Abstract
Many of the factors affecting the success of hematopoietic cell transplantation are still unknown. Here we show in mice that donor’s sleep deprivation reduces the ability of its hematopoietic stem cells (HSCs) to engraft and reconstitute the blood and bone marrow of an irradiated recipient by more than 50%. We demonstrate that sleep deprivation downregulates the expression of microRNA (miR)-19b, a negative regulator of the suppressor of cytokine signaling (SOCS) genes, which inhibit HSC migration and homing. Accordingly, HSCs from sleep-deprived mice have higher levels of SOCS genes expression, lower migration capacity in vitro and reduced homing to the bone marrow in vivo. Recovery of sleep after sleep deprivation restored the reconstitution potential of the HSCs. Taken together, this study provides insights into cellular and molecular mechanisms underlying the effects of sleep deprivation on HSCs, emphasizing the potentially critical role of donor sleep in the success of bone marrow transplantation.
While hematopoietic cell transplantation is a standard therapeutic procedure for various malignant and non-malignant diseases, the impact of sleep on hematopoietic cell transplantation has not been investigated. Sleep disorders affect metabolism, endocrine system and immune response1–4. We hypothesized that also hematopoietic stem cells (HSCs), are affected by sleep deprivation. These cells are used in hematopoietic cell transplantation procedures and are responsible for the lifelong production and maintenance of all blood and immune cells. Our hypothesis is supported by increasing evidence of a circadian regulation of HSCs and of the hematopoietic niche5–7. Circadian rhythms provide temporal organization to molecular, cellular, and biochemical processes and they may therefore be synchronizing HSCs functions with sleep. A relationship of this nature between sleep and the function of HSCs can be especially important, as more than 100 million people around the world, including potential bone marrow donors, suffer from disorders of sleep and wakefulness. Nevertheless, no studies, thus far, have linked donor sleep profile with hematopoietic cell transplantation outcomes. This work focuses on the effects of sleep deprivation on the functions of HSCs. We show that four hours of sleep deprivation in mice reduces the ability of HSCs to engraft and reconstitute the blood and bone marrow of an irradiated recipient by more than 50%. We attribute these effects to an attenuated responsiveness of the HSCs to migration-guiding cues ex vivo and in vivo. At the cellular level, we show that sleep deprivation downregulates the expression of microRNA (miR)-19b, which is a negative regulator of the suppressor of cytokine signaling (SOCS) genes that inhibit HSC migration and homing. Using a standard luciferase reporter assay, we validate in vitro the interaction between miR-19b and SOCS3 and demonstrate that sleep deprivation elevates SOCS3 levels. Using genetic manipulations, we demonstrate that in the absence of SOCS3, sleep deprivation does not significantly affect HSC migration. Finally, we show that growth hormone (GH) levels regulated by sleep alters miR-19b concentrations in HSCs and affects HSC migration, representing a possible mechanism mediating the effects of sleep deprivation on HSCs.
Results
HSCs from sleep-deprived mice have reduced reconstitution potential
To determine the effects of sleep deprivation on HSC transplantation potential, mice were allowed to sleep ad lib for four hours (Zeitgeber time (ZT) ZT0-ZT4; sleep) or were sleep-deprived for the same duration by gentle handling. The efficiency of the sleep deprivation protocol was monitored by electroencephalography (EEG) and electromyography (EMG; Fig. 1a). Gentle handling was chosen as the means of depriving mice of sleep, so as to avoid stress8. In line with previous studies8, there was no difference in plasma corticosterone (Cort) levels between the two experimental groups (Fig. 1b; Student’s t-test; p=0.6814; df=10; t=0.422). All animals were examined at the same circadian time, to rule out a potential direct circadian contribution to the observed effects.
Figure 1. HSCs isolated from sleep-deprived mice have reduced mid-term and long-term reconstitution potential in lethally irradiated hosts.
(a) Mice were sleep-deprived, by gentle handling, for four hours, immediately after light onset (ZT0-ZT4; n=8 per group). Rapid eye movement (REM) and non-REM (NREM) duration were determined using electroencephalography (EEG) and electromyography (EMG), individually plotted for each hour (sleep deprived points in red; mean±s.e.m). (b) Plasma of the sleep and sleep-deprived mice was analyzed by ELISA for corticosterone levels (n=8 per group; mean±s.e.m). (c) We isolated HSCs from mice that were allowed to sleep or sleep-deprived mice. The flow cytometry gating scheme for HSCs isolation is shown on a representative mouse. (d) To test the mid-term and long-term transplantation potential of the isolated HSCs we intravenously injected 300 HSCs mice into lethally irradiated congenic recipients (to distinguish between the donor and recipient cells we used CD45.1 mice as donors and CD45.2 as recipients). Peripheral blood (PB) samples were collected from the recipient mice at (e) eight weeks (peripheral blood), (f) 16 weeks and (g) bone marrow (BM) post-transplantation. Cells were analyzed for myeloid chimerism, as determined by the percentage of myeloid cells derived from the donor compared to the total number of myeloid cells in the indicated tissue (mean±s.e.m; Student’s t-test; ***p<0.0001; **p<0.001). In all experiments, HSCs were transplanted along with 1×106 bone marrow mononuclear cells to support mouse survival (d–g; n=12 mice per group). (h) Primary recipients received HSCs from sleep or sleep deprived donors as indicated in d–g. Then, at 16 weeks, 2.5 × 106 BM cells from primary recipients were transplanted into a new group of lethally irradiated CD45.2+ (secondary recipients) and donor-derived reconstitution was assessed in the secondary recipients (mean±s.e.m; Student’s t-test; p=0.83).
