Stable intronic sequence RNAs (sisRNAs) are present in Drosophila melanogaster, and a sisRNA modulates its host gene expression by repressing a long noncoding RNA during embryogenesis.
Abstract
Stable intronic sequence RNAs (sisRNAs) have been found in Xenopus tropicalis, human cell lines, and Epstein-Barr virus; however, the biological significance of sisRNAs remains poorly understood. We identify sisRNAs in Drosophila melanogaster by deep sequencing, reverse transcription polymerase chain reaction, and Northern blotting. We characterize a sisRNA (sisR-1) from the regena (rga) locus and show that it can be processed from the precursor messenger RNA (pre-mRNA). We also document a cis-natural antisense transcript (ASTR) from the rga locus, which is highly expressed in early embryos. During embryogenesis, ASTR promotes robust rga pre-mRNA expression. Interestingly, sisR-1 represses ASTR, with consequential effects on rga pre-mRNA expression. Our results suggest a model in which sisR-1 modulates its host gene expression by repressing ASTR during embryogenesis. We propose that sisR-1 belongs to a class of sisRNAs with probable regulatory activities in Drosophila.
Introduction
Noncoding RNAs (ncRNAs) have emerged as potent agents that regulate diverse processes during normal development (Matera et al., 2007; Lee, 2012; Rinn and Chang, 2012; Kung et al., 2013; Cech and Steitz, 2014). Introns are noncoding sequences interspersed between the coding exons in most genes of higher eukaryotes. During transcription, intronic RNAs are spliced from the precursor mRNAs (pre-mRNAs) by the splicing machinery (Wahl et al., 2009; Hoskins and Moore, 2012). Whereas mature mRNAs are exported to the cytoplasm, intronic RNAs remain in the nucleus, where they are rapidly degraded (Sharp et al., 1987; Clement et al., 1999). However, intronic RNAs can also be processed into functional ncRNAs, such as the small nucleolar RNAs (snoRNAs), small Cajal body-specific RNAs, and certain miRNAs (Liu and Maxwell, 1990; Leverette et al., 1992; Tycowski et al., 1993; Berezikov et al., 2007; Kim and Kim, 2007; Matera et al., 2007; Okamura et al., 2007; Ruby et al., 2007; Brown et al., 2008). They function in guiding modification of ribosomal RNA (rRNA) and small nuclear RNA or in regulating gene expression (Bushati and Cohen, 2007; Matera et al., 2007). Furthermore, intronic RNAs in plants and mammalian cell cultures may also function as long ncRNAs to modulate transcription and splicing (Heo and Sung, 2011; Guil et al., 2012; Yin et al., 2012; Zhang et al., 2013).
Recently, a class of ncRNAs called stable intronic sequence RNAs (sisRNAs) was discovered in the oocyte nucleus of the frog Xenopus tropicalis (Gardner et al., 2012). Soon after, sisRNAs were also found in human cell lines and the Epstein-Barr virus (Yin et al., 2012; Moss and Steitz, 2013; Zhang et al., 2013). Although the phenomenon is evolutionary conserved, the functions of sisRNAs during development remain poorly understood. Here we identify sisRNAs and present evidence for processing and a function of a sisRNA (sisR-1) in Drosophila melanogaster. We show that sisR-1 can be processed from the regena (rga) pre-mRNA, and nuclear sisR-1 is further processed at the 3′ end to form cytoplasmic sisR-1. During embryogenesis, sisR-1 represses the expression of a cis-natural antisense transcript (cis-NAT) called antisense transcript of rga (ASTR), and ASTR promotes the expression of the rga pre-mRNA. Therefore, our data suggest a regulatory loop whereby sisR-1 modulates rga pre-mRNA expression by repressing ASTR during embryogenesis in Drosophila.
Results and discussion
Identification of sisRNAs
The Drosophila early embryo constitutes an excellent system for identification of sisRNAs because it contains a store of mature and stable RNA molecules. During oogenesis, each egg chamber consists of a growing oocyte and 15 nurse cells surrounded by a sheet of somatic follicle cells (Fig. 1 A). Whereas the germinal vesicle is transcriptionally quiescent, the oocyte receives and stores RNAs from the transcriptionally active polyploid nurse cells (Spradling, 1993). Beginning from stage 10 of oogenesis, massive dumping of RNAs from the nurse cells to the oocyte occurs. A stage 10 egg chamber takes ∼10 h to develop into a stage 14 oocyte, during which time most of the stored RNAs persist (Lasko, 2012). Because zygotic transcription does not begin until 2 h after egg laying, most mature RNAs present in the 0–2 h embryo must have been stable for at least 10–12 h.
