Abstract
BACKGROUND AND OBJECTIVES:
Widespread newborn screening on a point-of-care basis could prevent bilirubin neurotoxicity in newborns with glucose-6-phosphate dehydrogenase (G6PD) deficiency. We evaluated a quantitative G6PD assay on a digital microfluidic platform by comparing its performance with standard clinical methods.
METHODS:
G6PD activity was measured quantitatively by using digital microfluidic fluorescence and the gold standard fluorescence biochemical test on a convenience sample of 98 discarded blood samples. Twenty-four samples were designated as G6PD deficient.
RESULTS:
Mean ± SD G6PD activity for normal samples using the digital microfluidic method and the standard method, respectively, was 9.7 ± 2.8 and 11.1 ± 3.0 U/g hemoglobin (Hb), respectively; for G6PD-deficient samples, it was 0.8 ± 0.7 and 1.4 ± 0.9 U/g Hb. Bland-Altman analysis determined a mean difference of –0.96 ± 1.8 U/g Hb between the digital microfluidic fluorescence results and the standard biochemical test results. The lower and upper limits for the digital microfluidic platform were 4.5 to 19.5 U/g Hb for normal samples and 0.2 to 3.7 U/g Hb for G6PD-deficient samples. The lower and upper limits for the Stanford method were 5.5 to 20.7 U/g Hb for normal samples and 0.1 to 2.8 U/g Hb for G6PD-deficient samples. The measured activity discriminated between G6PD-deficient samples and normal samples with no overlap.
CONCLUSIONS:
Pending further validation, a digital microfluidics platform could be an accurate point-of-care screening tool for rapid newborn G6PD screening.
What’s Known on This Subject:
Glucose-6-phosphate dehydrogenase deficiency remains a global as well as a North American burden for extreme hyperbilirubinemia and kernicterus and is often unpredictable during the first few days after birth. Newborn screening for this enzyme deficiency is not universally available but debated.
What This Study Adds:
Point-of-care screening, using digital microfluidics, provides accurate, low blood volume, and affordable technology for rapid newborn glucose-6-phosphate dehydrogenase enzyme screening that could guide clinicians before infants’ discharge from well-child nurseries and meet existing American Academy of Pediatrics’ recommendations.
Neonatal jaundice is a commonly occurring condition, with some degree of jaundice being noticed in up to 80% of otherwise healthy, term and late-preterm newborns.1,2 Generally, the jaundice has a benign outcome when screened for and treated in a timely manner. An unpredictable number of newborns, particularly those with glucose-6-phosphate dehydrogenase (G6PD) deficiency, can adversely progress with rapidity to extreme hyperbilirubinemia with signs of acute bilirubin encephalopathy. Extreme neonatal hyperbilirubinemia is effectively treated by phototherapy, with exchange transfusion as a backup treatment modality. As a result, kernicterus and exchange transfusions are minimally encountered in industrialized countries with functional and intact health care systems. Conversely, delay in seeking health care, rapid onset of progressive hyperbilirubinemia, and delayed or lack of access to effective phototherapy and/ or exchange transfusion (such as have been reported from developing or low- and middle-income countries) can result in an infant developing the potentially tragic consequences of lifelong choreoathetotic cerebral palsy, otherwise known as kernicterus.3–5
G6PD deficiency is an X-linked condition encountered among hundreds of millions of individuals. Originally confined to its indigenous distribution, including Africa, the Mediterranean Basin, the Middle East, and extending through Asia to the Far East, immigration patterns, the slave trade and modern ease of travel have made it a condition that may be encountered virtually worldwide. G6PD deficiency is a condition not only of low- and middle-income countries4 but also of high-income, industrialized countries.6 In the newborn, G6PD deficiency is associated with a high incidence of neonatal hyperbilirubinemia and especially with sudden, exponential, and unpredictable increases in the total serum bilirubin level to extreme concentrations. G6PD deficiency appears high on lists of known etiologies of newborns with kernicterus or extreme hyperbilirubinemia.7–10 Screening for G6PD deficiency in the newborn period has been recommended by the Working Committee of the World Health Organization to pinpoint affected newborns at high risk for extreme hyperbilirubinemia; the goal is to prevent exposure of these infants to known triggers of hemolysis and to facilitate the approach to appropriate medical facilities should hyperbilirubinemia develop and before the onset of bilirubin neurotoxicity.
