Abstract
Channelrhodopsins (ChRs) are light-gated cation channels. After blue-light excitation, the protein undergoes a photocycle with different intermediates. Here, we have recorded transient absorbance changes of ChR2 from Chlamydomonas reinhardtii in the visible and infrared regions with nanosecond time resolution, the latter being accomplished using tunable quantum cascade lasers. Because proton transfer reactions play a key role in channel gating, we determined vibrational as well as kinetic isotope effects (VIEs and KIEs) of carboxylic groups of various key aspartic and glutamic acid residues by monitoring their C=O stretching vibrations in H2O and in D2O. D156 exhibits a substantial KIE (>2) in its deprotonation and reprotonation, which substantiates its role as the internal proton donor to the retinal Schiff base. The unusual VIE of D156, upshifted from 1736 cm−1 to 1738 cm−1 in D2O, was scrutinized by studying the D156E variant. The C=O stretch of E156 shifted down by 8 cm−1 in D2O, providing evidence for the accessibility of the carboxylic group. The C=O stretching band of E90 exhibits a VIE of 9 cm−1 and a KIE of ∼2 for the de- and the reprotonation reactions during the lifetime of the late desensitized state. The KIE of 1 determined in the time range from 20 ns to 5 ms is incompatible with early deprotonation of E90.
Introduction
Channelrhodopsin-2 (ChR2) is a light-gated cation channel from the unicellular algae Chlamydomonas reinhardtii (1). Upon illumination, the cell membrane of any alien host expressing ChR2 is depolarized. This renders ChR2 a major tool in optogenetics where it is used to remotely induce action potentials in neuronal cells just by turning on the light (2,3).
The polypeptide of ChR2 folds into seven transmembrane helices with retinal as chromophore bound to a lysine residue of the apo-protein via a Schiff-base linkage. The dark state of ChR2 is composed of a 70:30 mixture of all-trans/13-cis retinal (4). After blue-light excitation, the retinal isomerizes from the all-trans to the 13-cis configuration, which eventually leads to ion permeation. Excitation of 13-cis retinal was concluded neither to be linked to proton transfer reactions nor to the formation of the conductive state (5), presumably because insufficient pKa and structural changes of the protein backbone are elicited by this retinal isomer. As a consequence of photonic excitation of all-trans retinal, the protein undergoes a photocycle passing the intermediate states P1500, P2390, P3520, and P4480 (6–8). (The subscripts indicate the temporal sequence of intermediate states in the photocycle, and the superscripts represent the wavelength of maximal absorption in the visible wavelength range.) It is stressed that these intermediate states were defined by their different electronic states of the retinal cofactor. Many more of the intermediate states are required to fully account for the complex structural changes that eventually lead to channel opening and closure—commonly denoted as “channel gating”. Here, time-resolved infrared (IR) spectroscopy contributes molecular sensitivity to resolve structural changes of the holo-protein, protonation states, and hydrogen-bonding of amino-acid side chains (9–11).
Despite the success of ChR2 and its variants in optogenetic applications, the coupling mechanism of the electronic changes in the retinal induced by photonic excitation and channel gating, to our knowledge, has not been clarified in molecular detail (12). The recently published crystal structure of a chimera of ChR1 and ChR2 from C. reinhardtii (C1C2) founded the basis for meaningful mechanistic studies and together with patch-clamp experiments suggested the presence of a pore framed by helices A, B, C, and G (13). Time-resolved electrophysiological experiments showed that channel opening occurs during the late P2390 state (200 μs) and channel closure during P3520 decay (10 ms). Pulsed electron double-resonance spectroscopic (14) and electron microscopic (15) studies on the trapped conductive state of (the C128T mutant of) ChR2 revealed light-induced rearrangements of helices B, F, and G that seem to persist in the closed (desensitized) state P4480 state (16).
The similarity to the proton pump bacteriorhodopsin (bR) suggested proton transfers to play key roles in the functional mechanism of ChR2. It was actually found that ChR2 behaves as a leaky proton pump (17). We observed in time-resolved experiments using a pH-indicating dye that proton release and uptake occurs during the P3520 formation and decay, respectively (18). Steady-state Fourier-transform infrared (FTIR) spectroscopic experiments helped to identify similar conformational changes and (de-) protonation events in ChR2 (6,19,20) and ChR1-ChR2 chimeras (21,22) from C. reinhardtii. Different conformational and protonation changes have been deduced from steady-state FTIR experiments on ChR1 from Chlamydomonas augustae (23–26). For ChR2 we have further shown by time-resolved FTIR spectroscopy that D253 acts as primary acceptor of the Schiff-base proton and D156 as primary donor (7). E90 has been identified to be a major part of the ion selectivity filter (6), and shown to deprotonate upon P4480 formation and reprotonate from the bulk during ground-state recovery (7). Consistently, while mutation of D156 or D253 has severe effects on the gating mechanism of ChR2 (D156A delays channel closure (27) and D253N prevents channel opening (7), mutation of E90 does not affect the photocycle kinetics but modulates ion selectivity (28,29)). Other groups stated that E90 deprotonates early in the photocycle and plays a direct role in the gating mechanism, a conclusion mostly supported by molecular dynamics simulations (30). While it is evident that electrostatics drive structural changes in proteins and that helix hydration facilitates ion conductance (5,12,30), a general consensus on how the observed proton transfer steps are linked to channel opening and closure, to our knowledge, has not yet been reached.
The exchange of H2O versus D2O as a solvent leads to delays in reaction steps, partially by solvent effects but primarily by deuteration of exchangeable hydrogens of the reactants. The increased mass of deuterium leads to a lower zero-point energy of the vibrational ground state, thereby increasing the energy barrier to break hydrogen bonds (31), which accounts for the pD value in D2O being 0.4 higher than the equivalent pH value in H2O (32). Two major effects are observed when deuterium substitutes hydrogen in a protein: the first is a frequency downshift of a vibration originating from an X-H group or coupled to such a group (vibrational isotope effect, VIE). Such a case applies to the C=O stretching vibration of carboxylic acids, which is coupled to the O-H bending vibration. The second is the delay of a reaction step of a protein that involves breaking and formation of bonds, which also involves hydrogen. It is quantified as the ratio of the rate (or time) constants recorded with the sample immersed in H2O and in D2O, respectively (33), and is known as the kinetic isotope effect (KIE). This parameter has proven of enormous impact in mechanistic studies of membrane proteins such as bacteriorhodopsin (34,35), photosystem II (36,37), cytochrome-c oxidase (38), lactose permease (39), and others.
Here, we determined the KIEs of the photoreaction of ChR2 by flash photolysis ultraviolet-visible (UV-vis) spectroscopy, namely, for the P1500→P2390, P2390→P3520, and P3520→P4480 transitions. We then introduce, to our knowledge, a novel flash-photolysis setup for nanosecond-time-resolved IR spectroscopy using tunable quantum cascade lasers (QCLs) as an IR source, to determine the KIE for specific deprotonation and reprotonation events. These results shed light on the role of D156 as the internal proton donor to the retinal Schiff base (22), and gauge the possibility that E90 deprotonates before the formation of the P4480 intermediate, as proposed by others (29,30).
Materials and Methods
Sample expression and purification
ChR2 and the D156E variant were heterologously expressed in Pichia pastoris and purified by affinity chromatography. The protein was solubilized in a solution of 0.2% dodecyl maltoside, 100 mM NaCl, and 20 mM HEPES at pH 7.4/pD 7.8 (see Radu et al. (19) and Nack et al. (20) for further details).
Flash photolysis in the UV-vis range
Time-resolved UV-vis experiments were performed using a commercial flash photolysis unit (LKS80; Applied Photophysics, Leatherhead, Surrey, UK). ChR2 was excited with a 10-ns laser pulse at a wavelength of 450 nm. The repetition frequency was set to 0.1 Hz and the energy density to 3 mJ/cm2. The measuring light was emitted from a Xenon arc lamp. The transient absorption changes were recorded on two different timescales. The arc lamp was run in pulsed mode to achieve good signal/noise in the early timescale (50 ns to 300 μs). A (quasi-) continuous operation of the lamp was used for the slower time range (300 μs to 10 s). Each kinetic trace was logarithmically averaged to reduce the data from 20,000,000 to 1000 points, and 20 of such traces were collected and averaged at six different wavelengths: 360, 380, 440, 480, 520, and 540 nm.