To test the transplantation potential of HSCs derived from mice that were allowed to sleep and sleep-deprived mice, 300 HSCs (lin−, c-kit+, Sca-1+, CD34−, CD150+; Fig 1c) were isolated by fluorescence-activated cell sorting (FACS) from the bone marrow of the two groups and intravenously transplanted into lethally irradiated recipient mice, along with 1×106 bone marrow-derived mononuclear cells to support recipient survival. Support cells were isolated from CD45.2+ mice (the recipients) that were allowed to sleep normally (the same pool of cells was used for both experimental groups). Reconstitution potential of transplanted HSCs was measured 8 and 16 weeks thereafter. The percentage of donor myeloid cells in the peripheral blood of the recipient was assessed, as a proxy for HSC chimerism9. Transplantation efficiency at 8 weeks and 16 weeks was lower for HSCs derived from sleep-deprived mice (Fig. 1d–g). At 16 weeks post-transplantation, donor myeloid chimerism was 26.42 ± 0.8% in mice that received HSCs from mice that were allowed to sleep compared to 11.62 ± 1% in mice that received HSCs from sleep-deprived mice (student’s t-test; t(22)=10.77; p<0.0001). Because all recipient mice underwent the same treatment prior to transplantation, we concluded that these effects were intrinsic to donor HSCs. To discern between short-term and long-term effects of sleep deprivation on the reconstitution potential of HSCs, we performed secondary transplantation experiments (Fig. 1h). In these experiments, the chimeric mice that received the initial bone marrow (BM) transplantation from either mice that were allowed to sleep or sleep-deprived mice served as the donors for the secondary transplantation. These secondary donors were not subjected to any sleep manipulation. Analysis of the donor myeloid chimerism in the secondary transplantation recipients did not identify any difference between the two groups. This suggested that sleep-deprivation induced a temporary change in the reconstitution potential of HSCs, which recovered by 16-weeks post-transplantation.
HSCs from sleep-deprived mice have reduced homing capacity
Reconstitution of the hematopoietic system following transplantation requires homing of HSCs from the peripheral blood to their supported niche in the bone marrow10,11. To determine whether the homing potential of HSCs was affected by sleep deprivation, donor HSCs were traced in vivo after transplantation. HSCs were isolated from the bone marrow of green fluorescence protein (GFP)-expressing transgenic mice that were either allowed to sleep ad lib. or were sleep-deprived for four hours. The GFP-expressing HSCs were then intravenously administered to lethally irradiated congenic, non-GFP-expressing recipient mice (2000 HSC per recipient). Twelve hours later, the mice were sacrificed and the number of GFP-expressing cells that successfully homed and localized to the bone marrow was determined. Fewer HSCs from sleep-deprived mice homed to the bone marrow during this time period, when compared to HSCs from control mice (Fig. 2a–b; 3.3 ± 1.4 % mice allowed to sleep; 1.7 ± 0.3 % sleep-deprived mice; student’s t – test; p<0.05). These findings show that HSCs from sleep-deprived mice are indeed impaired in their homing potential, a crucial component in the success of hematopoietic cell transplantation10,11.
Figure 2. HSCs from sleep-deprived mice have reduced homing capacity in vivo and in vitro.
(a) HSCs (2000 FACS-purified cells) derived from GFP-expressing, sleep-deprived mice were transplanted into lethally irradiated congenic mice that did not express GFP (control mice were allowed to sleep for the same duration). The donor HSCs were visualized 12 hours later in the recipient’s bone, under a fluorescence microscope. GFP expression was validated using anti-GFP staining and a representative image is shown in the insert. Scale 10µm (n=6–11 mice per group; mean±s.e.m). (b) The number of GFP-labeled cells in the recipient’s bone marrow was determined. Results are presented as the percentage of homing, assuming that two tibias and two femurs represent 20% of total mouse bone marrow (student’s t-test; p=0.021; t=2.56; df=15). (c) Migration towards SDF-1α was determined in vitro using a transwell migration assay. KLS cells were placed in the upper chamber and the chemoattractant (SDF-1α, 50, 100, or 200ng/mL) was immersed in the medium of the lower chamber. Migration across the membrane in response to the chemoattractant was determined using flow cytometry, after four hours of incubation, and compared to baseline migration in the absence of the chemoattractant. Results are presented as a migration index (50ng/mL SDF-1α: 9.6 ± 1.7% in the sleep group compared to 2.06 ± 0.4% in the sleep-deprived group; 100ng/mL SDF-1α: 14.5 ± 3.4% in the sleep compared to 3.6 ± 1.4% in the sleep-deprived mice; Repeated measures ANOVA- sleep: F(2,15)=10.18, p<0.0061; n=6 mice per group; mean ±s.e.m. (d) Plasma was collected and analyzed by ELISA for SDF-1α levels, from sleep or sleep-deprived mice. (e) CXCR4 levels on HSCs derived from sleep and sleep-deprived mice (mean fluorescent intensity (MFI)±s.e.m). (f) VLA4 (α4) levels on HSCs derived from sleep and sleep-deprived mice. (g) Cells were cultured with 0.5 or 10 nM LDV-FITC to evaluate VLA4 binding affinity. Binding was evaluated as FITC MFI on HSCs (mean±s.e.m). Statistical significance was determined by Student’s t-test. (h) Migration towards S1P was determined in vitro using a transwell migration assay as described in (c). S1P concentration is indicated. Repeated measure ANOVA (n=4 mice per group).
Migration of the HSCs towards guiding cues is required for their effective homing to the bone marrow. To determine whether the migration capacity of the HSCs is affected by sleep, we performed a transwell migration assay (Fig. 2c) and compared the number of HSCs migrating toward different concentrations of the migration guiding chemokine, stromal cell-derived factor α (SDF-1α). Migration towards low concentrations of SDF-1α (50–100ng/mL) was significantly decreased in HSCs derived from sleep-deprived mice, compared to HSCs derived from mice that were allowed to sleep. Interestingly, no differences in HSC migratory potential to a higher SDF-1α concentration (200ng/mL) were observed. This suggested that the HSCs of sleep-deprived mice do not lose their inherent ability to migrate towards SDF-1α, but rather, require a higher threshold of SDF-1α for migration. We could not explain this effect by changes in SDF-1α levels (Fig 2d) or in the levels of CXCR4, the cell-surface receptor for SDF-1α12, which were comparable between the experimental groups (Fig 2e). There was also no difference between the mice that were allowed to sleep and sleep-deprived mice in the levels of Very Late Antigen-4 (VLA4) integrin (alpha4 unit; Fig 2f) or its binding affinity (Fig 2g). Moreover, migration towards, sphingosine-1-phosphate (S1P), a chemotactic agent present in the blood and known to support HSCs egress from the bone marrow to the blood, was also unaffected by sleep deprivation (Fig 2h). These findings indicate that the overall motility potential of the cells was not affected by sleep deprivation, suggesting that the impaired localization of the HSCs to the bone marrow was mediated by specific change in the responsiveness to migration guiding cues, specifically SDF-1α.
