loss in Th17 cytokine production in SAA1/2-deficient settings during SFB induction, we speculate that other epithelial cell-derived factors likely contribute to the regulation of the Th17 effector program, which will be of great interest to study.
Innate Lymphoid Cell Regulation of Barrier Tissue Immunity
Tissue-resident RORγt-expressing innate lymphoid cells and “innate-like” T cells, e.g. γδ17 cells, can respond rapidly to cytokine stimulation, including IL-1β and IL-23, by producing their own cytokines, particularly IL-22. This occurs in the absence of antigen receptor stimulation in γδ17 cells (Cua and Tato, 2010; Martin et al., 2009; Sutton et al., 2009). For ILC3s, microbiota-induced local production of IL-23, IL-1β, and TL1a/TNFSF15 by CX3CR1+ mononuclear phagocytes is required for their production of IL-22 and intestinal barrier protection functions (Longman et al., 2014). We do not know whether RORγt+ SFB-specific T cells that migrate to the intestinal lamina propria require TCR signaling for induction of effector cytokines, or whether SAA1/2, in the appropriate microenvironment, suffices for this purpose. It will be important, in future studies, to identify the receptor(s) for SAA1/2 on Th17 cells and characterize the signaling mechanism required for the cytokine response.
Our findings suggest that SAA1 and SAA2 serve as sentinels for the tissue microenvironment, enlisting T helper cells with poised Th17 functions to protect or heal the epithelial barrier. It will be important to determine whether the SAAs are unique in their ability to amplify the Th17 cell transcriptional program and whether other induced mediators similarly influence other functional T cell subsets. In this regard, IL-18 and IL-33 were recently shown to amplify Th1 effector responses during infection with Salmonella or Chlamydia (O'Donnell et al., 2014). Efforts to dissect the upstream pathways and secretory outputs that impinge on T cell effector functions at steady state and in the face of multiple challenges will create novel opportunities to modulate tissue immunity.
A critical remaining question is how SFB are sensed within the ileum. Honda and colleagues (accompanying paper) showed that adhesion of microbes to epithelium is a necessary step for Th17 differentiation. It will be important to learn whether distinct signals received by ileal epithelial cells from bound SFB are critical for Th17 cell induction and subsequent cytokine production. Characterization of such signals may provide insights into how intestinal microbes contribute to local and systemic inflammation.
EXPERIMENTAL PROCEDURES
Mouse Strains and SFB
C57BL/6 mice were obtained from Taconic Farms or the Jackson Laboratory. All transgenic animals were bred and maintained in the animal facility of the Skirball Institute (NYU School of Medicine) in specific-pathogen-free (SPF) conditions. Il-23rgfp mice (Awasthi et al., 2009) were provided by M. Oukka and maintained on Jackson flora. Il-22 mutant mice were provided by Pfizer. Saa1/2 double knockout mice were previously described (Eckhardt et al., 2010) and were backcrossed for 8 generations onto the C57BL/6 background. SFB-specific Th17-TCRTg (7B8) mice were previously described (Yang et al., 2014) and maintained on the Il17agfp reporter (JAX; C57BL/6-Il17atm1Bcgen/J) and Ly5.1 background (JAX; B6. SJL-Ptprca Pepcb/BoyJ). EF1a: Lox-stop-lox-GFP-L10 mice were previously described (Stanley et al., 2013) and bred to Cd4 Cre (Taconic), Cd11c Cre (JAX) , Villin Cre (JAX), or Albumin Cre (JAX) mice and maintained with Jackson flora. Stat3flox/flox mice (JAX, B6.129S1 Stat3tm1Xyfu/J) were bred to Villin-Cre mice. Six- to 18-week old animals were used for in vivo experiments. All animal procedures were in accordance with protocols approved by the Institutional Animal Care and Use Committee of the NYU School of Medicine. Fecal pellets were provided by Y. Umesaki (Yakult) to establish a colony of SFB mono-colonized mice. Mice mono-colonized with SFB were housed in the NYU Medical School Skirball Institute gnotobiotic animal facility.
Acute SFB Colonization
Fecal pellets were collected from SFB-enriched Il-23r Rag2 DKO mice (SFBe), SFB mono-associated mice (pure SFB), or SFB-free JAX B6 mice (JAX), respectively. Fresh fecal pellets were homogenized through a 100 mm filter, pelleted at 3400 rpm for 10 minutes, and re-suspended in PBS. Each animal was administered 1/4 pellet by oral gavage.
Statistics
Two-tailed unpaired Student’s t-tests were performed to compare the results using Excel and Prism. We treated less than 0.05 of P-value as significant differences. * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001. All experiments were performed at least twice.
Supplementary Material
Highlights.
SFB-specific Th17 cells are primed in MLN and traffick throughout the GI tract.
IL-17A–producing Th17 cells are enriched in ileum, where SFB adheres to epithelium.
SAA1/2 from ileal epithelial cells contributes the induction of IL-17A in Th17 cells.
SFB-induced IL-22 production by ILC3 stimulates epithelial cells to make SAA1/2.