The time traces were analyzed by global exponential fitting and by maximum entropy lifetime distribution analysis (40). In the former method, an estimate for time constants and their corresponding amplitudes is obtained by fitting a limited number of discrete exponentials by nonlinear weighted least-squares. In the latter, a distribution of exponential amplitudes as a function of the time constant is estimated instead by the maximum entropy method. The regularized value, a scalar that controls the detail of the estimated distribution, was automatically determined using the L-curve method (40).
Time-resolved IR spectroscopy
Samples for IR spectroscopy were concentrated to 4 mg/mL, dried on a BaF2 window, and rehydrated over the vapor phase generated by a glycerol/water mixture (3:7 weight/weight). H/D exchange was achieved using a 2:5 (weight/weight) mixture of a deuterated glycerol and deuterium oxide mixture. The samples were measured at 24°C and at pH 7.4 and pD 7.8, respectively.
Pulsed laser excitation was performed under the same conditions as for time-resolved flash photolysis in the UV-vis range (see above). Time-resolved detection in the IR range was performed on a home-built spectrometer using tunable QCLs (an external cavity QCL run in continuous mode; Daylight Solutions, San Diego, CA). The monochromatic emission of the QCL was directed on the sample and the transmitted intensity was recorded by a photovoltaic MCT detector (KV104 Series, 50 MHz; Kolmar Technologies, Newburyport, MA). The detector output was amplified (KA-050 Series; Kolmar Technologies, Newburyport, MA) and divided into two separate channels. Both channels were digitized by picoscopes (4227 Series; PICO Technology, St. Neots, Cambridgeshire, UK) with the first running at a sampling frequency of 250 MHz and the second at 1 MHz. The time-resolution of the detection system, limited by the detector/preamplifier system response time, was determined to be 15 ns (5). A quantity of 50,000 pretrigger points were recorded (corresponding to 200 μs and 50 ms, respectively) before the exciting laser flash and were stored as reference intensity. Transient absorbance changes were recorded by taking the negative logarithm of the ratio of the intensity after the exciting laser pulse and the mean average intensity in the pretrigger range. The linear equidistant data points were logarithmically averaged and reduced to 100 points per decade. Each kinetic was 35 times recorded and averaged to improve the signal/noise. Then, the emission frequency of the QCL was tuned by 0.5 or 1 cm−1 steps for D2O and H2O, respectively. The experiment was repeated to cover the frequency range from 1600 to 1700 cm−1 in 200 steps. A second QCL head was used for the range of 1690–1780 cm−1. Due to the lower background absorption in this frequency range, the optical path length of the sample was doubled, which increased the change in absorption in these experiments. The overlapping spectral region was used to match signal amplitudes in both data sets, which resulted in a merged dataset covering the range of 1600–1780 cm−1. The data was multiplied by a Gaussian in Fourier space to generate spectra at 4 cm−1 nominal resolution. The recorded IR data were subjected to singular-value-decomposition analysis (see Lórenz-Fonfría et al. (7) for details). The QCL data set was reconstructed with six singular-value-decomposition components, respectively. Time-resolved rapid-scan FTIR experiments for samples rehydrated with H2O and D2O, respectively, were conducted using a Vertex 80v spectrometer (Bruker Optics, Ettlingen, Germany). The repetition rate of the pulsed laser was 0.01 Hz, and spectra were measured from 5 ms to 90 s. Transients from QCL and rapid-scan FTIR experiments were merged when monitoring changes later than 300 ms. Multiexponential fitting of transients at single wavenumbers was performed by ORIGINPRO (OriginLab, Northampton, MA). Time-resolved step-scan FTIR difference data measured in H2O were taken from Lórenz-Fonfría et al. (7) for comparison.
Results
The electronic and structural changes of ChR2 are induced by excitation of a 10-ns laser pulse. The corresponding photoreaction was recorded in the UV-vis and mid-IR ranges and the kinetics were analyzed from 50 ns on. Kinetic and vibrational isotope effects were determined by comparing experiments conducted on samples hydrated either with H2O or with D2O.
Deuterium KIEs in the visible wavelength range
The reaction intermediates of the photocycle of ChR2, as for any retinylidene protein, are characterized by the energy of the absorption maximum of retinal. The corresponding wavelength varies among intermediate states due to conformational changes of the retinal cofactor and/or changes in the protonation state of the retinal Schiff base. It has been shown that the transitions between the intermediates of ChR2 involve proton transfer reactions (7). Therefore, proton transfer may be a rate-limiting step in the photocycle kinetics of ChR2, expected to exhibit altered kinetics upon H/D exchange.
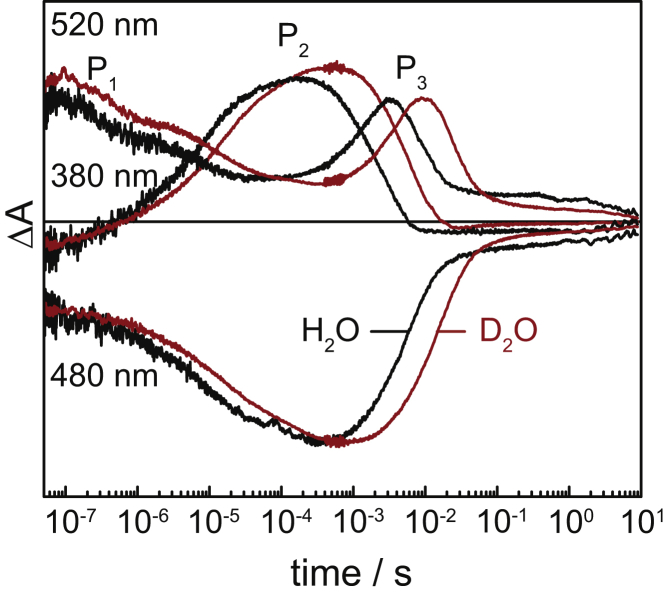
We probed transient absorption changes in the UV-vis range of ChR2 immersed in H2O (Fig. 1, black trace) and in D2O (red). Absorption changes were recorded at 380 nm, which are characteristic for the formation and decay of the blue-shifted P2390 state; at 520 nm, resolving the decay of the P1500 and the formation and decay of the P3520 red-shifted states; and at 480 nm, monitoring the recovery of the bleached ground state.
Figure 1.
Transient absorption changes in the UV-vis range after pulsed excitation of ChR2 in H2O (black traces) and D2O (red traces). The sample was thermostated at 25°C and at a pH 7.4 or pD 7.8, respectively. Ground-state recovery was probed at 480 nm, the red-shifted intermediates (P1500, P3520, and P4480) at 520 nm, and the blue-shifted intermediate (P3390) at 380 nm. To see this figure in color, go online.
We performed a global-fit analysis to obtain time constants, including kinetics recorded at 360, 380, 440, 480, 520, and 540 nm in H2O and in D2O. In addition, maximum entropy lifetime distribution analysis was applied to the kinetics of ChR2 in H2O, D2O, and in an equimolar mixture of H2O/D2O at two selected wavelengths at 380 nm (Fig. 2, bottom panel), and at 520 nm (Fig. 2, top panel), giving a visual perception of the magnitude of the KIE for each exponential component. The KIEs derived from lifetime distribution analysis will be used in the further discussion. The results of both fitting methods are summarized in Table 1 and compared to published data of the photocycle intermediates of the proton-pumping retinylidene protein bR (41–44).
Figure 2.
Maximum entropy lifetime distribution of transient absorption changes of ChR2 in H2O (black), 50% H2O/D2O (green), and D2O (red) measured at (top panel) 520 nm and (bottom panel) 380 nm. Components discussed in the text are labeled by their time constant. To see this figure in color, go online.
Table 1.