The present study tested and evaluated a digital microfluidics platform developed and programmed to measure G6PD activity from whole blood in a point-of-care setting, including all steps of analysis, from whole blood lysis, reagent preparation, sample processing, mixing, dilution, incubation, and fluorescence detection to waste handling; the objective was to determine how to best describe its potential as a screening device. Digital microfluidic technology enables all steps of analysis from sampling through waste handling on a disposable cartridge. Performance of immunoassays, enzyme assays, and DNA analyses by using small amounts of sample and reagents in a user-friendly, point-of-care device makes this technology potentially attractive to newborn screening in a global setting.11
Methods
Blood Samples
Discarded and deidentified whole blood samples (N = 98) were obtained under an institutional review board–approved protocol from newborn and older children (cared for at Lucile Packard Children’s Hospital) and adult patients (cared for at Stanford Hospital). The discarded samples were made available from random samples tested at the Stanford clinical laboratory, and the known G6PD-deficient samples were from the Hillview RBC Special Studies Laboratory. Gender of the sample sources was documented. Each sample was collected in EDTA-coated tubes and stored at 4°C until analysis. All the samples were quantitatively analyzed for G6PD activity within 72 hours of collection by using the standard diagnostic method at the Stanford Hillview laboratory. Within 24 hours of collection, the same samples were analyzed by using the digital microfluidic platform for G6PD activity at the Stevenson Laboratory at Stanford. Based on previous experiences, it is known that red blood cell G6PD activity is stable for >21 days in blood collected in EDTA and stored at 4°C.12 All of our samples were assayed well within these limits. The samples analyzed within 72 hours of collection reflected technician time available in the Hillview laboratory.
Description of Digital Microfluidic Analyzer and Sample-Input Cartridge
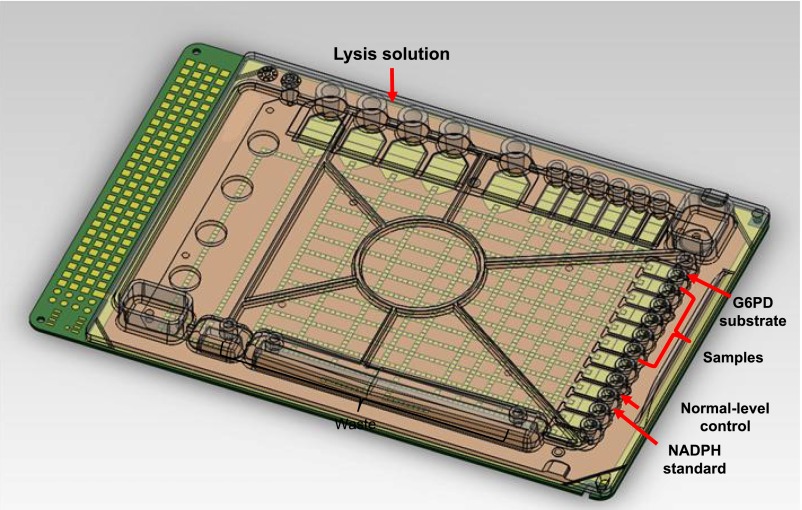
A disposable, single-use digital microfluidic cartridge for G6PD assays was designed (Fig 1) with input reservoirs for all assay reagents, calibrants, controls and samples, and a set of reservoirs for holding samples, reagents for G6PD, and waste located around the edges of the cartridge. Droplets are dispensed from the liquid in the reservoirs by appropriate electrode activation sequences.11 Before running the assays, silicone oil is used to fill the working space in the cartridge to prevent droplet evaporation and lubricate droplet motion.13
FIGURE 1.
Digital microfluidic cartridge layout. A single-use cartridge is loaded with reagents, calibrants, control samples, and up to 6 patient samples.
A digital microfluidic analyzer functions to maintain the cartridge temperature, supply voltage to drive the droplets, and perform optical detection of the assays. The temperature is maintained at 37°C over the entire cartridge and for the duration of the assay. The analyzer controls individual electrodes on the cartridge to perform all droplet operations, including dispensing, mixing, incubation, and sample disposal. The custom-built optical detector on the device features 2 channels: 1 channel for fluorescence readings at 360 nm excitation/460 nm emission and 1 channel for absorbance readings from 490 from 710 nm.