Sleep deprivation decreases miR19b and increases SOCS3 mRNA in HSCs
Post-transcriptional changes within the HSCs derived from mice that were allowed to sleep or following sleep deprivation were analyzed in efforts to identify intracellular changes occurring in the HSCs. We were looking for changes that could explain the altered HSC in vitro migration to SDF-1α, in vivo homing following transplantation, and the overall reduced transplantation potential. MicroRNAs (miRs) are non-coding RNA molecules that repress transcriptional outputs in a sequence-dependent manner13. It was previously shown that sleep loss alters miR expression in the brain14. We profiled the expression of a panel of 376 highly conserved and well- known miRs. Data were normalized to U6 expression, which had similar CT values. Values for other controls, snoRNA-202 and snoRNA-135, were also comparable; therefore, differences in miR expression were not attributable to changes in controls. Of the 376 miRs in the array, 98 miRs were amplified (CT<35). Comparative analysis of expression levels between HSCs derived from sleep-deprived mice or mice that were allowed to sleep, revealed an overall down-regulation of miRs (Fig. 3a; public deposit at GEO; GSE48144), where 67% of the normally amplified miRs were down-regulated in HSCs from sleep-deprived mice, while 8% were further amplified in HSCs derived from sleep-deprived mice. Comparable levels were observed between the two groups for 24% of the tested miRs. Single TaqMan probes were used for specific miRs to validate the array (Supplementary. Fig 1). In the down-regulated set of miRs, miR-152 and miR-361 were downregulated by 8-fold in cells from sleep-deprived mice. However, these miRNAs were amplified with high CT values suggesting that they were not abundant in HSCs and we therefore did not focus on these miRNAs. Nevertheless, several members of the miR-17 family were downregulated. The miR-17 family includes miR-17, miR-18a, miR-19a, miR-20a, 19b-1, miR-92a-1, which are transcribed from the same polycistron and in the same genomic context (in mice it is chromosome 14). In both samples, miR-17, miR-19b and miR-92 were the most abundant miRNAs in the cluster (CT<25), and miR-18a/b were the least abundant (CT>35). miR-19b displayed the greatest fold change (four-fold decrease) in the cluster upon sleep deprivation and we therefore focused on that miRNA.
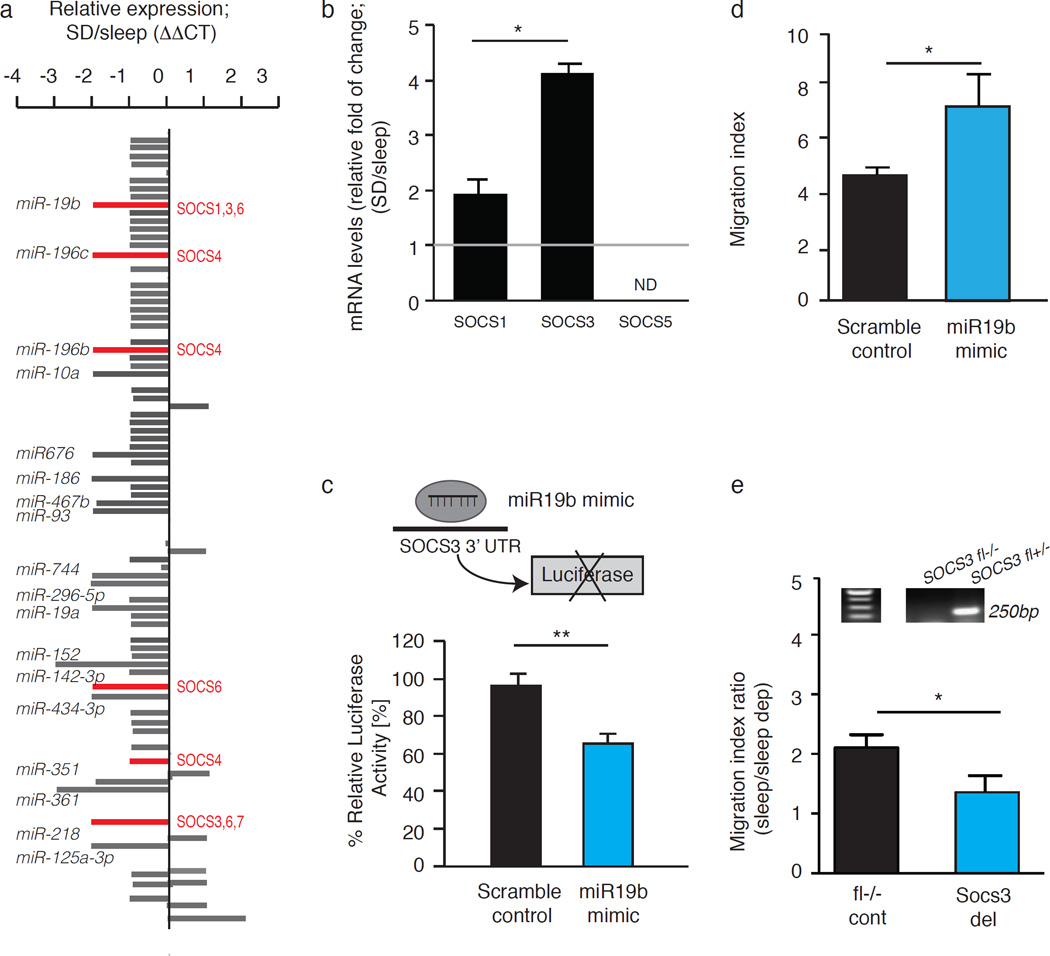
Figure 3. Sleep deprivation decreases the levels of miR19b and increases the levels of SOSC3 in HSCs.