Time constants for transitions of photocycle intermediates
| ChR2 Transitions | τ (H2O) | τ (D2O) | KIE of ChR2 | bR Transitions | KIE of bR |
|---|---|---|---|---|---|
| P1500 → P2390 | 8 μs (9.6 μs) | 14 μs (17 μs) | 1.8 (1.8) | L → M | 4.6–5.6 |
| P2390 → P3520 | 1.5 ms (2.3 ms) | 5 ms (7.0 ms) | 3.3 (3.0) | M → N | 1.7–1.8 |
| P3520 → P4480 | 6 ms (6.4 ms) | 19 ms (18 ms) | 3.2 (2.8) | N → O | 1.5–2.0 |
| P44 → ChR2a | 19 s | 49 s | 2.6 | O → bR | 1.5–2.0 |
Time constants were derived from lifetime distribution and multiexponential global fitting (in parentheses) of transient absorption changes of ChR2 at 360, 380, 440, 480, 520, and 540 nm after blue-light (450 nm) excitation. The sample was measured in H2O and D2O and the KIE is given by the ratio of the time constants. The KIEs characterizing the transitions of the various intermediate states of the bacteriorhodopsin (bR) photocycle were taken from the literature (35,41–44). Because the rise of the M intermediate in bR is multiexponential, the KIE for the formation of M is given for the component with the largest amplitude. The KIE of the recovery of ground-state ChR2 was determined by rapid-scan FTIR as described by Lórenz-Fonfría et al. (7).
Derived from multiexponential fitting of the rapid-scan FTIR spectroscopic data at 1242 cm−1.
In H2O, the rise of P2390 is multiexponential, with a dominant phase of τ = 8 μs (70% of the amplitude); the decay is clearly monoexponential, with τ = 2 ms (Fig. 2, top panel, black trace). P3520 also rises monoexponentially with a τ = 1.5 ms, and decays monoexponentially with τ = 6 ms (Fig. 2, bottom panel, black trace). The absorption of the P1500 state decays in several phases (Fig. 2, bottom panel, black trace). We assign its fast decaying component with τ = 300 ns to a transition between two P1500 intermediates that is silent in the E123T variant of ChR2 (5). P1500 further decays, with τ = 8 μs matching the main rise of the P2390 state. Finally, the P4480 decays with τ ≈ 19 s; this was determined by rapid-scan FT-IR at 1242 cm−1, which reflects the ground-state recovery (7). Although weak in intensity, the continuous light used to detect absorbance changes in the visible, may lead to photoconversion of the P4480 state back to the initial ground state, particularly when the lifetime is very long. At this stage, the much lower energy of IR radiation is certainly more appropriate when the time constant of the decay of the P4480 state is determined (see below).
KIEs were derived from the ratio of the time constants of ChR2 dissolved in H2O and in D2O. A KIE of 1.8 was determined for the P1500→P2390 transition (Table 1). This is in contrast to bR in purple membrane, which exhibits a much larger KIE of 5.6 for the main step in the formation of the M state, despite the fact that the retinal Schiff base gets deprotonated in both intermediate states. The KIE for P3520 formation and decay are 3.3 and 3.2, respectively. These may be compared to the M→N and N→O state transitions of bR showing weaker KIEs of 1.7 and 1.5, respectively. The ground-state recovery kinetics of ChR2 exhibited with 2.6, which indicates a proton transfer reaction to be rate-limiting in the recovery of ChR2 (Table 1).
Time-resolved IR absorbance changes of ChR2
Visible spectroscopy is well suited to identify intermediates of the ChR2 photocycle based on the electronic changes in the retinal cofactor, but structural changes of the surrounding opsin moiety are spectrally silent. To address this, the photoreaction of ChR2 was recorded by transient absorption spectroscopy in the mid-IR range. A, to our knowledge, novel spectrometer, employing tunable QCLs, was developed, where kinetic traces are recorded with a temporal resolution of 15 ns.
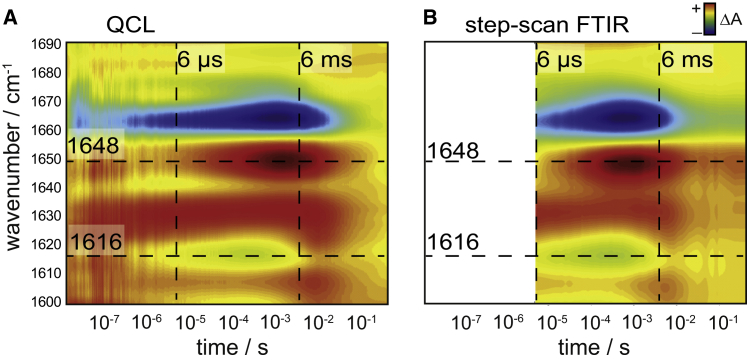
The tunability of the QCL allows us to measure across a broad frequency range. As an example of the fidelity of the recorded data, Fig. 3 A provides an overview of the photoinduced transient absorbance changes of ChR2 in the amide I region (predominantly C=O stretching vibration of the peptide bond in the spectral range between 1600 and 1690 cm−1). The QCL data are validated by time-resolved FTIR spectroscopy using the step-scan technique (Fig. 3 B; data were taken from Lórenz-Fonfría et al. (7)). The latter data have been recorded at a lower time resolution (6 μs). Overall, the data exhibit close agreement. Clearly, the QCL approach provides higher time resolution, but a broader spectral range is covered by step-scan FTIR spectroscopy (30,45).
Figure 3.
Contour plot of transient absorbance changes of ChR2 in the amide I region (of 1600–1690 cm−1) recorded with the, to our knowledge, novel QCL spectrometer (A) and by step-scan FTIR spectroscopy (B). The latter data have been taken from Lórenz-Fonfría et al. (7). The logarithmic time axis covers the range from 20 ns to 300 ms. Negative (blue) and positive (red) absorption changes. (Dashed horizontal lines) Bands at specific wavenumbers (1648 and 1616 cm−1); (dashed vertical lines) time-specific points (6 μs and 6 ms) as discussed in the text (see also Fig. 4). To see this figure in color, go online.
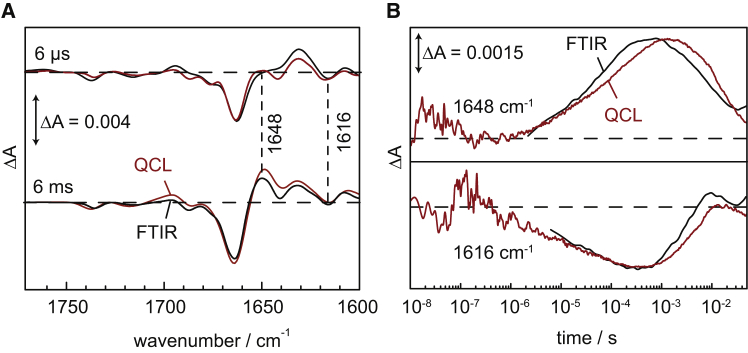
For a detailed inspection, we extracted difference spectra at 6 μs and 6 ms after the exciting laser pulse (dashed vertical lines in Fig. 3) as recorded by QCL (Fig. 4 A, red spectrum) and step-scan FTIR spectroscopy (black spectrum), respectively. The comparison reveals close agreement between both spectroscopic methods. Minor deviations in the region at ∼1630, 1640, and 1700 cm−1 are the result of the higher spectral resolution of the QCL setup (4 vs. 8 cm−1 of the FTIR data). Two time traces at 1648 and 1616 cm−1 (dashed horizontal lines in Fig. 3) were extracted to show the transient absorption changes recorded with the dispersive QCL setup and by step-scan FTIR spectroscopy (Fig. 4 B). Overall, the kinetics agree well with minor deviations to be accounted for the different spectral resolutions used (see above).
Figure 4.
Comparison of the time-resolved IR data recorded by QCL (red) and step-scan FTIR spectroscopy (black). (A) Extracted IR difference spectra at 6 μs and 6 ms. (B) Kinetics of absorbance changes (ΔA) at 1616 and 1648 cm−1. (Dashed horizontal lines) Zero line (ΔA = 0). The FTIR data have been scaled to the amplitude of the QCL data to facilitate the comparison. To see this figure in color, go online.