Reagents
G6PD controls were obtained from Trinity Biotech (Bray, Ireland). Tris buffer, bovine serum albumin, sodium carbonate, sodium bicarbonate, Tween-20, magnesium chloride, maleimide, β-nicotinamide adenine dinucleotide phosphate (NADP), D-glucose-6-phosphate, and β-nicotinamide adenine dinucleotide phosphate hydrate (NADPH) were obtained from Sigma-Aldrich Corporation (St Louis, MO). Molecular grade water was obtained from Fisher Scientific (Pittsburgh, PA), and 5 cSt silicone oil was obtained from Gelest, Inc (Morrisville, PA).
The G6PD assay consisted of lysis solution (1.0% Tween-20 in distilled water) for diluting the samples and lysing the red blood cells, substrate solution (2.0-mM D-glucose-6-phosphate, 2.6-mM NADP, 2.4-mM magnesium chloride, and 26-mM maleimide in 100-mM Tris buffer at pH7.8 and containing 0.05% Tween-20), and fluorescence standard (100-μM NADPH in 50% (v/v) Tris buffer and 50% (v/v) lysis solution). The controls (lyophilized blood hemolysate with specified levels of G6PD activity) were reconstituted in a 0.05% (w/v) Tween-20 solution. The substrate and standard were stored at −80°C, and the lysis solution and control samples were stored at 4°C. The cartridge was inserted into the analyzer deck as described previously,11 and reagents and samples were loaded onto the cartridge via a pipette. Briefly, for each assay, 5 μL of whole blood was pipetted directly into the cartridge. After loading the samples and reagents, all subsequent assay steps were performed by the analyzer. The protocols for performing the assays on the microfluidic cartridges are briefly described in the following section. Unless otherwise noted, all droplets were 300 nL in volume.
G6PD Fluorescence Assay on a Digital Microfluidic Cartridge
A digital microfluidic cartridge was loaded with 6 patient blood samples. One droplet of each of the 6 whole blood samples and 1 droplet of normal-level control were diluted at a ratio of 1:9 by mixing with a double-sized droplet of lysis solution, splitting off and discarding a double-sized droplet, and mixing the remaining 300-nL droplet with a second double-sized droplet of lysis solution. This sequence involves 2 consecutive 3-fold serial dilutions resulting in a 9-fold final dilution of the samples. The resulting 900-nL droplet was then further split into 2 fractions of 300 and 600 nL, respectively. The 600-nL droplet was routed to the detector, and the hemoglobin (Hb) concentration in the sample was determined by measuring absorbance at 540 nm. The 300-nL droplet was further diluted in lysis solution to a final ratio of 1:27 by mixing with a double-sized droplet of lysis solution and splitting off and discarding a double-sized droplet. The remaining droplet was then mixed with a droplet of substrate, and the G6PD in the samples converted NADP to NADPH. Fluorescence of the NADPH in the assays was measured at 360-nm excitation and 460-nm emission kinetically at 3 time points and was related directly to the G6PD activity via the rate of NADPH generation by using a linear calibration curve created by measuring the fluorescence of the NADPH standard on each cartridge. The reported activity was normalized by using the Hb concentration. It took 17 minutes to run all 6 samples simultaneously. This time can be considerably reduced, but turnaround time was not the primary objective of the study and therefore no effort was made to shorten it.
G6PD Quantitative Assay at the Stanford Red Cell Special Studies Laboratory
Collected blood was washed 3 times in isotonic NaCl and then filtered through a cellulose slurry to remove leukocytes and platelets. A 1:20 destromatized hemolysate was then prepared by suspending 0.1 mL of 50% filtered red blood cell suspension into 0.9 mL of a hemolyzing solution. The standard premade hemolyzing solution contained the following: 3.3 mL of 3-mM NADP, 0.05 mL of mercaptoethanol, 10 mL of 10% EDTA pH 7.4, and distilled water added to a final volume of 1 L. G6PD activity was measured according to a modification of the World Health Organization assay,12 whereby the rate of NADP reduction to NADPH is measured when hemolysate is added to a reaction mixture containing glucose-6-phosphate. Specifically, 50 μL of a 1:20 hemolysate was added to 1.0 mL of reaction mixture containing 100-mM Tris-HCl, pH 8.0, 10-mM magnesium chloride, and 0.3-mM NADP. The reaction was started by adding 100 μL of 10-mM glucose-6-phosphate. The rate of NADP reduction to NADPH was measured by calculating the increase in optical density at 340 nm at 25°C for 10 minutes. Because enzyme activity is dependent on temperature, a correction factor is necessary to adjust for these variations when the assays are run at different temperatures, such as in this study. In the case of G6PD, the corrected activity at 25°C compared with 37°C is 0.559.