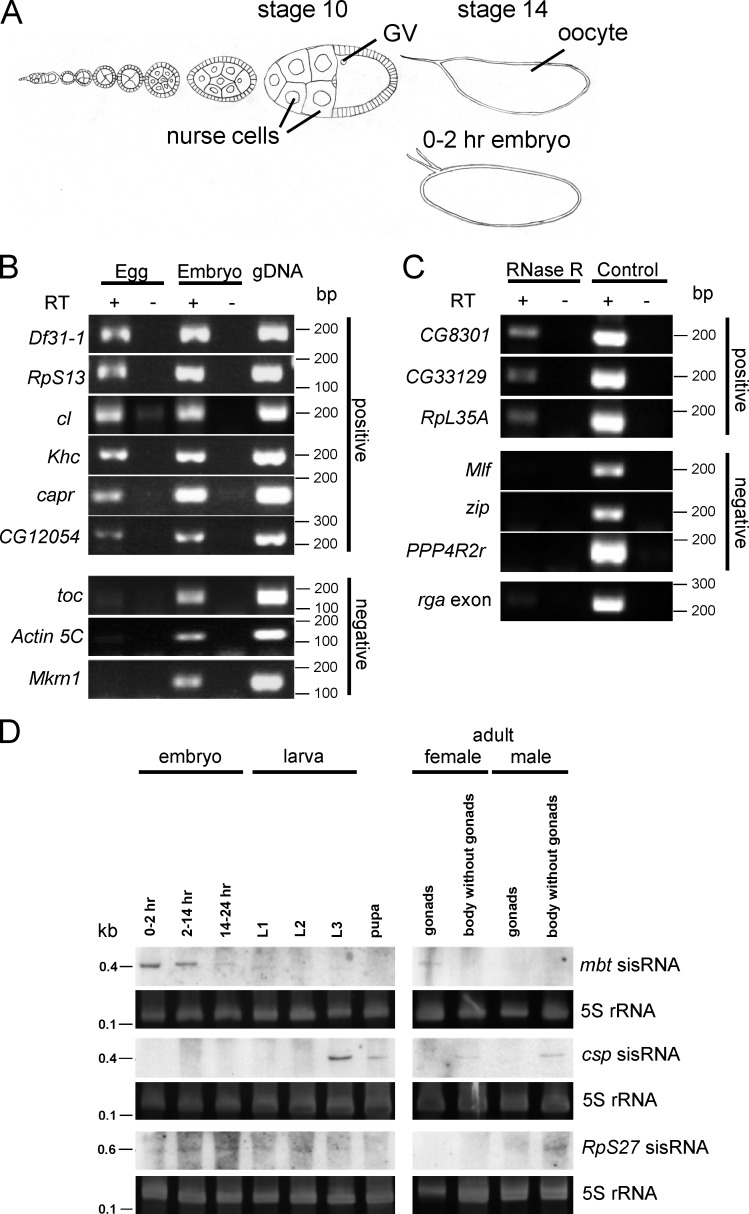
Figure 1.
Identification of sisRNAs. (A) Diagram of an ovariole showing stages in the development of the oocyte and its accompanying nurse cells. Also shown is a 0–2-h embryo. GV, germinal vesicle. (B) RT-PCR showing expression of some sisRNAs in unfertilized eggs. (C) RT-PCR showing expression of some RNase R-resistant sisRNAs in unfertilized eggs. rga exon was used as a positive control for RNase R activity. (D) Northern blots showing expression of mbt, csp, and RpS27 sisRNAs during development and in adult somatic tissues and gonads. The gels were stained with SYBR Gold to visualize 5S rRNA as a loading control. L1, first instar larvae; L2, second instar larvae; L3, third instar larvae.
