Abstract
Understanding cellular structure is key to understanding cellular regulation. New developments in super-resolution fluorescence imaging, electron microscopy, and quantitative image analysis methods are now providing some of the first three-dimensional dynamic maps of biomolecules at the nanometer scale. These new maps—comprehensive nanometer-scale cellular cartographies—will reveal how the molecular organization of cells influences their diverse and changeable activities.
In one of the most classic books on data visualization and analysis, Exploratory Data Analysis, the mathematician John Tukey argues that the “greatest value of a picture is when it forces us to notice what we never expected to see” (Tukey, 1977). The early history of cell biology is filled with examples that illustrate this point. Pioneers in biological EM, including George Palade, Albert Claude, and Keith Porter, made fundamental discoveries in cell biology by inventing and then using new ways of imaging (Farquhar, 2012). They learned by observing, by exploring the basic yet unknown structures of the cell. Working only 10 years after the invention of the electron microscope, they developed new stains, fixation methods, and thin sample techniques that allowed them and others to observe cells for the first time at the nanometer scale. What followed was the discovery of many new structures, including the ribosome, the endoplasmic reticulum, mitochondrial cristae, synaptic structures, and other intracellular bodies. This rapid pace of discovery in the late 1940s and 1950s necessitated the founding of a new journal first called The Journal of Biophysical and Biochemical Cytology and later renamed The Journal of Cell Biology (Farquhar, 2012). Central to this early period of modern cell biology was the drive to understand cells by observing their basic internal structures or organelles.
Today, new tools in imaging are allowing cell biology to enter into a new molecular phase of structural exploration. Here, I discuss the possibilities and challenges of using these new imaging methods to assemble a complete molecular picture of the dynamic cell—one of the great challenges for cell biology over the next few decades. When complete molecular pictures of the cell can be produced, we can expect, as George Palade discussed in his Nobel lecture, that new and exciting biology will be “suggested by the structural organization [of the cell] itself (Palade, 1975; Heuser, 2014).” Similar to the way that elucidating the structure of DNA explained the elegant mechanism of heredity, we can anticipate that determining the structures, patterns, and dynamics of the cell will fundamentally illuminate how cells function.
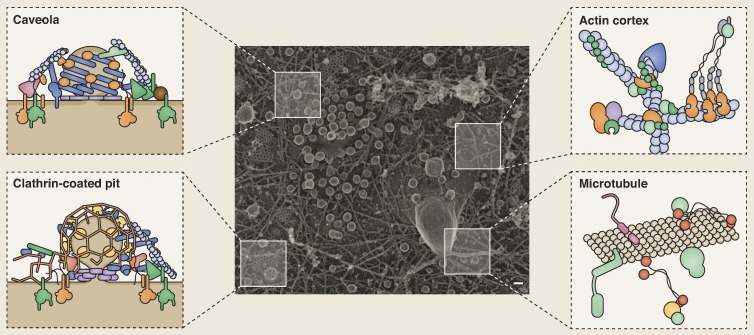
Although much is known about the structure of cells, we still have only a rough idea about the distributions of proteins within that structure (Fig. 1). Without a detailed view, we have little chance of understanding cellular systems. This is the case because whereas the physics of biomolecules (diffusion, binding interactions, concentrations, and reaction rates) is seen to largely govern cell behavior, this view is incomplete. The cell is far from a test tube. Cells originate from other cells as highly structured and interconnected objects with a preset complex, polarized, compartmentalized, and sometimes plastic structure. Many molecules are physically constrained, clustered, and anchored. This inherited molecular organization isolates components and limits or enhances their activities. Thus, to a large degree, cells generate their complex behaviors by manipulating the organizations, dynamics, and partnerships of their biomolecules (proteins, nucleotides, lipids, small chemicals, sugars, etc.) at the nanometer scale.
Figure 1.
Visualizing the molecular architecture of the cell. A transmission EM image of a platinum replica of the inner plasma membrane of a HeLa cell. Many cellular features can be seen, including clathrin-coated pits and plaques, caveolae, the actin cortex, microtubules, and other vesicles. Although we know many of the molecular components (depicted in the schematics), a complete molecular cartography of the plasma membrane would identify the location, structure, and relationships of all proteins that make up the diverse features of even this small region of the cell. Bar, 77 nm. Transmission EM image courtesy of Kem Sochacki (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD).