(a) miRNA profiles of HSCs from sleep and sleep-deprived mice were analyzed by multiplex quantitative real-time PCR after HSC sorting by FACS. The ΔΔCt method was used, with U6, sno-202 and sno-135 as normal controls. MicroRNA levels underwent > 4-fold changes and potentially targeted SOCS genes are highlighted. (b) qPCR analysis of SOCS1, SOCS3 and SOCS5 levels. Results are presented as fold change normalized to GAPDH (ND=not detected; 3 independent repeats, n=4 mice per group; SOCS3: 4.13± 0.5-fold and SOCS1: 1.9± 0.8-fold). (c) A schematic presentation of the luciferase reporter system in 293T cells that were transfected with a microRNA mimic or scramble control and pMir Target construct containing the 3'UTR of SOCS3 and an RFP reporter. Luciferase activity was measured 48 hours after transfection and normalized to RFP and then to the values measured for vector alone (student’s t-test; t(4)=8.313; p=0.0011; mean±s.e.m). (d) Migration of HSCs, transfected with 50nM miR-19b mimic or scramble control, towards 50ng/mL SDF-1α. Migration was compared to baseline migration in the absence of chemoattractant (migration index). (e) Mx-1 cre mice were crossed with SOCS3-floxed mice. Mx-1 cre mice express Cre recombinase under the control of an inducible Mx1 promoter, active after induction with polyinosinic-polycytidylic acid (pI:pC). When bred with SOCS3-floxed mice (carrying SOCS3 gene flanked by loxP recognition sites), the expression of Cre recombinase causes SOCS3 gene deletion. Upper panel: Genomic DNA was extracted from FACS-sorted KLS cells and analyzed using PCR. The PCR product obtained from the SOCS fl+ locus is 250 bp band indicates cre-mediated deletion of SOCS3 (SOCS3del). Lower panel: Migration of KLS cells isolated from sleep mice and from mice that were deprived of sleep for four hours. We compared two sets of mice, mice that express mx-1-cre and SOCS3- floxed gene and their littermates that do not express mx-1-cre. This allows controlling for potential effect of pI:pC injection required to induce cre expression. Migration towards 50 ng/mL SDF-1 α was determined for cells from sleep or sleep-deprived mice. The migration index was determined for each group and the ratio between the migration index of the control and the SOCS-3 deleted mice was calculated (mean±s.e.m).
The highly conserved SOCS3, a protein previously implicated in HSC migration15–17 was predicted using Targetscan, to be a miR-19b target. Thus we compared the levels of SOCS genes expression in our experimental groups reasoning that decreases in miR levels can result in the stabilization of otherwise degraded transcripts. A two-fold increase in SOCS1 and a four-fold increase in SOCS3 mRNA levels were observed in HSCs from sleep-deprived mice, relative to HSCs from mice that were allowed to sleep (Fig. 3b). We did not detect a significant difference in mRNA levels of another miR-19b target, Pten, and we therefore focused on SOCS genes and specifically analyzed the interaction of miR-19b with SOCS3.
To validate the interaction between miR-19b and SOCS3, we performed a standard luciferase reporter assay. We used 293T cells transfected with a reporter containing the 3’UTR of SOCS3 (Fig. 3c). In this test, if the manipulated miR (miR19b) binds the 3’UTR of the target gene (SOCS3) it reduces luciferase activity. A significant reduction in luciferase activity was measured among 293T cells transfected with a miR-19b mimic for 48 h, demonstrating that miR-19b interacts with the 3’UTR of SOCS3. Taken together, these findings indicate that sleep deprivation decreases miR-19b levels and up-regulates SOCS3 gene expression levels in HSCs.
To establish a functional connection between these changes in miR-19b and altered HSC migration capacity, we transfected KLS (c-kit+, Lin−, Sca+) cells with miR-19b mimic for 36 hours and tested their migration capacity towards SDF-1α. We used KLS cells because of the technical limitation of performing such manipulation on HSCs. In line with evidence obtained in other cells types18, miR-19b mimic increased HSC migration towards SDF-1α (Fig. 3d; Student’s t-test p=0.0195). This evidence indicates that the change in miR-19b levels can explain at least partially, the reduced migration capacity of HSCs following sleep deprivation.
The involvement of SOCS3 in HSCs migration has been manifested before15–17. Yet, to directly test whether the effects of sleep deprivation on SOCS3 levels, could affect the motility of these cells, we used a Cre recombinase under the control of the Mx-1 promoter to induce the deletion of SOCS3 gene in multiple cell types including bone marrow cells. We found that HSCs from mice that had reduced levels of SOCS3 expression did not demonstrate a significant sleep deprivation-associated impairment of migratory capacity when compared to controls (Fig 3e; Student’s t-test p=0.017). Although there are some limitations to this experimental paradigm, these findings suggest that the effect of sleep deprivation on HSC migration is mediated at least in part via SOCS3.
Growth hormone affects miR-19b expression and cell migration
The effects of sleep deprivation on the HSCs raise questions regarding the identity of the signal from the sleeping brain to the hematopoietic system. One potential candidate is growth hormone (GH), which is secreted during sleep. GH has been shown to regulate lymphocyte migration19 and increase mobilization of HSCs in humans20. GH receptors (GHR) are expressed in all murine hematopoietic tissues, especially in the bone marrow21. In line with previous evidence22, we found that the sleep-dependent increase in GH levels was attenuated in sleep-deprived mice (Fig. 4a; One-way ANOVA F(2,22)=8.44; p=0.0019; Tukey post-hoc comparisons p<0.01 for the light onset compared to the group of mice allowed to sleep; p<0.05 for mice allowed to sleep compared to sleep deprived mice). To determine whether GH can mediate the observed changes in miRNA levels, HSCs were incubated with growth hormone for 30 min and RNA was extracted immediately thereafter. We focused on the expression of miRs encoded by the miR-17 cluster, which includes miR-19b. GH treatment led to elevation in the levels of miR-19b and other members of the cluster, suggesting that growth hormone may regulate the expression of this polycistron during sleep (Fig 4b). Moreover, GH treatment had a direct, dose-dependent (bell-shaped response) effect on the migration capacity of HSCs (Fig 4c). Low doses of GH increased migration, whereas high doses of GH had the opposite effect, by reducing HSCs migration. These findings show that GH levels regulated by sleep alters miR-19b concentrations in HSCs and affects HSC migration, representing a possible mechanism mediating the effects of sleep deprivation on HSCs.
Figure 4. Growth hormone alters the expression of miR-19b and cell migration.