Results
G6PD was measured on 98 whole blood samples from newborn infants 1 to 3 days old (n = 52), older children aged 4 days to 6 months (n = 17), and adult patients (n = 29) who served as controls. Of these 98 samples, 32 were from female patients, and 66 were from male patients. Five (n = 5) newborn male infants (1–3 days old) were identified as G6PD deficient. Six (n = 6) older children (aged 4 days–6 months) were identified as G6PD deficient; 5 of these 6 children were male. Thirteen of the adults were identified as G6PD deficient (12 male patients). Patients with documented enzymatic G6PD sufficiency, according to standard enzymatic assays, served as control subjects. Of this cohort, 24 samples from male subjects were designated G6PD deficient, whereas the remainder (control) had normal activity. The G6PD-deficient and normal samples were divided into 2 distinct subgroups with no overlap. The percentages of G6PD-deficient and normal samples should not be regarded as representative cohorts but only representative of a convenience population.
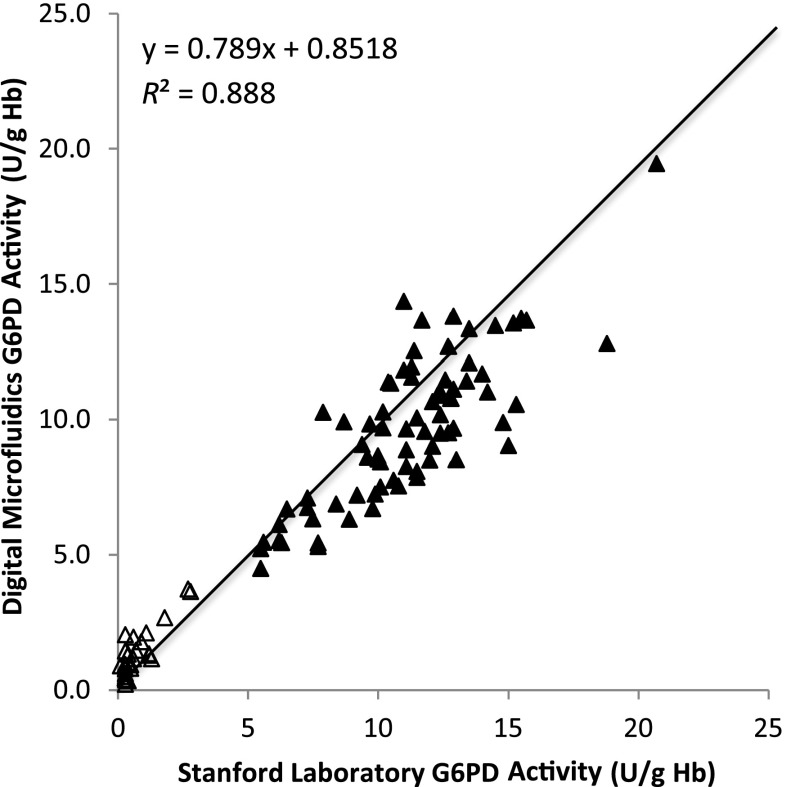
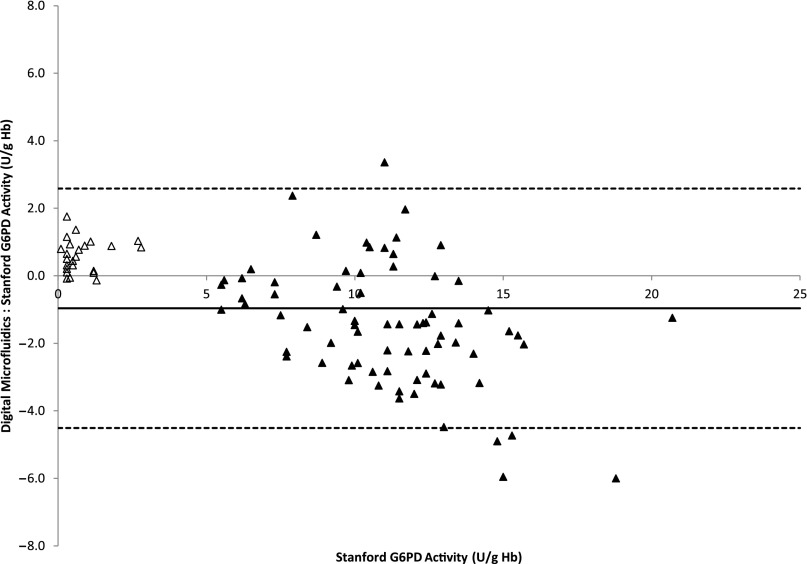
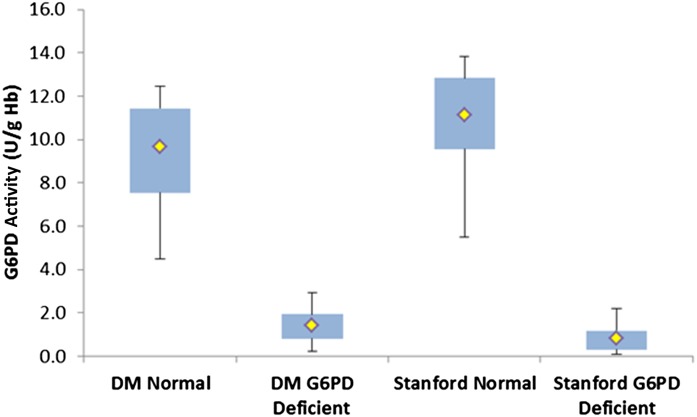
Comparison of G6PD activity obtained by using the digital microfluidic cartridge and the Stanford Diagnostic Laboratory method are presented in Fig 2. The solid line is not a fit but drawn to depict a 45° line with a slope of 1. Mean ± SD G6PD activity for normal samples using the digital microfluidic cartridge and the Stanford method were 9.7 ± 2.8 and 11.1 ± 3.0 U/g Hb, respectively; for the G6PD-deficient samples, it was 1.4 ± 0.9 and 0.8 ± 0.7 U/g Hb. The lower and upper limits for the digital microfluidic platform were 4.5 to 19.5 U/g Hb for normal samples and 0.2 to 3.7 U/g Hb for G6PD-deficient samples. The lower and upper limits for the Stanford method were 5.5 to 20.7 U/g Hb for normal samples and 0.1 to 2.8 U/g Hb for G6PD-deficient samples. Figure 3 illustrates that the mean difference between these 2 methods was –0.96 ± 3.5 U/g Hb. No data points were rejected for outliers. Figure 4 summarizes all the data in a box plot to highlight the discrimination of G6PD-deficient samples from normal samples by using digital microfluidics; this metric assumes more importance for newborn screening. The box represents the 75th and 25th percentiles, and the whiskers represent the minimum and maximum G6PD activity values. The yellow dot represents the mean activity in each case.
FIGURE 2.
Scatter plot of comparisons of G6PD activity obtained from the digital microfluidic method versus the standard laboratory method at Stanford University.