In the past, nanometer-scale information of molecular positions was limited to immunogold EM or cryogenic EM (cryo-EM) methods (Roth et al., 1978; Lučič et al., 2013). Although incredibly useful, immunogold methods are tricky. Issues of antibody availability, penetration, specificity, the size of the gold particles, inherent background issues, working in a monochromatic contrast regimen, and the obscuring of the biological object by the label itself create difficulties in making this method universal and robust. With cryo-EM, proteins can be directly imaged in a rapidly frozen sample of the cell. This method is incredibly powerful, and proteins can be observed in three dimensions without additional contrast agents in an environment that is as close to their living states as possible (Asano et al., 2015). In cryo-EM, however, the unique shapes of the molecules must be known. Furthermore, the method works best for large or purified protein complexes. These issues have slowed the widespread use of these tools to map the complete structural proteome of cells.
How can the goal of mapping the entire molecular architecture of the cell be achieved? The development of super-resolution fluorescence (SRF) methods over the last 20 years has provided a way forward (Hell, 2003; Huang et al., 2009; Liu et al., 2015). Although still far from the resolution limit of the electron microscope, super-resolution light-based methods offer many advantages. The most fundamental advantage of light-based methods over EM is that living cells can be imaged. This means that cells can be studied as they grow, divide, change, and interact with their environment. These methods also allow the use of multiple color probes in the same sample, robust statistical tests to evaluate positions and correlations of molecules, genetically encoded tags, and a distinctive contrast regimen that has high signal over the background (Liu et al., 2015). Many new structures and behaviors are being discovered with these constantly improving methods (Kanchanawong et al., 2010; Xu et al., 2013; Li et al., 2015). Nanometer-scale images are again transforming our understanding of the organization of cells. Furthermore, the commercialization and standardization of many of these microscopes and analysis methods have allowed this type of imaging to be applied across more of the cellular landscape.
Although much development has occurred for SRF methods, several advances are still needed to fully leverage these tools for global cellular analysis. First, we need better probes. Point localization (PALM, STORM, dSTORM), point-spread function-shaping methods (STED), or structured illumination methods (SIM) have the physical ability to image with nanometer precision, but doing this over long time scales, particularly in living cells in three dimensions, will require a substantial jump in the brightness, stability, ease of inactivation, and spectral options of available fluorophores. All probes have an inherent size. Standard antibodies in particular are large (∼10 nm) and place the fluorophore nanometers away from their intended target. New generations of smaller probes, such as ScFvs antibodies and nanobodies, are exciting options (Helma et al., 2015). We additionally need ways of specifically tagging or labeling the entire proteome. Direct genome engineering offers a promising way forward. Currently, adding small fluorescent or affinity tags to proteins at their genomic locations offers the best option to visualize proteins at their native expression levels (Doyon et al., 2011). This has been done in yeast, and it is now possible in many other systems (Huh et al., 2003; Chong et al., 2015). Imaging two, three, or even four components in the same sample is also important to understanding how proteins and organelles interact. Spectral methods that would allow multiple probes to be imaged simultaneously are likely necessary for this type of analysis. Lastly, the most resolved SRF methods work only in thin slices of the cell. Advancing these methods to image the entire 3D volume of the cell gently, isotropically, and quickly at nanometer resolution is a major technical challenge for the next few years (Liu et al., 2015). Also, although SRF methods are promising, they visualize proteins on a black featureless background. EM can provide a rich cellular backdrop to position these proteins in relationship to the general cellular architecture. Thus, the combination of light and EM methods is particularly exciting (de Boer et al., 2015). Several correlative light and EM methods are now bridging the gap and providing the best information from both methods (Kukulski et al., 2012; Sochacki et al., 2014). With these advances it is possible to imagine determining the position of hundreds/thousands of protein types in a cell.