To identify candidate sisRNAs, we examined RNAs from 0–2-h embryos by deep sequencing. Total RNA was extracted, depleted of rRNA, and subjected to deep sequencing. Reads that were mapped to the introns of each gene were retrieved bioinformatically and analyzed manually. From a list of genes with intronic reads, candidate sisRNAs were identified using the following criteria. First, the intronic reads should display a distinct peak on the genome browser. Second, with reference to the FlyBase annotation (Dmel Release 6), the intronic reads should not derive from retained introns of alternatively spliced isoforms. Finally, the reads should not map to intronic or overlapping protein-coding genes and ncRNAs (including mirtrons, snoRNAs, and long ncRNAs). Using these criteria, we identified 34 candidate sisRNAs in the 0–2-h embryo (Table S1).
Because we used 0–2-h embryos for deep sequencing, which might be contaminated with some late-stage embryos and therefore pre-mRNAs from zygotic transcription, we verified our candidates using unfertilized eggs, which contain a pool of stable and mature RNAs. By RT-PCR, we verified the presence of 31 out of 34 candidates (>90%; Fig. 1 B and Fig. S1 A). We also examined the possibility that some of these sisRNAs may be RNase R resistant, implying that they may be circular. We tested six sisRNAs, and three appeared to have some populations of them being RNase R resistant (Fig. 1 C), suggesting the presence of both linear and circular sisRNAs. We further examined the expression patterns of three sisRNAs by Northern blotting. Expression of the mushroom bodies tiny (mbt) sisRNA was detected primarily in the ovaries and early embryos and dropped gradually as the embryos developed (Fig. 1 D, Fig. S1 B), suggesting that its expression is more restricted to the female germ line. A sisRNA from the cysteine string protein (csp) gene is highly abundant in third instar larvae, pupae, and adult somatic tissues (Fig. 1 D). However, we could not detect its expression in the early embryos, suggesting that its abundance was below the detection level of Northern blotting. Finally, we observed expression of Ribosomal protein S27 (RpS27) sisRNA at a low level during development and in adult male somatic tissues (Fig. 1 D). Our data demonstrate that the expression of sisRNAs is not oocyte specific and that individual sisRNAs display extensive variation in their expression patterns during development and in adult tissues.
sisRNA (sisR-1) from the rga locus
We focused on a candidate sisRNA from the fourth intron of the rga gene locus (Fig. 2 A). The rga gene encodes for the NOT2 protein, a component of the CCR4-NOT deadenylase complex (Frolov et al., 1998). We performed strand-specific RT-PCR using primers within the intronic and exonic peaks (Fig. 2 A) and found that the sisRNA is transcribed from the coding strand (Fig. 2 B). To determine whether the intronic peak represents a sisRNA of discrete size, we performed Northern blotting using two probes that target different regions of the intron. Probe A spans the region within the intronic peak, whereas probe B derives from the region that lacks reads and served as a negative control (Fig. 2 A). We detected a band of ∼300 nucleotides with probe A using RNA from 0–2-h embryos, whereas probe B gave no signal (Fig. 2 C). These results demonstrated that the sequences seen on the browser derive from a discrete sisRNA. Furthermore, expression was also detected in ovaries and unfertilized eggs (Fig. 2 C and Fig. S1 A), suggesting that the sisRNA is maternally deposited. We named this sisRNA sisR-1 for the first sisRNA to be characterized in D. melanogaster. Interestingly, sisR-1, similar to its cognate rga mRNA, is ubiquitously expressed in embryos, larvae, pupae, and adult gonadal and somatic tissues and appears to be developmentally regulated (Fig. 2 D and Fig. S2, A and B).