ACKNOWLEDGEMENTS
We thank Sebastian Amigorena, Kenneth Cadwell, Suzan R. Schwab, and Sang V. Kim for valuable discussion, Wenjun Ouyang for providing control and anti-IL-22 neutralizing antibody, Jeffrey M. Friedman and Ana Dominguez for the EF1-lox-stop-lox-GFP-L10 mice, and Frederick De Beer for providing the Saa mutant mice. We thank Maria Ciofani for her contributions to the RNAseq studies. We thank Richard M. Myers at HudsonAlpha Institute for Biotechnology for RNAseq and TRAPseq studies. This work was supported by fellowships from the TOYOBO Bioscience foundation (T.S.), Human Frontier Science Program (T.S.), Cancer Research Institute (W.H.), and NIH T32 CA009161_Levy (W.H.); and by the Howard Hughes Medical Institute (D.R.L.), the Helen and Martin Kimmel Center for Biology and Medicine (D.R.L.) and National Institutes of Health grant R01DK103358 (R.B. and D.R.L.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.S. and W.H. designed and performed most experiments and analyzed the data. J.A.H. performed experiments related to ILC3 activation, J.L. performed organoid culture studies, A.C. and S.J.G. assisted with in vivo experiments, and Y.Y. generated TCR transgenic mice and provided cells for RNAseq analyses. E.M., R.B., and W.H. analyzed RNAseq and Ribosome TRAPseq data. J.Z. performed studies related to IL-23R regulation of SFB-induced epithelial responses. T.S., W.H., J.A.H. and D.R.L. wrote the manuscript. D.R.L. supervised the research and participated in experimental design.
REFERENCES
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, Bettelli E, Kuchroo VK, Oukka M. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers RE, MacFarlane DG, Whicher JT, Dieppe PA. Serum amyloid-A protein concentration in rheumatoid arthritis and its role in monitoring disease activity. Ann Rheum Dis. 1983;42:665–667. doi: 10.1136/ard.42.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Zeng X, Prinz I. The natural and the inducible: interleukin (IL)-17-producing gammadelta T cells. Trends in immunology. 2013;34:151–154. doi: 10.1016/j.it.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cray C, Zaias J, Altman NH. Acute phase response in animals: a review. Comparative medicine. 2009;59:517–526. [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nature reviews Immunology. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- Derebe MG, Zlatkov CM, Gattu S, Ruhn KA, Vaishnava S, Diehl GE, MacMillan JB, Williams NS, Hooper LV. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. eLife. 2014;3:e03206. doi: 10.7554/eLife.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt ER, Witta J, Zhong J, Arsenescu R, Arsenescu V, Wang Y, Ghoshal S, de Beer MC, de Beer FC, de Villiers WJ. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol. 2010;10:133. doi: 10.1186/1471-230X-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eken A, Singh AK, Treuting PM, Oukka M. IL-23R+ innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal immunology. 2014;7:143–154. doi: 10.1038/mi.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuzawa-Carballeda J, Vargas-Rojas MI, Cabral AR. Autoimmune inflammation from the Th17 perspective. Autoimmun Rev. 2007;6:169–175. doi: 10.1016/j.autrev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Gagliani N, Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limon P, Paiva RS, Ching T, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015 doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese MC, Van den Bosch F, Roberson SA, Bojin S, Biagini IM, Ryan P, Sloan-Lancaster J. LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebo-controlled, proof-of-concept study. Arthritis Rheum. 2010;62:929–939. doi: 10.1002/art.27334. [DOI] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nature protocols. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annual review of immunology. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;(108 Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, Klinakis A, Charo IF, Jung S, Gommerman JL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugering A, Ross M, Sieker M, Heidemann J, Williams IR, Domschke W, Kucharzik T. CCR6 identifies lymphoid tissue inducer cells within cryptopatches. Clinical and experimental immunology. 2010;160:440–449. doi: 10.1111/j.1365-2249.2010.04103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature reviews Immunology. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Martineau AR, James WY, Hooper RL, Barnes NC, Jolliffe DA, Greiller CL, Islam K, McLaughlin D, Bhowmik A, Timms PM, et al. Vitamin D3 supplementation in patients with chronic obstructive pulmonary disease (ViDiCO): a multicentre, double-blind, randomised controlled trial. The Lancet Respiratory medicine. 2015;3:120–130. doi: 10.1016/S2213-2600(14)70255-3. [DOI] [PubMed] [Google Scholar]
- Neufert C, Pickert G, Zheng Y, Wittkopf N, Warntjen M, Nikolaev A, Ouyang W, Neurath MF, Becker C. Activation of epithelial STAT3 regulates intestinal homeostasis. Cell Cycle. 2010;9:652–655. doi: 10.4161/cc.9.4.10615. [DOI] [PubMed] [Google Scholar]
- O'Donnell H, Pham OH, Li LX, Atif SM, Lee SJ, Ravesloot MM, Stolfi JL, Nuccio SP, Broz P, Monack DM, et al. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity. 2014;40:213–224. doi: 10.1016/j.immuni.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara R, Murphy EP, Whitehead AS, FitzGerald O, Bresnihan B. Acute-phase serum amyloid A production by rheumatoid arthritis synovial tissue. Arthritis research. 2000;2:142–144. doi: 10.1186/ar78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nature reviews Immunology. 2014;14:783–795. doi: 10.1038/nri3766. [DOI] [PubMed] [Google Scholar]
- Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods in molecular biology. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- Stanley S, Domingos AI, Kelly L, Garfield A, Damanpour S, Heisler L, Friedman J. Profiling of Glucose-Sensing Neurons Reveals that GHRH Neurons Are Activated by Hypoglycemia. Cell metabolism. 2013;18:596–607. doi: 10.1016/j.cmet.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Current opinion in immunology. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annual review of immunology. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.