Along with the technical aspects of physically producing these images, there is an equally challenging aspect of image analysis. Studying the location and relationships of molecules in a quantitative and standardized way that can be correlated to features of the cell (organelles, cell shape, cell behaviors, and local environment) is not a straightforward task (Fig. 1). Additionally, the diversity within a single cell population in terms of the possible morphologies, cell cycle states, or spatial positions (number of neighbors or physical location for example) is large. These variables could dramatically alter the distribution of molecules. Thus, developing unbiased automated ways to classify the important key features of a protein’s location in relationship to these variables is needed and would be incredibly powerful. Because it is difficult to infer generalities from a single cell, it is also important to develop high-throughput and high-content SRF methods. By imaging hundreds or thousands of cells and many cell types, it will be possible to determine general features of protein distributions and their relationship to cellular states and functions. To accomplish this, SRF mapping methods would need to be combined with standardized image processing and automated analysis pipelines. Finally, imaging single cells over long time scales to track these features as cells grow and change, even in complex tissues, is an important challenge. Orthogonal methods such as nanometer-localized mass spectroscopy, FRET, or FCS methods could be needed to fully realize the goal of mapping the global molecular structure of cells. Of specific note is the emerging method of soft x-ray tomography, which has the potential to image entire unstained and hydrated cells in three dimensions at the nanometer scale.
The imaging I have discussed will only map the physical location of molecules (Fig. 1). Protein structures drive protein actions. The structural state of a protein is mostly invisible to the methods discussed above. New probes that highlight unique structural states such as state-specific nanobodies, biosensors, or FRET methods will be needed to illuminate not just the position, but also the state (and activity) of target molecules (Irannejad et al., 2013; Helma et al., 2015). Finally, the plethora of nonprotein molecules in the cell (lipids, nucleotides, small molecules, and sugars) remains mostly unmarked by existing methods. We need a new generation of probes to visualize these important biomolecules.
We are near to a time when we will be able to visualize the complete molecular structure of a cell. Several organized initiatives are now underway to work toward these and similar goals. The Human Protein Atlas and the Allen Institute for Cell Science, for example, are working to map the molecular architecture of cells in a systematic way (Uhlén et al., 2015). These scientific initiatives, along with work from many individual laboratories and institutions, are providing the first comprehensive maps of cellular components (Huh et al., 2003; Larson et al., 2014; Chong et al., 2015). These programs have similar motivations to the human genome and brain connectome mapping projects. Global molecular maps of cells will have wide reaching and fundamental contributions to biology and human health. For example, tracking how the molecular cartography of cells changes as they progress into a cancerous state or neurons acquire neurodegenerative phenotypes could lead to new understandings of these complex diseases. At present, most of biology is in the dark as to the global location, level, and structural state of proteins important for many cellular functions. Furthermore, what these components are associating with and how these associations drive their activities are unclear.
We need to place structural biology, biochemistry, genetics, and biophysics into the overarching dynamic architecture of the physical cell. This will contextualize the mechanistic insights gained from different fields into a global cellular model. As the pioneering cell biologist E.B. Wilson wrote in the 1920s, “We assume, as our fundamental working hypothesis that the specificity of each kind of cell depends essentially upon what we call its organization, i.e. upon the construction of the cell-machine” (Wilson, 1925). We are now at the brink of visualizing this “cell-machine” with all its diverse and varied parts. These maps, a molecular cartography of cells, combined with data from the biochemical and biophysical sciences, are a necessary starting point to build a comprehensive predictive model of the basic unit of life.
Acknowledgments
I would like to thank Keir Neuman, Jonathan Silver, Kem Sochacki, Agila Somasundaram, and Adam Trexler for helpful comments on this manuscript. Illustration was provided by Neil Smith (www.neilsmithillustration.co.uk).
J.W. Taraska is supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
The author declares no competing financial interests.