FIGURE 3.
Bland-Altman plot in which the solid line is the mean difference between the 2 methods, and the dotted lines represent the 95% confidence interval. Open triangles indicate deficient samples, and solid triangles indicate normal samples.
FIGURE 4.
Box plot of G6PD activity comparison. The box represents 75th and 25th percentiles, and the whiskers represent the minimum and maximum G6PD activity values. Open triangles indicate deficient samples, and solid triangles indicate normal samples; the yellow dot represents the mean activity in each case. Assays performed with digital microfluidics (DM) are compared with the assay performed at the Stanford University Laboratories.
Discussion
Although the World Health Organization has recommended G6PD screening to identify newborns at high risk in population groups with a male frequency >3% to 5%,14 implementation for regional or community-wide newborn screening has been inconsistent among nations and geographic locations.6,15 Screening on a national basis, however, has been limited only to a few countries. Among these countries, the impact of newborn screening regarding prevention of kernicterus seems encouraging as recently reviewed16 but has not been rigorously tested other than through historical data. One of the major reasons for nonimplementation of screening programs is the cost of the project for a universal quantitative enzyme assay. Although the reagents necessary are not expensive, manpower and administrative overhead remain the limiting factors. Mass screening with an affordable digital microfluidic platform offers a solution to this problem, as the device completely automates all the steps in performance of the assay. For this analysis, we specifically excluded female patients because there were only a few female subjects who were not genotyped. Male subjects were accurately categorized because they divide into 2 distinct groups: deficient and normal. Female subjects, on the other hand, would divide into 3 genetic categories: homozygous normal, heterozygotes, and homozygous deficient. Because of nonrandom X chromosome inactivation, heterozygotes would not divide into a distinct group, but there would be a continuum of G6PD activity, from low (deficient) through intermediate to normal.17 It has been difficult to identify heterozygotes by using biochemical methods, although values in the intermediate range suggest the possibility of heterozygosity. The present pilot study thus shows the feasibility of the device for practical clinical use in a point-of-care setting for quantitative enzyme assay.