Deuterium isotope effects in the carboxylic region
After validation of the QCL setup, time-resolved IR difference spectroscopy was used to study the absorption changes between 1690 and 1775 cm−1 range (Fig. 5). In this frequency range, transient protonation and hydrogen-bonding changes of acidic amino acids (aspartic and glutamic acids) have been previously reported for ChR2, albeit at lower time resolution (5–7,29,30).
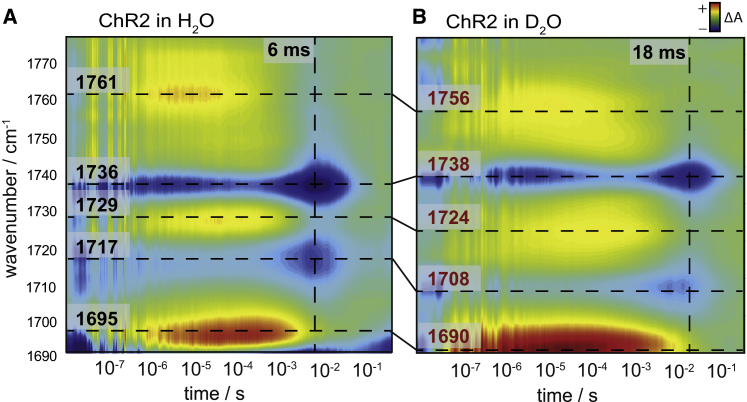
Figure 5.
Contour plot of the IR absorption differences of ChR2 in H2O (A) and D2O (B) in the carboxylic region from 1690 to 1775 cm−1. Positive (red) and negative (blue) absorbance changes. (Dashed horizontal lines) Maxima and minima of difference bands at particular wavenumbers as discussed in the text. (Dashed vertical lines) Time for maximal accumulation of the P3520 state. To see this figure in color, go online.
Two of the isotope effects induced by the H/D exchange must be discriminated in the analysis of time-resolved data. First, the vibrational frequency of C=O stretching modes are downshifted by 5–20 cm−1 due to the increase in reduced mass induced by the heavy isotope (46). The frequency downshift of the C=O stretch comes about due to the coupling to the O-H bending vibration that is lost when the hydroxyl of the carboxylic group is deuterated (46). Second, the increased mass of the isotope leads to a deceleration of the kinetics of the vibrational bands that are associated with proton transfer reactions (47). Thus, the analysis of isotope effects in time-resolved IR experiments is complex.
Fig. 5 shows the time-resolved IR differences of ChR2 in H2O (left panel) and in D2O (right panel), respectively. Dashed horizontal lines in the contour plot indicate specific bands at the wavenumber of their maximum absorbance change. The vertical line indicates the time at which the P3520 state of the ChR2 photocycle has maximal accumulation. This photocycle intermediate state is of particular relevance as ion permeation place during its lifetime.
The intense positive band (red contour in Fig. 5 A) at 1695 cm−1, to which the C=O vibration of the primary acceptor of the Schiff-base proton, D253, contributes (7), is downshifted in frequency by 5 cm−1 upon H/D exchange. Because this vibrational band contains spectral contributions from overlapping vibrations other than the C=O stretch of D253, the determination of the KIE of its rise and decay kinetics is pointless. The negative and positive vibrational bands at 1717 and 1729 cm−1 (Fig. 5 A) are shifted by 9 and 5 cm−1 wavenumbers in D2O (Fig. 5 B), respectively. These two bands have been assigned to changes in hydrogen bonding of the C=O group of protonated E90 (6,7,29). The negative band at 1736 cm−1 was assigned to the C=O vibration of the primary proton donor to the Schiff base D156 (7). This band is slightly upshifted by 2 cm−1 in D2O (Fig. 5 B), which is an unusual response of the carboxylic C=O stretching vibration to deuteration. The positive band at 1761 cm−1, assigned to the C=O stretch of D156 in the P1500 intermediate (7) and more specifically to a late P1500 state (5), is downshifted by 4 cm−1. Therefore, although the frequency of the C=O stretch of D156 in the dark state is hardly affected in D2O (see below), it shows a normal frequency downshift during the lifetime of the P1500 intermediate, indicative of deuteration of the O-H group of the carboxylic group of D156. The assignment of the difference bands (6,7) and their vibrational isotope effects are compiled in Table 2.
Table 2.
Deuterium vibrational isotope effects (VIEs) in the carboxylic region of the IR difference spectra of ChR2
| ν in H2O | ν in D2O | VIEs (Δν) | Assignment |
|---|---|---|---|
| 1717 cm−1 | 1708 cm−1 | −9 cm−1 | (−) E90 |
| 1729 cm−1 | 1724 cm−1 | −5 cm−1 | (+) E90 |
| 1736 cm−1 | 1738 cm−1 | +2 cm−1 | (−) D156 |
| 1761 cm−1 | 1756 cm−1 | −5 cm−1 | (+) D156 |
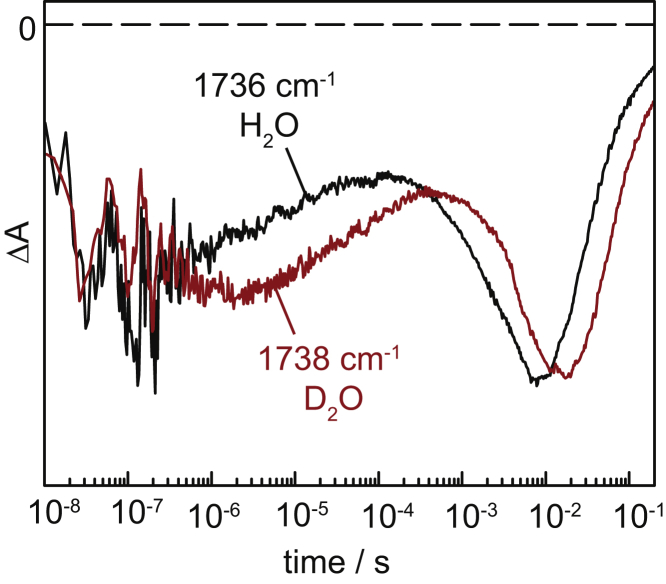
The transient absorbance changes at 1736 and 1738 cm−1, corresponding to the C=O stretch of D156 in ground-state ChR2, are plotted in Fig. 6 (black trace for experiments in H2O and red trace in D2O). The decay and the subsequent recovery in the millisecond time range, which were assigned to deprotonation and reprotonation of D156 (7), exhibit a KIE of 2.7 and 2.4, respectively. These KIEs agree well with those determined for the rise and decay of the P3520 state as derived from time-resolved UV-vis spectroscopy (see Table 1), within the standard deviation of ∼10–15% of the resulting time constants. This finding is in coherence with our previous assignment of D156 to act as the proton donor to the retinal Schiff base (7). Furthermore, the fact that a substantial KIE is observed after H/D exchange strongly suggests that the side chain of D156 is accessible to H/D exchange.
Figure 6.
Transient absorption changes of the C=O stretching vibrational band of D156 (proton donor to the retinal Schiff base) probed at 1736 cm−1 in H2O (black) and 1738 cm−1 in D2O (red). To see this figure in color, go online.