(a) Mice were either sacrificed with light onset, after 1.5h or 3h of sleep deprivation or after 1.5h or 3h of sleep, and plasma GH levels were determined by ELISA (one-way ANOVA followed by Tukey’s multiple comparisons test; mean±s.e.m). (b) KLS cells were treated with GH (100ng/mL- dose was determined based on previous studies33,34) for 30 min and RNA was extracted using Trizol for analysis of miRs levels. All values were statistically significant within each group (technical replicas) and are reported as fold of change relative to control that was used as a reference (mean±s.e.m). (c) Cells from control mice were pre-incubated for 30 min with GH (0, 5, 25, 50 or 100ng/mL), washed and the migration of KLS to increasing concentrations of SDF-1α was determined on transwell migration plates. Statistical analyses was analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test; ***p<0.01; n=4 mice per group.
Sleep rebound restores the transplantation potential of HSCs
Sleep is intensified after sleep deprivation and can often reverse processes impaired by sleep deprivation23. We tested whether recovery sleep can reverse the impaired transplantation potential of HSCs derived from sleep-deprived mice. Lethally-irradiated congenic mice were transplanted with HSCs derived from (i) mice that were sleep-deprived for four hours and allowed to recover their sleep for two hours (ii) mice that were allowed to sleep for six hours or (iii) mice that were sleep-deprived for six hours. Bone marrow chimerism of recipient mice was analyzed at 16 weeks post-transplantation (Fig 5). Two hours of recovery sleep reversed the impaired transplantation capacity induced by sleep deprivation. Thus, HSCs from mice that were allowed to sleep and in the sleep rebound group were equally potent in their transplantation potential. These findings suggest that controlling sleep can affect hematopoietic transplantation outcomes.
Figure 5. Sleep rebound restores the reconstitution potential in lethally irradiated hosts.
Chimerism in the bone marrow of mice transplanted with HSC derived from mice that were allowed to recover their sleep for 2h, mice deprived of sleep for 6h and mice allowed to sleep for 6h. Analysis of was performed at 16 weeks after transplantation (F(2,15)=192.9; p<0.0001). *p<0.05; **p<0.01; ***p<0.001. Mean ± s.e.m; n=6 mice per group.
Discussion
This study demonstrates that sleep deprivation impairs the homing capacity and the reconstitution potential of transplanted HSCs. We show that this is a temporary effect, as secondary transplantation was not affected by the sleep deprivation. We attribute these effects of sleep deprivation on the transplantation potential to the reduced homing of the HSC to their niche in the bone marrow as manifested in the in vivo homing assay. We further support the negative effects of sleep deprivation on HSCs migration by demonstrating that the ex-vivo migration capacity towards SDF-1α of the cells from sleep-deprived mice was reduced. We show that this was not due to changes in the receptor for SDF-1α, CXCR-4, or changes in the levels of other integrins such as VLA-4, but rather due to a change in the intracellular potential of the cells to respond to migration guiding cues. We specifically show that cells derived from sleep-deprived mice have lower levels of miR-19b and increased SOCS3 levels, which was previously shown to affect migration to SDF-1α and stem cells trafficking15–17. We then used a genetic (Mx-1 cre) manipulation to reduce SOCS3 levels in HSCs. We demonstrate that in the absence of SOCS3, sleep deprivation had no significant effect on the migration potential of HSCs, further supporting the role of SOCS3 in attenuating HSC migration following sleep-deprivation. Finally, we show that sleep rebound reverses the effects of sleep deprivation on the transplantation potential of HSCs. Taken together our findings indicate that even short-term sleep deprivation of a donor can affect the success of hematopoietic cell transplantation.
These findings are especially relevant, as sleep-deprivation is common even in healthy populations. Moreover, bone marrow donors may experience acute sleep deprivation, partially due to the anticipation of the donation procedure, and partially due to the effect of pharmacological treatments. For example, it was shown that granulocyte colony-stimulating factor (G-CSF) suppresses sleep intensity manifested by reduced amount of slow-wave sleep and a reduction in the electroencephalographic delta power24. Yet, in this study we did not examine the effects on stem cell mobilization procedures. Nor did we analyze the potential effects of sleep deprivation on the routine circulation of HSCs between the bone marrow and peripheral blood.
As sleep is a complex phenomenon, it is difficult to determine the factors that mediate the effects of sleep on the reconstitution potential of the HSC. Sleep affects almost every physiological and behavioral system (metabolism, heart rate, endocrine system, immune system, etc2,7,25–27). It is therefore unlikely that a single factor mediates all the effects of sleep on the hematopoietic system. Nevertheless, we propose GH as one possible mediator, particularly because its levels are regulated by sleep and, as we show, it can alter the levels of the miR-17 family, including miR-19b, in HSCs. These findings fall in line with previous studies indicating that GH regulates lymphocyte migration19 as well as the mRNA expression and kinetics of suppressors of cytokine signaling (SOCS3) in several cell types28,29. Accordingly, GH is used to increase mobilization of HSCs in humans20 and can be potentially applied to overcome the adverse effects of sleep-deprivation on human hematopoietic cell transplantation efficacy.
It is less likely that circadian mediators are responsible for the effects described in this study, as all experiments were performed at the same circadian phase and the sleep deprivation was for only four hours. Moreover, CXCR4/SDF1α levels, which have been previously implicated to mediate some of the circadian effects on HSCs5–7, were not affected in our experiments. We suggest that under normal conditions, both sleep and circadian factors affect HSC trafficking; circadian factors regulate SDF-1α levels30, while sleep controls intracellular SOCS levels. Long-term sleep deprivation (several days), or deprivation of specific sleep stages, has been shown to have additional, substantial effects on the hematopoietic system4. For example, a decrease in total cellularity of the bone marrow and peripheral blood4, along with a decrease in the absolute number of hematopoietic stem/progenitor cells and colony-forming units, have been previously observed in mice that were deprived of REM sleep for 72 hours4.
The presented work demonstrated that sleep deprivation increases SOCS3 levels in HSCs. SOCS genes regulate the JAK/STAT pathway, which regulates the functions of various cytokines, including those used in mobilization and stimulation of the hematopoietic and immune systems, such as G-CSF, erythropoietin, thrombopoietin, interferons, and numerous interleukins31. Sleep-deprivation results in lower levels of miR19b, stabilizing SOCS3 mRNA. Thus, we propose that a reduction in responsiveness to multiple cytokines, induced by an increase in SOCS levels, reduces HSC migration capacity and their homing from the blood to the bone marrow after transplantation. It is also possible that sleep deprivation alters additional aspects of the HSCs interaction with their bone marrow niche (e.g the multilineage differentiation of the grafted cells). Thus, the specific molecular mechanism and the signals mediating the effects of sleep on the HSCs will require further detailed investigation.