There are several screening tests for G6PD deficiency, and most rely on qualitative assays for enzyme activity. The fluorescent spot test (Trinity Biotech PLC, Bray, Ireland) and the CareStartTM Rapid Diagnostic Tests (Access Bio Inc, Somerset, NJ) are the most frequently used qualitative screening tests. The enzyme activity cutoff point is low (2.1 and 2.7 U/g Hb, respectively), and only those patients with severe deficiency would be detected. Almost all female heterozygotes would be missed and reported as “normal.” In a study of African-American neonates, G6PD findings in the “deficient group” were 2.7 ± 1.1 U/g Hb (range: 0.4–6.6 U/g Hb). A considerable number of infants would not have been identified had only a qualitative test been used.18 In the event a qualitative test is used, confirmatory testing may then be informative. The test we used is superior in that we performed a quantitative enzyme assay. This method allows individuals with levels that are higher, but still in the G6PD-deficient range, to be detected. In addition, in those with intermediate G6PD levels, heterozygosity can be suspected, although the only definitive test for heterozygosity is by using polymerase chain reaction analysis.
One of the technical limitations of the study is that although the assay was run at 37°C on the digital microfluidic cartridge, the conventional assay is performed at room temperature on most bench studies, including the Stanford laboratory. Temperature difference is known to affect enzyme activity. Therefore, all the activity values obtained from the digital microfluidic platform were multiplied by 0.559, the factor required to normalize the activity measured at 37°C to that measured at room temperature.12 A positive correlation was found between the 2 platforms. Clearly, both the digital microfluidic method as well as the standard laboratory technique can discriminate G6PD-deficient samples from normal samples, as has been shown in previous studies of male neonatal populations.19,20 Although there were some differences in enzyme activity results between the 2 methods, these discrepancies were minimal, of no clinical significance, and did not affect the classification of samples tested in the G6PD-deficient or normal groups.
Preliminary method comparison results reported for determining G6PD activity highlight the potential for digital microfluidics to be an effective G6PD screening tool that enables fully automated and user-friendly screening for causes leading to neonatal hyperbilirubinemia. The only manual steps are loading of sample and reagents; all other operations, including lysis, reagent preparation, sample processing, incubation, and waste disposal, were completely automated. Eventually, we propose to test every newborn for bilirubin and G6PD deficiency on the same cartridge. In a hospital setting, we expect that multiple specimens can be batched due to the higher daily birth rate. In other settings with lower number of births, single-sample cartridges can be used. The digital microfluidic system can be used to control both a single-sample cartridge and a multiple-sample cartridge because, fundamentally in digital microfluidics, individual droplets are independently controlled, allowing unlimited scalability of number of droplet operations; thus, cartridges can be used for single-sample or multisample testing with the same software-controlled droplet operations.21 The multiplexed platform can also provide comprehensive data on neonatal jaundice through additional point-of-care screening for total serum bilirubin (results not presented here). In addition, the device has the potential to include analysis of the UGT1A1 (TA)n promoter polymorphism (UGT1A1*28) associated with Gilbert syndrome and hyperbilirubinemia in G6PD-deficient neonates.22 The digital microfluidic platform has also been shown to multiplex enzymatic assays for newborn screening for lysosomal storage disorders in public health laboratories using 40 samples on a slightly different cartridge.23,24 The use of digital microfluidics for G6PD deficiency screening has potential as a point-of-care screening tool even in the field setting because the user is only required to load samples and reagents while all other steps are performed by the cartridge with no further need for human interaction.21
Conclusions
This study highlights the potential for digital microfluidics to be an effective point-of-care screening tool for G6PD deficiency, suitable even in resource-limited settings. Addition of comprehensive bilirubin testing and screening for UGT1A1 gene polymorphisms associated with neonatal hyperbilirubinemia could further enhance the potential value of this technology in pinpointing neonates at high risk for hyperbilirubinemia. Point-of-care screening is crucial to conditions such as hyperbilirubinemia, whereby a quick treatment response is necessary and devastating health consequences for the newborn can occur if treatment was delayed due to wait times for results from a central laboratory.