VIE in the D156E mutant
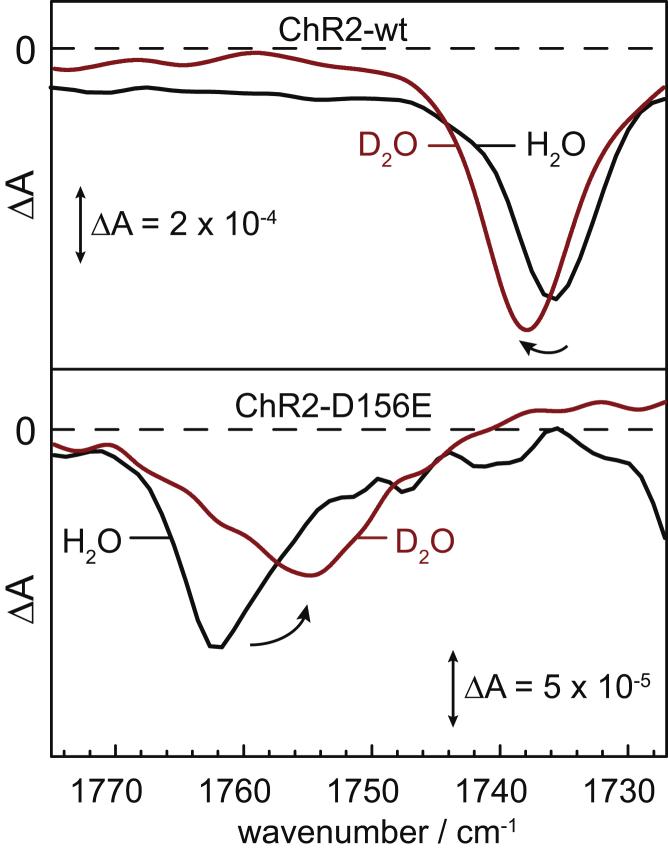
To further corroborate the exchangeability of the proton of the carboxylic group of D156, the aspartic acid at position 156 was replaced by glutamic acid. This conservative replacement does not impair functionality (7). Because of the delayed decay of P3520 (reprotonation of E156), it is possible to observe the C=O stretch frequency of E156 by FTIR difference spectroscopy under continuous illumination (Fig. 7, lower panel). As previously reported (7,19,20), the replacement leads to an upshift of the C=O vibration in the dark state from 1736 cm−1 (D156) to 1763 cm−1 (E156) in H2O (black traces in Fig. 7). The longer glutamate side chain likely locates the carboxylic group in a conformation less favorable for H-bonding with C128 and, thus, the frequency of the C=O stretch is upshifted. In D2O, the C=O stretch of E156 exhibits a spectral downshift by 8 cm−1, from 1763 cm−1 to 1755 cm−1 (bottom panel of Fig. 7), which provides evidence that the carboxylic group at position 156 undergoes H/D exchange. Thus, it is accessible to the externally added D2O during some steps of the photocycle.
Figure 7.
IR difference spectra in the carboxylic region of wild-type ChR2 (top panel) and the D156E variant (bottom panel). For the wild-type, spectra at 6 ms in H2O and at 18 ms in D2O after the exciting laser pulse were extracted from the QCL data. The spectrum for the D156E variant was recorded under continuous illumination at 450 nm, representing mostly the P3520 state. The negative band was assigned to the C=O stretching vibration of protonated D156 (top panel) and E156 (bottom panel), respectively. Vibrational isotope effect leads to a spectral upshift of 2 cm−1 in wild-type ChR2 (top panel). The D156E variant exhibits a vibrational downshift of 8 cm−1 in D2O, typical for H/D exchange (bottom panel). To see this figure in color, go online.
KIE of proton transfer reactions involving E90
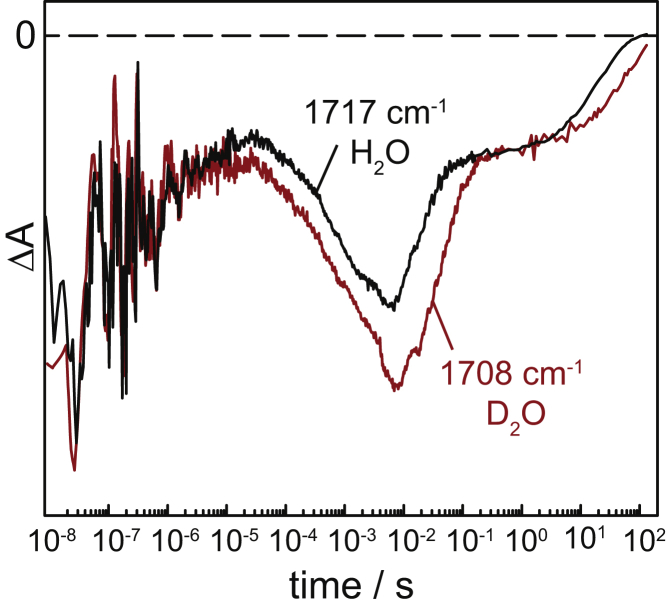
E90 is the other carboxylic residue that is protonated in dark-state ChR2 and transiently deprotonated during the ChR2 photocycle. The frequency of the C=O stretch of the carboxylic of E90 is at 1717 cm−1 in H2O, which is shifted to 1708 cm−1 in D2O (6,29,48). Despite this significant vibrational isotope effect, the transients at these frequencies (Fig. 8) are very similar from 15 ns to 5 ms (KIE = 1). This experimental observation agrees well with our previous conclusion that the absorption changes at 1717 cm−1 in the submicrosecond range arise from changes in hydrogen-bonding rather than from deprotonation changes of E90 (7). This conclusion is also in agreement with the wild-type-like photocycle kinetics (28) and channel activity of the E90A mutant (13,28).
Figure 8.
Transient absorption changes of wild-type ChR2 at 1717 cm−1 in H2O (black) and at 1708 cm−1 in D2O (red). These frequencies correspond to the C=O stretching vibration of E90 in water and in deuterium oxide, respectively. Absorbance changes from 300 ms to 90 s were recorded by time-resolved rapid-scan FTIR spectroscopy and merged to the time-resolved QCL data. To see this figure in color, go online.
At times longer than 10 ms, the kinetics of E90 at 1717 cm−1 is clearly delayed in D2O. This time range corresponds to the decay of the P3520 state, which is bifurcated to repopulate the initial ground state (75% of the molecules) and to pass the P4480 state (25% of the molecules). Only during the lifetime of the latter, is E90 deprotonated (7), a process that exhibits a KIE of 2.2 (Fig. 8). Reprotonation of E90 takes place in the transition from P4480 to the initial ground state, which occurs in ∼10–20 s (6,7) and shows a KIE of 2.1. Both results indicate a late de- and reprotonation of E90 in the ChR2 photocycle.
Discussion
The first indications of proton transfer reactions in channelrhodopsin have been derived from time-resolved UV-vis spectroscopic experiments. In analogy to other microbial rhodopsins, a blue-shifted photocycle intermediate was observed in the photocycle of ChR2 whose rise is associated with the deprotonation of the Schiff-base linkage of the retinal to K257 (8,49). This interpretation has been corroborated for ChR1 from C. augustae by resonance Raman spectroscopy (25). Transient proton release and uptake by ChR2 to and from the bulk aqueous medium were detected by pH-sensitive dyes (18). More recently, D253 and D156 have been identified as acceptors and donors to the retinal Schiff base, respectively (7). These proton transfer reactions do not only account for the residual proton-pumping activity of ChR2 (17) but it appears that the channel function of ChR2 is also highly dependent on these (12). As a prominent example for the latter, the reprotonation of D156 is a rate-limiting channel closure, as its replacement by a nonprotonatable amino acid delayed the lifetime of the open state from 10 ms to >150 s (27). Exchanges of D253 to nonprotonatable amino acids abolished ion conductance (7,13,50). E90 was shown to decrease its initial high pKa during the photocycle (29), but deprotonation takes place upon formation of the desensitized (nonconductive) P4480 state (7). This observation explains why the replacement of E90 to nonprotonatable amino acids retains ion conductivity with barely affected kinetics (28,29). In contrast, recently published molecular dynamics simulations and time-resolved step-scan FTIR results suggested a downward movement of E90, coupled to the deprotonation of E90 in the submicrosecond time range, which opens the pore (30). This causes an influx of water though the opened pore and a tilt of helix B that fully opens the channel (30). We showed here that the C=O stretching vibration of E90 (at 1717 cm−1 in H2O and at 1708 cm−1 in D2O) exhibited a significant KIE only after P3520 formation. This result supports our earlier conclusion that the deprotonation of E90 occurs within the lifetime of the P4480. It is evident, however, that E90 plays a central role in ion selectivity of the channel because replacements lead to drastic alterations in cation selectivity (29). As a matter of fact, the replacement of the uncharged E90 to the positively charged lysine or arginine residues converts the cation channel into an anion channel (51).