The current study provides evidence that sleep deprivation impairs hematopoietic cell transplantation potential. The potential implication of sleep deprivation of the donor may be crucial for the success of the fragile transplantation procedure. Our findings that sleep rebound can reverse the effects of sleep deprivation suggest that simple behavioral procedures can improve the success of bone marrow transplantation.
Materials and Methods
Animals
All procedures were approved by the Stanford University Institutional Animal Care and Use Committee and were in compliance with the Guide for the Care and Use of Laboratory Animals. The donor mice were 8–12-week-old C57BL/6-CD45.1+ or green fluorescence protein (GFP)-transgenic C57BL/6-CD45.1+ mice. The recipient mice were 8–12-week-old C57BL/6-CD45.2 male mice. Animals were derived and maintained at Stanford University's Research Animal Facility or obtained from Jackson Laboratories. Environmental conditions were maintained at 24 ± 1°C, humidity 40–60%, and light cycle 12h/12h. Food and water were provided ad libitum. Animals were euthanized using CO2. All groups were treated similarly to avoid potential difference in stress levels.
Polysomnographic recording and data analysis
Surgery
A custom-made electroencephalogram (EEG) and electromyogram (EMG) implant was surgically implanted, under intraperitoneal ketamine/xylazine anesthesia (80 and 16 mg/kg, respectively), using a small animal stereotaxic frame (David Kopf Instruments, CA, USA). EEG signals were recorded from electrodes placed over the frontal (AP, −2 mm; ML, ±1 mm) and temporal (AP, 3 mm; ML, 2.5 mm) cortices. EMG signals were recorded from two electrodes inserted in the neck musculature. The implants were affixed to the skull with C&B Metabond (Parkell; Edgewood, NY) and dental acrylic. After the surgical procedure, animals were allowed to recover for at least 14 days. They were then acclimated to a flexible EEG/EMG connection cable for 7 days, within individual recording chambers and habituated daily to handling.
Data acquisition
EEG and EMG signals, detected by the surgically implanted electrodes, were collected using commercial hardware (Embla; Broomfield, CO), digitized at 256 Hz and visualized using sleep recording software Somnologica-3 (Medcare, Reykjavik, Iceland).
Scoring
Sleep was scored using sleep analysis software (SleepSign for Animal; Kissei Comtec America). All scoring was performed manually, based on the visual signature of the EEG and EMG waveforms in 4 sec epochs. Two independent investigators verified scoring criteria as follows: waking was defined as a predominance of fast, desynchronized waves and a high-EMG amplitude. Non-rapid eye movement (NREM) was defined as synchronized, high-amplitude, low-frequency (0.4–4 Hz) EEG and highly reduced EMG activity compared with wakefulness, with no phasic bursts. REM sleep was defined as that with a pronounced theta rhythm (4–9 Hz) and a flat EMG signal (muscle atonia).
Sleep deprivation
After baseline sleep recording, mice were sleep-deprived for the duration indicated in each experiment (4 or 6h), starting at light onset (ZT0). Mice were sleep-deprived by gentle handling, to avoid stress responses.
Analysis of corticosterone levels
Mice were sacrificed immediately at the end of four hours of sleep or four hours of sleep deprivation. Serum corticosterone levels were determined using a standard ELISA assay for corticosterone (Millipore; Billerica, MA), as per the standard procedure recommended by the manufacture.
Flow cytometry
Mice were euthanized, using CO2, and bone marrow was harvested into PBS containing 2% fetal calf serum (FCS). Lineage staining was performed with fluorochrome-conjugated monoclonal antibodies (eBioscience) against Ter119 (15-5921), Gr1 (15-5931), Mac1 (15-0112), B220 (15-0452), CD3 (15-0031), CD4 (15-0041), and CD8 (15-0081) (all in 1:200 dilution). KLS (c-Kit+, Lin−, Sca1+) subset delineation was performed with antibodies against c-Kit (47-1171; 1:100), and Sca-1 (17-5981;eBioscience; 1:100). KLS proportions were evaluated in blood and bone marrow. HSCs were determined using antibodies against CD150 (25-1502; 1:100) and CD34 (11-0341; eBioscience; 1:25). VLA4 (103705, Biolegend; 1:100). Cells were analysed with the LSR II (BD Biosciences) or FACSAria II (BD Biosciences).
HSC transplantation
Bone marrow cells samples were enriched for c-kit-positive cells prior to staining, using anti-CD117-conjugated magnetic beads (AutoMACS, Miltenyi, Germany), according to the manufacturer’s instructions. Cells were then stained with the antibodies listed above. Cells were sorted with a FACSAria II (BD Biosciences). A total of 300 sorted putative HSCs purified from sleep controls or sleep-deprived mice and 106 helper bone marrow-derived mononucleated cells (to support the survival of the mice during recovery) were transplanted into lethally irradiated (9.5 Gy γ-irradiation delivered in a split dose), 8–12-week-old congenic recipient mice via intravenous injection into the retro-orbital sinus. Peripheral blood was drawn from the tail vein of recipient mice at 8 weeks posttransplantation, and red blood cells were sedimented with 2% dextran and lysed with ammonium-chloridepotassium (ACK) lysing buffer (150 mM NH4Cl, 1mM KHCO3, and 0.1 mM EDTA) for 5 min on ice. Recipient mice were euthanized at 16 weeks post-transplantation and whole blood was obtained by collecting the perfusate from the heart and bone marrow was harvested by crushing bones. Donor-derived hematopoietic cells were identified by flow cytometry, using antibodies against CD45.1 (eBioscience) and host-derived hematopoietic cells were identified using anti-CD45.2 antibodies (eBioscience). There was no difference in the relative distribution of the donor cells between the blood and bone marrow at 16 weeks. There was no difference between the two groups in mouse survival.
Secondary transplantation
To demonstrate the self-renewing capacity of the donor-derived long-term repopulating cells, 2.5 × 106 BM cells from primary recipients (who received HSCs from sleep or sleep–deprived mice) were transplanted into lethally irradiated CD45.2+ secondary recipients and donor-derived reconstitution was assessed. Of note, secondary transplant donors were not sleep-deprived prior to transplantation.