Acknowledgments
We acknowledge Dr Sharon Geaghan in the initial phases of this study. We thank Dr David K. Stevenson for his support and the use of his research laboratory and Dr Janelle Aby, director of the Well Baby Nursery at Lucile Packard Children’s Hospital. We appreciate Dr Glader’s laboratory technologist (including Carolyn Wong) who conducted the gold standard G6PD assay at the Stanford Red Cell Special Studies Laboratory. The authors specifically thank Martin E. Castillo Cuadrado who recruited patients, collected the whole blood samples, and conducted the experiments on the digital microfluidic analyzer; he also assisted in the manuscript preparation. Ronald J. Wong supervised the point-of care bench assays performed by Mr Castillo Cuadrado; he also provided editorial assistance in the preparation of the manuscript as well expert supervision to operationalize this translational research.
Glossary
- G6PD
glucose-6-phosphate dehydrogenase
- Hb
hemoglobin
- NADPH
β-nicotinamide adenine dinucleotide phosphate hydrate
- NADP
β-nicotinamide adenine dinucleotide phosphate
Footnotes
Dr Bhutani conceptualized the study, supervised data collection, and revised the final manuscript; Dr Kaplan participated in the study as an advisor, reviewed the data collection, and revised the final manuscript; Dr Glader assisted in the conceptual design of the study, supervised data collection through the assay at Stanford Hillview Laboratory, and revised the final manuscript as submitted; Dr Cotten conceptualized the study; Dr Kleinert conceptualized the technical design, developed the assay, and generated the program protocols for performing the glucose-6-phosphate dehydrogenase assays on the digital microfluidic cartridges. Dr Pamula conceptualized and designed the study, directed development of the digital microfluidic system, and drafted the initial manuscript; he had no influence on the clinical design of the study. All authors approved the final manuscript as submitted.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FINANCIAL DISCLOSURE: Dr Kleinert was an employee of Advanced Liquid Logic, Inc, and Dr Pamula is an employee of Baebies, Inc, and has stock ownership. The other authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R44HD062316. All phases of this study were supported by National Institutes of Health grant R44HD062316. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: Dr Kleinert was an employee of Advanced Liquid Logic, Inc at the time of the study and is now an employee at Illumina, Inc which acquired Advanced Liquid Logic, Inc, and Dr Pamula is an employee of Baebies, Inc, and has stock ownership. The other authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Keren R, Luan X, Friedman S, Saddlemire S, Cnaan A, Bhutani VK. A comparison of alternative risk-assessment strategies for predicting significant neonatal hyperbilirubinemia in term and near-term infants. Pediatrics. 2008;121(1). Available at: www.pediatrics.org/cgi/content/full/121/1/e170 [DOI] [PubMed] [Google Scholar]
- 2.Bhutani VK, Stark AR, Lazzeroni LC, et al. Initial Clinical Testing Evaluation and Risk Assessment for Universal Screening for Hyperbilirubinemia Study Group . Predischarge screening for severe neonatal hyperbilirubinemia identifies infants who need phototherapy. J Pediatr. 2013;162(3):477–482.e1 [DOI] [PubMed] [Google Scholar]
- 3.Hameed NN, Na’ Ma AM, Vilms R, Bhutani VK. Severe neonatal hyperbilirubinemia and adverse short-term consequences in Baghdad, Iraq. Neonatology. 2011;100(1):57–63 [DOI] [PubMed] [Google Scholar]
- 4.Olusanya BO, Emokpae AA, Zamora TG, Slusher TM. Addressing the burden of neonatal hyperbilirubinaemia in countries with significant glucose-6-phosphate dehydrogenase deficiency. Acta Paediatr. 2014;103(11):1102–1109 [DOI] [PubMed] [Google Scholar]