The C=O stretching vibration of carboxylate side chains of aspartic and glutamic acids is diagnostic for proton transfer reactions involving these residues. We developed and applied here, to our knowledge, a novel, time-resolved setup for IR absorption spectroscopy on the basis of tunable QCLs, which provides a time resolution of 15 ns and exquisite sensitivity. This setup is considered very versatile, and we expect its application to mechanistic studies of proteins of larger size and higher complexity.
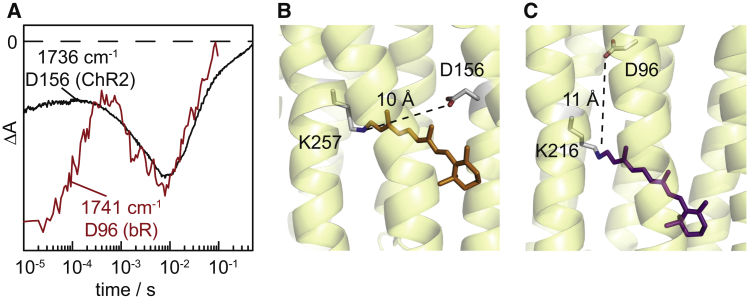
D156 of ChR2 was shown to be the internal proton donor to the retinal Schiff base (SB) (7,30). Thus, it is interesting to compare the kinetics of the de- and reprotonation of D156 in ChR2 with D96 (Fig. 9 A), the proton donor to the retinal SB in bR. The pKa of the latter residue is >12 (52) and drops during the photocycle down to 7.1 (53). The frequencies of the C=O stretches of D156 and D96 appear with 1736 and 1741 cm−1, respectively, in a range that is typical for the carbonyl oxygen accepting a single hydrogen bond. Thus, the C=O probe a local vicinity similar in electrostatics for both residues. Despite the fact that the two residues localize at very different spatial positions within the respective proteins, the distances to the SB are with 10 Å (Fig. 9 B) and 11 Å (Fig. 9 C) also similar. Although proton transfers take place during intermediate state transitions that involve structural rearrangements, it is not surprising that the proton transfer from the respective donors to the retinal SB take place in the same time range of 1–100 ms (Fig. 9 A).
Figure 9.
(A) Transient absorption changes of the C=O stretching vibrations of the primary proton donors of D156 of ChR2 and D96 of bR. The kinetics recorded at 1736 cm−1 correspond to D156 of ChR2 (black trace) and is redrawn from Fig. 6. For comparison, the kinetics at 1741 cm−1 of D96 of bR (red) is replotted from Zscherp et al. (53). (B) Structure of channelrhodopsin (PDB:3UG9) including the distance of the internal proton donor D156 to the retinal Schiff base. (C) Structure of bacteriorhodopsin (PDB:1C3W) including the distance of the internal proton donor D96 to the retinal Schiff base. The distances given are from the N of the retinal Schiff base to the carboxylic OD2 of D96 in bR and to the carboxylic OD1 of D195 in the C1C2 chimera (corresponding to D156) of ChR2, respectively. To see this figure in color, go online.
Despite the coincidence in kinetics, and additional experimental evidence as reviewed recently in Lórenz-Fonfría and Heberle (12), the role of D156 as the SB proton donor has been challenged (22,30), as well as the timing of E90 deprotonation (30). Therefore, additional experimentation is demanded and the determination of deuterium isotope effects represents a means to gain insight into the role and timing of the proton transfers in the photocycle of ChR2.
Using time-resolved UV-vis spectroscopy, we determined only a small KIE (<2) for the rise of the P2390 state, the photocycle intermediate with deprotonated SB. This result is in stark contrast to bR where the rise of the M state exhibits a much larger KIE of ∼5 (Table 1). It is concluded from this result that deprotonation of the Schiff base is not rate limiting in ChR2, but it is in bR. Furthermore, the P2390-to-P3520 as well as the P3520-to-P4480 transitions show a stronger KIE in ChR2 (3.3 and 3.2, respectively) than corresponding transitions in bR (KIE < 2). This indicates that proton transfer is rate-limiting during the lifetime of the late intermediates of the ChR2 photocycle. Although proton-pumping bR and ion-conducting ChR2 share structural and mechanistic similarities, the different KIEs indicate distinct differences in their proton transfer pathways. This is in agreement with different residues playing the role of the SB proton acceptor/donor: D253/D156 in ChR2 (equivalent to residues D212/D115 of bR), while the SB proton acceptor/donor pair in bR is D85/D96 (equivalent to residues E123/H134 of ChR2).
The comparison of the time-resolved absorption changes bands in the carboxylic region show that all observed C=O vibrations are sensitive toward H/D exchange (Fig. 5) but to variable degrees. The KIE for the deprotonation/protonation of D156 (Fig. 6) is very similar to the KIEs of formation and decay of the P3520 state as detected in the visible (Table 1). This result further supports our previous identification of D156 as the primary proton donor (6). This assignment has recently been challenged by Ito et al. (22) and by Kühne et al. (30). These authors argued from the H/D insensitivity of the C=O stretching vibrational band of D156 (6,22,30,48) that the O-H group of the carboxylic group of D156 is not exchangeable, refuting the role of D156 as proton donor to the retinal Schiff base (22,30).
In fact, we observed here that the C=O stretch of D156 is sensitive to H/D exchange, albeit with an unusual frequency upshift by 2 cm−1 (Fig. 7, top panel), instead of the typically observed downshift by 5–10 cm−1 of this vibration. It is pointed out that the C=O stretch of D156 exhibited a more typical downshift by 5 cm−1 (from 1761 to 1756 cm−1) during the lifetime of the P1500 intermediate (Fig. 5). The typical H/D-induced downshift by 8 cm−1 (from 1763 to 1755 cm−1) is apparent in the D156E variant, demonstrating that the carboxylic group at position 156 is indeed accessible to H/D exchange. Thus, our results resolve the major argument against the assignment of D156 as the proton donor to the Schiff base.
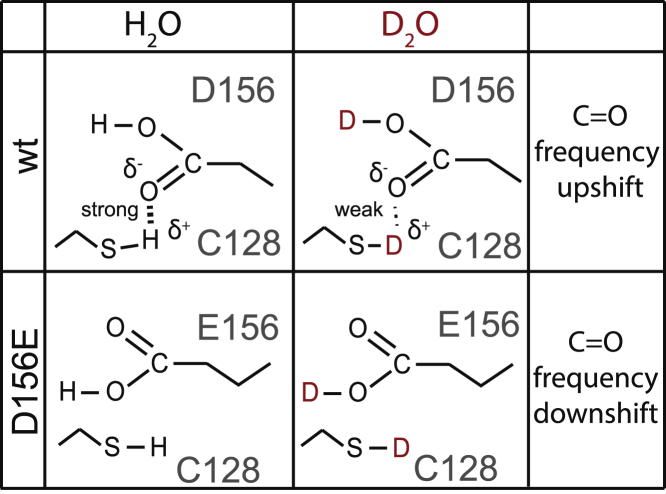
What is left to be discussed is the scenario of the hydrogen-bonded network surrounding the carboxylic group of D156 that accounts for the observed 2 cm−1 frequency upshift of the C=O stretching vibration after H/D exchange, as well as the 27 cm−1 (in H2O) upshift upon exchange from Asp to Glu. It is well known that the frequency of the C=O stretch of a carboxylic group is modulated by H-bonding (46,54–56), not only due to direct H-bonding to the C=O group, but also to the O-H of the carboxylic group.