Footnotes
Abbreviations used in this paper:
- cryo-EM
- cryogenic EM
- SRF
- super-resolution fluorescence
References
- Asano S., Fukuda Y., Beck F., Aufderheide A., Förster F., Danev R., and Baumeister W.. 2015. A molecular census of 26S proteasomes in intact neurons. Science. 347:439–442. 10.1126/science.1261197 [DOI] [PubMed] [Google Scholar]
- Chong Y.T., Koh J.L., Friesen H., Duffy S.K., Cox M.J., Moses A., Moffat J., Boone C., and Andrews B.J.. 2015. Yeast proteome dynamics from single cell imaging and automated analysis. Cell. 161:1413–1424. (published erratum appears in Cell 2015. 162:221) 10.1016/j.cell.2015.04.051 [DOI] [PubMed] [Google Scholar]
- de Boer P., Hoogenboom J.P., and Giepmans B.N.. 2015. Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods. 12:503–513. 10.1038/nmeth.3400 [DOI] [PubMed] [Google Scholar]
- Doyon J.B., Zeitler B., Cheng J., Cheng A.T., Cherone J.M., Santiago Y., Lee A.H., Vo T.D., Doyon Y., Miller J.C., et al. 2011. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Biol. 13:331–337. 10.1038/ncb2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M.G. 2012. A man for all seasons: reflections on the life and legacy of George Palade. Annu. Rev. Cell Dev. Biol. 28:1–28. 10.1146/annurev-cellbio-101011-155813 [DOI] [PubMed] [Google Scholar]
- Hell S.W. 2003. Toward fluorescence nanoscopy. Nat. Biotechnol. 21:1347–1355. 10.1038/nbt895 [DOI] [PubMed] [Google Scholar]
- Helma J., Cardoso M.C., Muyldermans S., and Leonhardt H.. 2015. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 209:633–644. 10.1083/jcb.201409074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J.E. 2014. Some personal and historical notes on the utility of “deep-etch” electron microscopy for making cell structure/function correlations. Mol. Biol. Cell. 25:3273–3276. 10.1091/mbc.E14-05-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Bates M., and Zhuang X.. 2009. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78:993–1016. 10.1146/annurev.biochem.77.061906.092014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Irannejad R., Tomshine J.C., Tomshine J.R., Chevalier M., Mahoney J.P., Steyaert J., Rasmussen S.G., Sunahara R.K., El-Samad H., Huang B., and von Zastrow M.. 2013. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 495:534–538. 10.1038/nature12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W., Schorb M., Kaksonen M., and Briggs J.A.. 2012. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 150:508–520. 10.1016/j.cell.2012.05.046 [DOI] [PubMed] [Google Scholar]
- Larson B.T., Sochacki K.A., Kindem J.M., and Taraska J.W.. 2014. Systematic spatial mapping of proteins at exocytic and endocytic structures. Mol. Biol. Cell. 25:2084–2093. 10.1091/mbc.E14-02-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Shao L., Chen B.C., Zhang X., Zhang M., Moses B., Milkie D.E., Beach J.R., Hammer J.A. III, Pasham M., et al. 2015. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 349:aab3500 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lavis L.D., and Betzig E.. 2015. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell. 58:644–659. 10.1016/j.molcel.2015.02.033 [DOI] [PubMed] [Google Scholar]
- Lučič V., Rigort A., and Baumeister W.. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 202:407–419. 10.1083/jcb.201304193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. 1975. Intracellular aspects of the process of protein synthesis. Science. 189:347–358. 10.1126/science.1096303 [DOI] [PubMed] [Google Scholar]
- Roth J., Bendayan M., and Orci L.. 1978. Ultrastructural localization of intracellular antigens by the use of protein A-gold complex. J. Histochem. Cytochem. 26:1074–1081. 10.1177/26.12.366014 [DOI] [PubMed] [Google Scholar]
- Sochacki K.A., Shtengel G., van Engelenburg S.B., Hess H.F., and Taraska J.W.. 2014. Correlative super-resolution fluorescence and metal-replica transmission electron microscopy. Nat. Methods. 11:305–308. 10.1038/nmeth.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J.W. 1977. Exploratory Data Analysis. Addison-Wesley Pub. Co., Reading, MA. 688 pp. [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Wilson E.B. 1925. The Cell in Development and Heredity. Third edition The Macmillan Company, New York. 1232 pp. [Google Scholar]
- Xu K., Zhong G., and Zhuang X.. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]