- 5.Gamaleldin R, Iskander I, Seoud I, et al. Risk factors for neurotoxicity in newborns with severe neonatal hyperbilirubinemia. Pediatrics. 2011;128(4). Available at: www.pediatrics.org/cgi/content/full/128/4/e925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhutani VK, Zipursky A, Blencowe H, et al. Neonatal hyperbilirubinemia and Rhesus disease of the newborn: incidence and impairment estimates for 2010 at regional and global levels. Pediatr Res. 2013;74(suppl 1):86–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson L, Bhutani VK, Karp K, Sivieri EM, Shapiro SM. Clinical report from the pilot USA Kernicterus Registry (1992 to 2004). J Perinatol. 2009;29(suppl 1):S25–S45 [DOI] [PubMed] [Google Scholar]
- 8.Kuzniewicz MW, Wickremasinghe AC, Wu YW, et al. Incidence, etiology, and outcomes of hazardous hyperbilirubinemia in newborns. Pediatrics. 2014;134(3):504–509 [DOI] [PubMed] [Google Scholar]
- 9.Sgro M, Campbell D, Shah V. Incidence and causes of severe neonatal hyperbilirubinemia in Canada. CMAJ. 2006;175(6):587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manning D, Todd P, Maxwell M, Jane Platt M. Prospective surveillance study of severe hyperbilirubinaemia in the newborn in the UK and Ireland. Arch Dis Child Fetal Neonatal Ed. 2007;92(5):F342–F346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millington DS, Sista R, Eckhardt A, et al. Digital microfluidics: a future technology in the newborn screening laboratory? Semin Perinatol. 2010;34(2):163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beutler E. Red Cell Metabolism: A Manual of Biochemical Methods. 3rd ed. Orlando, FL: Grune & Stratton; 1984 [Google Scholar]
- 13.Sista RS, Eckhardt AE, Wang T, et al. Digital microfluidic platform for multiplexing enzyme assays: implications for lysosomal storage disease screening in newborns. Clin Chem. 2011;57(10):1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO Working Group . Glucose-6-phosphate dehydrogenase deficiency. Bull World Health Organ. 1989;67(6):601–611 [PMC free article] [PubMed] [Google Scholar]
- 15.Luzzatto L. Glucose 6-phosphate dehydrogenase deficiency: from genotype to phenotype. Haematologica. 2006;91(10):1303–1306 [PubMed] [Google Scholar]
- 16.Kaplan M, Hammerman C, Bhutani VK. Parental education and the WHO neonatal G-6-PD screening program: a quarter century later [published online ahead of print July 16, 2015]. J Perinatol. 10.1038/jp.2015.77 [DOI] [PubMed] [Google Scholar]
- 17.Watchko JF, Kaplan M, Stark AR, Stevenson DK, Bhutani VK. Should we screen newborns for glucose-6-phosphate dehydrogenase deficiency in the United States? J Perinatol. 2013;33(7):499–504 [DOI] [PubMed] [Google Scholar]
- 18.Kaplan M, Hoyer JD, Herschel M, Hammerman C, Stevenson DK. Glucose-6-phosphate dehydrogenase activity in term and near-term, male African American neonates. Clin Chim Acta. 2005;355(1–2):113–117 [DOI] [PubMed] [Google Scholar]
- 19.Kaplan M, Herschel M, Hammerman C, Hoyer JD, Stevenson DK. Hyperbilirubinemia among African American, glucose-6-phosphate dehydrogenase-deficient neonates. Pediatrics. 2004;114(2). Available at: www.pediatrics.org/cgi/content/full/114/2/e213 [DOI] [PubMed] [Google Scholar]
- 20.Algur N, Avraham I, Hammerman C, Kaplan M. Quantitative neonatal glucose-6-phosphate dehydrogenase screening: distribution, reference values, and classification by phenotype. J Pediatr. 2012;161(2):197–200 [DOI] [PubMed] [Google Scholar]
- 21.Sista R, Hua Z, Thwar P, et al. Development of a digital microfluidic platform for point of care testing. Lab Chip. 2008;8(12):2091–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan M, Renbaum P, Levy-Lahad E, Hammerman C, Lahad A, Beutler E. Gilbert syndrome and glucose-6-phosphate dehydrogenase deficiency: a dose-dependent genetic interaction crucial to neonatal hyperbilirubinemia. Proc Natl Acad Sci USA. 1997;94(22):12128–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sista RS, Wang T, Wu N, et al. Multiplex newborn screening for Pompe, Fabry, Hunter, Gaucher, and Hurler diseases using a digital microfluidic platform. Clin Chim Acta. 2013;424:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins PV, Campbell C, Klug T, Rogers S, Raburn-Miller J, Kiesling J. Lysosomal storage disorder screening implementation: findings from the first six months of full population pilot testing in Missouri. J Pediatr. 2015;166(1):172–177 [DOI] [PubMed] [Google Scholar]