The frequency of the C=O stretching vibration can upshift by >12 cm−1 and downshift by >70 cm−1, depending on the H-bonding configuration according to quantum-mechanical calculations (46,55). Differences between the strength of H-bonds and D-bonds (57) to the C=O group might induce different shifts in the C=O vibration. An H-bond between the S-H group of cysteine and the C=O of a carboxylic group, as was proposed for the DC gate (12,20), is expected to downshift the C=O frequency by 17 cm−1 according to calculations performed on protonated carboxylic acids in vacuo (55). In E156, this H-bond is possibly disrupted due to the longer side chain of Glu with respect to Asp, accounting for the 27 cm−1 higher C=O frequency in the D156E variant (Fig. 10). It should be noted that S-D groups are expected to be less polarized than S-H groups, as deduced by the 2.5 lower extinction coefficient of S-D stretches with respect S-H stretches (58,59). Under this reasonable assumption, deuteration of C128 will reduce the strength of its H-bond with D156, presumably upshifting the C=O vibration and compensating the downshift caused by the deuteration at the hydroxyl group of D156 (Fig. 10).
Figure 10.
Illustration of the hydrogen-bonding interaction of C128 and D156 (DC gate) in H2O and D2O. This model explains the different VIEs of D156 and E156 upon H/D exchange. To see this figure in color, go online.
Recent FTIR spectroscopic experiments recorded on the C1C2 chimera and its variants, in which the corresponding residues of D156 and C128 have been replaced (21), agree very well with these data on ChR2. Thus, we consider the correlation of FTIR experiments that resolve structural changes of ChR2 with the dark-state structure of C1C2, as determined by x-ray crystallography (13), to be valid and meaningful. Deviations in the photocycle kinetics of the two variants of channelrhodopsin, ChR2 and C1C2, have been noted (21), but are irrelevant as long as time-resolved approaches succeed in being used for separate photocycle states, as was shown here.
Atypical shifts in the carboxylic region, as shown here for the C=O stretching vibration of D156 in dark-state ChR2, have also been reported for the C=O stretching vibration of D96 in the L intermediate of bR (47). In this latter case, the C=O frequency of D96 in the ground state showed a normal 9 cm−1 downshift in D2O (from 1741 to 1732 cm−1), but in the L intermediate the frequency exhibited a mere 1 cm−1 downshift (from 1748 to 1747 cm−1) (47). Maeda et al. (47) suggested a putative H-bond of the hydroxyl group of D96 in the L intermediate as the cause for the anomalous shift upon deuteration—a suggestion that, to our knowledge, still lacks confirmation.
In any case, anomalous shifts in the frequency of the C=O stretch of carboxylic groups of proteins upon deuteration might indicate an unusual arrangement of H-bonds, whose characterization are best addressed by applying sophisticated quantum-mechanics/molecular-mechanics calculations (56,60,61).
Author Contributions
T.R. and B.-J.S. performed experiments; T.R., B.-J.S., and V.A.L.-F. analyzed data; and R.S. contributed with materials and equipment. All authors discussed the data, J.H. conceived the research, and T.R., V.A.L.-F., and J.H. wrote the article.
Acknowledgments
The authors thank I. Wallat, K. Hoffmann, and J. Wonneberg for excellent sample preparation and technical support. T.R. is grateful to N. Krause for introducing the preparation of protein samples for IR spectroscopy.
This work was supported by the Deutsche Forschungsgemeinschaft under grants No. SFB-1078/B3 to J.H. and SFB-1078/B4 to R.S.
Editor: Joseph Mindell.
Footnotes
Tom Resler and Bernd-Joachim Schultz contributed equally to this work.
References
- 1.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunaydin L.A., Yizhar O., Hegemann P. Ultrafast optogenetic control. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 3.Han X. In vivo application of optogenetics for neural circuit analysis. ACS Chem. Neurosci. 2012;3:577–584. doi: 10.1021/cn300065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nack M., Radu I., Heberle J. The retinal structure of channelrhodopsin-2 assessed by resonance Raman spectroscopy. FEBS Lett. 2009;583:3676–3680. doi: 10.1016/j.febslet.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 5.Lórenz-Fonfría V.A., Schultz B.J., Heberle J. Pre-gating conformational changes in the ChETA variant of channelrhodopsin-2 monitored by nanosecond IR spectroscopy. J. Am. Chem. Soc. 2015;137:1850–1861. doi: 10.1021/ja5108595. [DOI] [PubMed] [Google Scholar]
- 6.Ritter E., Stehfest K., Bartl F.J. Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J. Biol. Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lórenz-Fonfría V.A., Resler T., Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamann C., Kirsch T., Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 9.Kötting C., Gerwert K. Proteins in action monitored by time-resolved FTIR spectroscopy. ChemPhysChem. 2005;6:881–888. doi: 10.1002/cphc.200400504. [DOI] [PubMed] [Google Scholar]
- 10.Mäntele W. Reaction-induced infrared difference spectroscopy for the study of protein function and reaction mechanisms. Trends Biochem. Sci. 1993;18:197–202. doi: 10.1016/0968-0004(93)90186-q. [DOI] [PubMed] [Google Scholar]
- 11.Gerwert K. Molecular reaction-mechanisms of proteins as monitored by time-resolved FTIR spectroscopy. Curr. Opin. Struct. Biol. 1993;3:769–773. doi: 10.1515/BC.1999.115. [DOI] [PubMed] [Google Scholar]
- 12.Lórenz-Fonfría V.A., Heberle J. Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Kato H.E., Zhang F., Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krause N., Engelhard C., Bittl R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by EPR spectroscopy. FEBS Lett. 2013;587:3309–3313. doi: 10.1016/j.febslet.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 15.Müller M., Bamann C., Kühlbrandt W. Projection structure of channelrhodopsin-2 at 6 Å resolution by electron crystallography. J. Mol. Biol. 2011;414:86–95. doi: 10.1016/j.jmb.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 16.Sattig T., Rickert C., Bamann C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew. Chem. Int. Ed. Engl. 2013;52:9705–9708. doi: 10.1002/anie.201301698. [DOI] [PubMed] [Google Scholar]
- 17.Feldbauer K., Zimmermann D., Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. USA. 2009;106:12317–12322. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nack M., Radu I., Heberle J. Kinetics of proton release and uptake by channelrhodopsin-2. FEBS Lett. 2012;586:1344–1348. doi: 10.1016/j.febslet.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 19.Radu I., Bamann C., Heberle J. Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 2009;131:7313–7319. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- 20.Nack M., Radu I., Heberle J. The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 2010;9:194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 21.Inaguma A., Tsukamoto H., Furutani Y. Chimeras of channelrhodopsin-1 and -2 from Chlamydomonas reinhardtii exhibit distinctive light-induced structural changes from channelrhodopsin-2. J. Biol. Chem. 2015;290:11623–11634. doi: 10.1074/jbc.M115.642256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito S., Kato H.E., Kandori H. Water-containing hydrogen-bonding network in the active center of channelrhodopsin. J. Am. Chem. Soc. 2014;136:3475–3482. doi: 10.1021/ja410836g. [DOI] [PubMed] [Google Scholar]
- 23.Ogren J.I., Mamaev S., Rothschild K.J. Retinal chromophore structure and Schiff base interactions in red-shifted channelrhodopsin-1 from Chlamydomonas augustae. Biochemistry. 2014;53:3961–3970. doi: 10.1021/bi500445c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lórenz-Fonfría V.A., Muders V., Heberle J. Changes in the hydrogen-bonding strength of internal water molecules and cysteine residues in the conductive state of channelrhodopsin-1. J. Chem. Phys. 2014;141:22D507. doi: 10.1063/1.4895796. [DOI] [PubMed] [Google Scholar]
- 25.Muders V., Kerruth S., Schlesinger R. Resonance Raman and FTIR spectroscopic characterization of the closed and open states of channelrhodopsin-1. FEBS Lett. 2014;588:2301–2306. doi: 10.1016/j.febslet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Ogren J.I., Yi A., Rothschild K.J. Proton transfers in a channelrhodopsin-1 studied by Fourier transform infrared (FTIR) difference spectroscopy and site-directed mutagenesis. J. Biol. Chem. 2015;290:12719–12730. doi: 10.1074/jbc.M114.634840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamann C., Gueta R., Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 28.Ruffert K., Himmel B., Eulenburg V. Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 2011;410:737–743. doi: 10.1016/j.bbrc.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Eisenhauer K., Kühne J., Gerwert K. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühne J., Eisenhauer K., Bartl F. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew. Chem. Int. Ed. Engl. 2015;54:4953–4957. doi: 10.1002/anie.201410180. [DOI] [PubMed] [Google Scholar]
- 31.Bell R.P. Recent advances in study of kinetic hydrogen isotope-effects. Chem. Soc. Rev. 1974;3:513–544. [Google Scholar]
- 32.Glasoe P.K., Long F.A. Use of glass electrodes to measure acidities in deuterium oxide. J. Phys. Chem. 1960;64:188–190. [Google Scholar]
- 33.Krishtalik L.I. The mechanism of the proton transfer: an outline. Biochim. Biophys. Acta. 2000;1458:6–27. doi: 10.1016/s0005-2728(00)00057-8. [DOI] [PubMed] [Google Scholar]
- 34.Heberle, J., and N. A. Dencher. 1992. Proton Transfer in Hydrogen-Bonded Systems. T. Bountis, editor. Plenum Press, New York. 187–197.