Homing experiment
2000 FACS-purified HSCs derived from GFP-transgenic C57BL/6− CD45.1+ mice were intravenously transplanted into congenic C57BL/6-CD45.2 mice, whose bones were collected 12 hours thereafter.
Immunohistochemistry
Bones were incubated in 4% PFA for 24 h, washed in PBS and transferred to 400mM EDTA for 20 days, at 4°C, protected from light. The decalcified bones were cryoprotected overnight, at 4°C, in 30% sucrose. Bones were sliced to a thickness of 10µm and the number of cells expressing endogenous GFP was determined. For verification, bones were stained with anti-GFP antibody (1:1000; Abcam; Cambridge, MA; ab13970).
Binding of FITC-conjugated LDV
The activation of integrin-α4β1 was determined by α4β1 integrin (VLA-4) coupled to Fluorescein isothiocyanate (FITC) (LDV-FITC) binding. Cells derived from BM of sleep-deprived mice and mice that were allowed to sleep were incubated for 30 min at 37°C with 0.5 or 10 nM LDV-FITC and analyzed with flow cytometry.
Mx-1 and SOCS3 deletion experiment
Mx-1 male 8–10 weeks old mice mice were bred with SOCS-3 floxed mice. Cre-negative mice were used as a control for potential effects of the breeding background and the effects of intraperitoneal injections of 250 µg poly I-C (Invitrogen), which was given to induce Mx-1 mediated deletion. Poly I-C was injected 3 times at 24 h intervals.
microRNA profiling
Total RNA was isolated from FACS-purified HSCs, using TRIzol reagent (Invitrogen), and reverse transcribed using MegaPlex RT primer pool A (Applied Biosystems). Profiling of miRNAs expression was performed using the TaqMan Array Rodent Card A v2.0 (Applied Biosystems, CA, USA). The 384-well rodent array pool A plate contains four wells of U6 and snoRNA as internal controls, 335 individual highly conserved mouse miRNAs in each well, and 45 wells of two other snoRNAs (sno-202, sno-135) and miRNAs of rat and other species. Given limited amounts of RNA (<50ng) that can be recovered from sorted HSCs, RNA was pre-amplified using TaqMan PreAmp Master Mix and PreAmp Primers (2×), as per the manufacture's protocol.
Quantitative PCR
cDNA was synthesized with SuperScript® II Reverse Transcriptase (Invitrogen), using an anchored oligo dT primer. The samples were amplified, in duplicates, by qPCR, using TaqMan gene expression assays (Applied Biosystems) on an ABI 7900 HT system. Values were normalized to GAPDH. The following primers were used: Ghr Mm00439093; SOCS5 Mm01232423; SOCS1 Mm00782550; SOCS3 Mm01249143; hsa-miR-19a 000395; hsa-miR19b 000396 (all from Life Technologies)
In vitro transmigration
For chemotaxis assays, isolated KLS cells were suspended in migration medium comprised of RPMI-1640 medium, supplemented with 0.5% BSA, and then seeded in 8 µm pore transwell plates (Corning-Costar Corp). Murine SDF-1α (50, 100, or 200ng/mL; Sigma-Aldrich) or S1P (10 or 100nM; Sigma-Aldrich) was added to the lower chamber (specific concentration is indicated for each figure). KLS cells (1×104 or 5×104 cells) were placed in the upper well and incubated at 37°C for 4 hours and the number of cells in the lower wells was determined by flow cytometry. The migration index was calculated as the fold of increase in migration to the chemoattractant relative to baseline migration (without chemoattractant). Levels of SDF-1α and S1P were selected based on previous reports32,33.
Luciferase Assay
293T cells (ATCC) were transfected with the 3'UTR SOCS-3 luciferase/RFP construct (Origene; SC215826), miR-19 mimics and scrambled controls (based on cel-miR-67, mature sequence: UCACAACCUCCUAGAAAGAGUAGA; Dharmacon), using DharmoFECT 1. Cells were analyzed 48hrs later, using the Luciferase Assay kit (Promega). Transfection efficiency was normalized to RFP signals.
Statistical analysis
Significance levels of the data were determined using Prism5 (GraphPad Software, La Jolla, CA). Experiments were analyzed by two-tailed Student’s t-test or by one-way or two-way analysis of variance (ANOVA), as indicated for each experiment. ANOVA was followed by a post-hoc test. Alpha was set at 0.05.
Supplementary Material
Acknowledgments
We would like to thank Samantha Wong, Bayarsaikhan Chuluun, Catherine Carswell-Crumpton, Ada Diane Eban-Rothschild, Charles Chang, Hilla Azulay-Debby, Tamar Ben-Shannan, Nathanael Green and Chris Wong for their help. AR is supported by EMBO and the Rothschild Fellowships. WWP is supported by the Stanford Medical Scientist Training Program. II is supported by CIRM scholar supported by CIRM grant TG2-01159. ILW is supported for studies in this manuscript by Ludwig Institute and NIH grants -U01-HL099999-03, R01 CA86065 and R01 HL058770. LdL is supported by grants from NIMH and the Klarman Family Foundation.
Footnotes
Author contributions:
AR and WWP conceived the project, designed and performed the experiments, analyzed the data and wrote the manuscript. II designed, performed and analyzed the molecular experiments and wrote the manuscript. DC coordinated and helped designing the sleep recordings and analysis. PB helped with the experiments and their design. HCH, ILW and LDL helped design the experiments and edited the manuscript.
Competing financial interests
None of the authors has competing interests.
Accession codes
The miRNA microarray data have been deposited in the GEO database with accession code GSE48144.