- 35.le Coutre J., Gerwert K. Kinetic isotope effects reveal an ice-like and a liquid-phase-type intramolecular proton transfer in bacteriorhodopsin. FEBS Lett. 1996;398:333–336. doi: 10.1016/s0014-5793(96)01254-9. [DOI] [PubMed] [Google Scholar]
- 36.Jenson D.L., Evans A., Barry B.A. Proton-coupled electron transfer and tyrosine D of photosystem II. J. Phys. Chem. B. 2007;111:12599–12604. doi: 10.1021/jp075726x. [DOI] [PubMed] [Google Scholar]
- 37.Klauss A., Haumann M., Dau H. Alternating electron and proton transfer steps in photosynthetic water oxidation. Proc. Natl. Acad. Sci. USA. 2012;109:16035–16040. doi: 10.1073/pnas.1206266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson A.L., Chakrabarty S., Brzezinski P. Proton-transport mechanisms in cytochrome c oxidase revealed by studies of kinetic isotope effects. Biochim. Biophys. Acta. 2011;1807:1083–1094. doi: 10.1016/j.bbabio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnova I., Kasho V., Kaback H.R. Role of protons in sugar binding to LacY. Proc. Natl. Acad. Sci. USA. 2012;109:16835–16840. doi: 10.1073/pnas.1214890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lórenz-Fonfría V.A., Kandori H. Bayesian maximum entropy (two-dimensional) lifetime distribution reconstruction from time-resolved spectroscopic data. Appl. Spectrosc. 2007;61:428–443. doi: 10.1366/000370207780466172. [DOI] [PubMed] [Google Scholar]
- 41.Brown L.S., Needleman R., Lanyi J.K. Origins of deuterium kinetic isotope effects on the proton transfers of the bacteriorhodopsin photocycle. Biochemistry. 2000;39:938–945. doi: 10.1021/bi9921900. [DOI] [PubMed] [Google Scholar]
- 42.Korenstein R., Sherman W.V., Caplan S.R. Kinetic isotope effects in the photochemical cycle of bacteriorhodopsin. Biophys. Struct. Mech. 1976;2:267–276. doi: 10.1007/BF00535372. [DOI] [PubMed] [Google Scholar]
- 43.Cao Y., Brown L.S., Lanyi J.K. Relationship of proton release at the extracellular surface to deprotonation of the Schiff base in the bacteriorhodopsin photocycle. Biophys. J. 1995;68:1518–1530. doi: 10.1016/S0006-3495(95)80324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S.Y. Light-induced currents from oriented purple membrane: I. Correlation of the microsecond component (B2) with the L-M photocycle transition. Biophys. J. 1990;57:943–950. doi: 10.1016/S0006-3495(90)82614-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radu I., Schleeger M., Heberle J. Time-resolved methods in biophysics. 10. Time-resolved FT-IR difference spectroscopy and the application to membrane proteins. Photochem. Photobiol. Sci. 2009;8:1517–1528. doi: 10.1039/b9pp00050j. [DOI] [PubMed] [Google Scholar]
- 46.Takei K., Takahashi R., Noguchi T. Correlation between the hydrogen-bond structures and the C=O stretching frequencies of carboxylic acids as studied by density functional theory calculations: theoretical basis for interpretation of infrared bands of carboxylic groups in proteins. J. Phys. Chem. B. 2008;112:6725–6731. doi: 10.1021/jp801151k. [DOI] [PubMed] [Google Scholar]
- 47.Maeda A., Sasaki J., Lanyi J.K. Structures of aspartic acid-96 in the L and N intermediates of bacteriorhodopsin: analysis by Fourier transform infrared spectroscopy. Biochemistry. 1992;31:4684–4690. doi: 10.1021/bi00134a022. [DOI] [PubMed] [Google Scholar]
- 48.Neumann-Verhoefen M.K., Neumann K., Wachtveitl J. Ultrafast infrared spectroscopy on channelrhodopsin-2 reveals efficient energy transfer from the retinal chromophore to the protein. J. Am. Chem. Soc. 2013;135:6968–6976. doi: 10.1021/ja400554y. [DOI] [PubMed] [Google Scholar]
- 49.Ernst O.P., Sánchez Murcia P.A., Hegemann P. Photoactivation of channelrhodopsin. J. Biol. Chem. 2008;283:1637–1643. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 50.Sineshchekov O.A., Govorunova E.G., Spudich J.L. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wietek J., Wiegert J.S., Hegemann P. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 52.Száraz S., Oesterhelt D., Ormos P. pH-induced structural changes in bacteriorhodopsin studied by Fourier transform infrared spectroscopy. Biophys. J. 1994;67:1706–1712. doi: 10.1016/S0006-3495(94)80644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zscherp C., Schlesinger R., Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc. Natl. Acad. Sci. USA. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dioumaev A.K. Infrared methods for monitoring the protonation state of carboxylic amino acids in the photocycle of bacteriorhodopsin. Biochem. Mosc. 2001;66:1269–1276. doi: 10.1023/a:1013135419529. [DOI] [PubMed] [Google Scholar]
- 55.Nie B., Stutzman J., Xie A. A vibrational spectral maker for probing the hydrogen-bonding status of protonated Asp and Glu residues. Biophys. J. 2005;88:2833–2847. doi: 10.1529/biophysj.104.047639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welke K., Watanabe H.C., Elstner M. QM/MM simulations of vibrational spectra of bacteriorhodopsin and channelrhodopsin-2. Phys. Chem. Chem. Phys. 2013;15:6651–6659. doi: 10.1039/c3cp44181d. [DOI] [PubMed] [Google Scholar]
- 57.Scheiner S. Calculation of isotope effects from first principles. Biochim. Biophys. Acta. 2000;1458:28–42. doi: 10.1016/s0005-2728(00)00058-x. [DOI] [PubMed] [Google Scholar]
- 58.Bare G.H., Alben J.O., Bromberg P.A. Sulfhydryl groups in hemoglobin. A new molecular probe at the α1β1 interface studied by Fourier transform infrared spectroscopy. Biochemistry. 1975;14:1578–1583. doi: 10.1021/bi00679a005. [DOI] [PubMed] [Google Scholar]
- 59.Noguchi T., Fukami Y., Inoue Y. Fourier transform infrared study on the primary donor P798 of Heliobacterium modesticaldum: cysteine S-H coupled to P798 and molecular interactions of carbonyl groups. Biochemistry. 1997;36:12329–12336. doi: 10.1021/bi970853c. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe H.C., Welke K., Elstner M. Structural model of channelrhodopsin. J. Biol. Chem. 2012;287:7456–7466. doi: 10.1074/jbc.M111.320309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe H.C., Welke K., Elstner M. Towards an understanding of channelrhodopsin function: simulations lead to novel insights of the channel mechanism. J. Mol. Biol. 2013;425:1795–1814. doi: 10.1016/j.jmb.2013.01.033. [DOI] [PubMed] [Google Scholar]