References
- 1.Besedovsky L, Lange T, Born J. Sleep and immune function. Pflugers Arch. 2012;463:121–137. doi: 10.1007/s00424-011-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krueger JM, Majde JA, Rector DM. Cytokines in immune function and sleep regulation. Handb. Clin. Neurol. 2011;98:229–240. doi: 10.1016/B978-0-444-52006-7.00015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant PA, Trinder J, Curtis N. Sick and tired: Does sleep have a vital role in the immune system? Nat. Rev. Immunol. 2004;4:457–467. doi: 10.1038/nri1369. [DOI] [PubMed] [Google Scholar]
- 4.Guariniello LD, Vicari P, Lee KS, de Oliveira AC, Tufik S. Bone marrow and peripheral white blood cells number is affected by sleep deprivation in a murine experimental model. J. Cell. Physiol. 2012;227:361–366. doi: 10.1002/jcp.22743. [DOI] [PubMed] [Google Scholar]
- 5.Tsinkalovsky O, et al. Circadian expression of clock genes in purified hematopoietic stem cells is developmentally regulated in mouse bone marrow. Exp. Hematol. 2006;34:1249–1261. doi: 10.1016/j.exphem.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel A, Kalinkovich A, Shivtiel S, Kollet O, Lapidot T. Stem cell regulation via dynamic interactions of the nervous and immune systems with the microenvironment. Cell Stem Cell. 2008;3:484–492. doi: 10.1016/j.stem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 8.Longordo F, Fan J, Steimer T, Kopp C, Lüthi A. Do mice habituate to ‘gentle handling?’ A comparison of resting behavior, corticosterone levels and synaptic function in handled and undisturbed C57BL/6J mice. Sleep. 2011;34:679–681. doi: 10.1093/sleep/34.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 10.Suárez-Álvarez B, López-Vázquez A, López-Larrea C. Mobilization and homing of hematopoietic stem cells. Adv. Exp. Med. Biol. 2012;741:152–170. doi: 10.1007/978-1-4614-2098-9_11. [DOI] [PubMed] [Google Scholar]
- 11.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 12.Haribabu B, et al. Regulation of human chemokine receptors CXCR4. Role of phosphorylation in desensitization and internalization. J. Biol. Chem. 1997;272:28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- 13.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci. Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kershaw NJ, et al. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013;20:469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 2009;30:592–602. doi: 10.1016/j.it.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pello OM, et al. SOCS up-regulation mobilizes autologous stem cells through CXCR4 blockade. Blood. 2006;108:3928–3937. doi: 10.1182/blood-2006-02-006353. [DOI] [PubMed] [Google Scholar]
- 18.Wu Q, et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savino W, Smaniotto S, Mendes-da-Cruz DA, Dardenne M. Growth hormone modulates migration of thymocytes and peripheral T cells. AnnNY. Acad. Sci. 2012;1261:49–54. doi: 10.1111/j.1749-6632.2012.06637.x. [DOI] [PubMed] [Google Scholar]
- 20.Carlo-Stella C, et al. Use of recombinant human growth hormone (rhGH) plus recombinant human granulocyte colony-stimulating factor (rhG-CSF) for the mobilization and collection of CD34+ cells in poor mobilizers. Blood. 2004;103:3287–3295. doi: 10.1182/blood-2003-07-2428. [DOI] [PubMed] [Google Scholar]
- 21.Dardenne M, Mello-Coelho V, Gagnerault MC, Postel-Vinay MC. Growth hormone receptors and immunocompetent cells. AnnNY. Acad. Sci. 1998;840:510–517. doi: 10.1111/j.1749-6632.1998.tb09589.x. [DOI] [PubMed] [Google Scholar]
- 22.Kimura F, Tsai CW. Ultradian rhythm of growth hormone secretion and sleep in the adult male rat. J. Physiol. 1984;353:305–315. doi: 10.1113/jphysiol.1984.sp015337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hipólide DC, et al. Paradoxical sleep deprivation and sleep recovery: effects on the hypothalamic-pituitary-adrenal axis activity, energy balance and body composition of rats. J. Neuroendocrinol. 2006;18:231–238. doi: 10.1111/j.1365-2826.2006.01412.x. [DOI] [PubMed] [Google Scholar]
- 24.Schuld A, et al. Effects of granulocyte colony-stimulating factor on night sleep in humans. Am. J. Physiol. 1999;276:R1149–R1155. doi: 10.1152/ajpregu.1999.276.4.R1149. [DOI] [PubMed] [Google Scholar]
- 25.Lange T, Dimitrov S, Born J. Effects of sleep and circadian rhythm on the human immune system. AnnNY. Acad. Sci. 2010;1193:48–59. doi: 10.1111/j.1749-6632.2009.05300.x. [DOI] [PubMed] [Google Scholar]
- 26.Lapidot T, Kollet O. The brain-bone-blood triad: traffic lights for stem-cell homing and mobilization. Hematology Am. Soc. Hematol. Educ. Program. 2010;2010:1–6. doi: 10.1182/asheducation-2010.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Méndez-Ferrer S, Battista M, Frenette PS. Cooperation of beta(2)- and beta(3)-adrenergic receptors in hematopoietic progenitor cell mobilization. AnnNY. Acad. Sci. 2010;1192:139–144. doi: 10.1111/j.1749-6632.2010.05390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tollet-Egnell P, Flores-Morales A, Stavréus-Evers A, Sahlin L, Norstedt G. Growth hormone regulation of SOCS-2, SOCS-3, and CIS messenger ribonucleic acid expression in the rat. Endocrinology. 1999;140:3693–3704. doi: 10.1210/endo.140.8.6878. [DOI] [PubMed] [Google Scholar]
- 29.Kasagi Y, Tokita R, Nakata T, Imaki T, Minami S. Human growth hormone induces SOCS3 and CIS mRNA increase in the hypothalamic neurons of hypophysectomized rats. Endocr. J. 2004;51:145–154. doi: 10.1507/endocrj.51.145. [DOI] [PubMed] [Google Scholar]
- 30.Lucas D, Battista M, Shi PA, Isola L, Frenette PS. Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell. 2008;3:364–366. doi: 10.1016/j.stem.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward AC, Touw I, Yoshimura A. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood. 2000;95:19–29. [PubMed] [Google Scholar]
- 32.Tao W, Hangoc G, Cooper S, Broxmeyer HE. SDF-1alpha/CXCL12 enhances retroviral-mediated gene transfer into immature subsets of human and murine hematopoietic progenitor cells. Gene Ther. 2004;11:61–69. doi: 10.1038/sj.gt.3302127. [DOI] [PubMed] [Google Scholar]
- 33.Soriano SF, et al. Functional inactivation of CXC chemokine receptor 4-mediated responses through SOCS3 up-regulation. J. Exp. Med. 2002;196:311–321. doi: 10.1084/jem.20012041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taub DD, et al. Growth hormone promotes human T cell adhesion and migration to both human and murine matrix proteins in vitro and directly promotes xenogeneic engraftment. J. Clin. Invest. 1994;94:293–300. doi: 10.1172/JCI117320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.