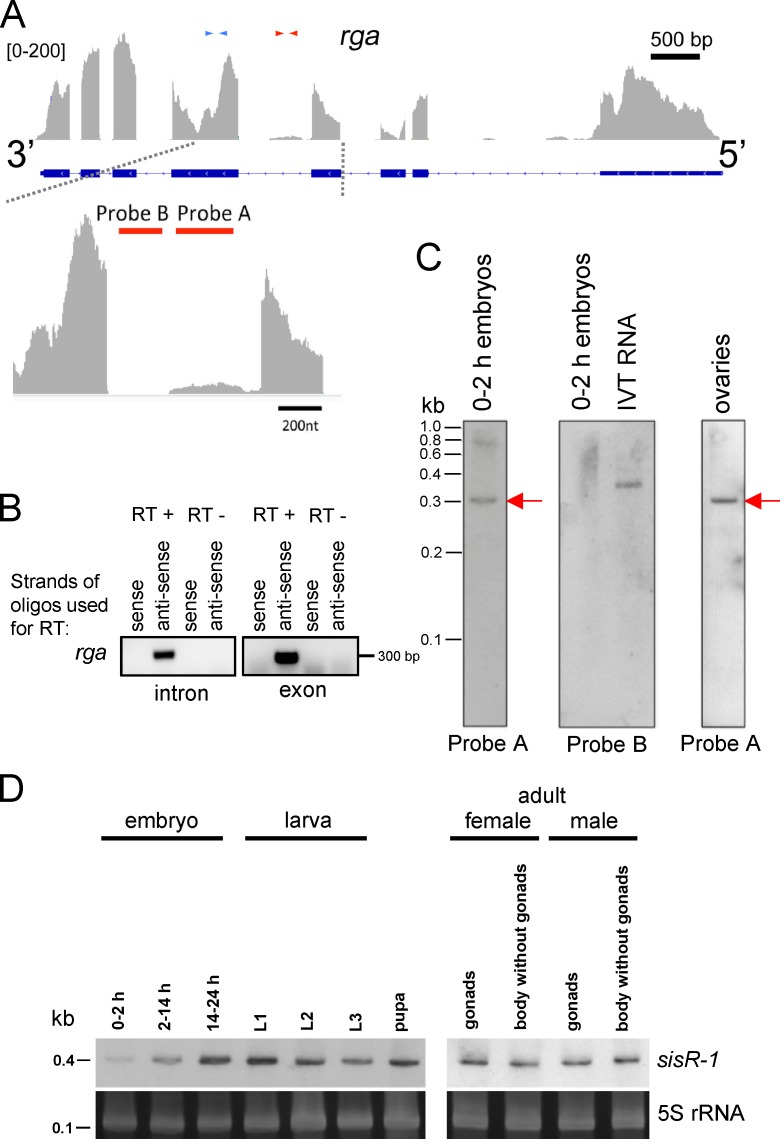
Figure 2.
sisRNA (sisR-1) from the regena locus. (A) Genome browser view of exonic and intronic sequences in the rga locus. Blue and red arrows indicate exonic and intronic primers used for RT-PCR. The bottom panel shows an enlarged view of the intronic peak. Red bars indicate the regions to which probes A and B hybridize. (B) Strand-specific RT-PCR using primers in the intron and exon of rga. (C) Northern blots using probes A and B to detect expression of sisR-1 in 0–2 h embryos and ovaries are shown. A band of ∼300 nt was detected with probe A (red arrows). Probe B gave no detectable signal in RNA from 0–2-h embryos but did hybridize with 10 pg of in vitro transcribed (IVT) RNA. (D) Northern blots showing expression of sisR-1 during development and in adult somatic tissues and gonads. The gels were stained with SYBR Gold to visualize 5S rRNA as a loading control. L1, first instar larvae; L2, second instar larvae; L3, third instar larvae.
To determine the stability of sisR-1, we examined RNA from ovaries that had been treated with actinomycin D to inhibit transcription. Incubation for 30 min was sufficient to inhibit transcription as assayed by 5-ethynyluridine staining (Fig. S2 C; Jao and Salic, 2008). We observed little or no change in sisR-1 levels after 120 min of actinomycin D treatment compared with a partial loss of an unstable species of RNA, gypsy retrotransposon mRNA (Fig. S2 D), which is negatively regulated by the piwi-interacting RNA pathway (Sarot et al., 2004). This result indicated that sisR-1 was relatively more stable than gypsy mRNA. In X. tropicalis, the abundance of sisRNAs was estimated to be ∼1% of the cognate mRNAs (Gardner et al., 2012). We performed the same analysis based on the peak heights of sisR-1 and rga mRNA on the browser (Fig. 2 A) and found that the amount of sisR-1 is ∼5% that of rga mRNA. We also estimated the relative abundance of sisR-1, lariats, and pre-mRNAs in 2–14-h embryos. By Northern blotting we could only detect the presence of sisR-1 but not the lariats and pre-mRNAs (Fig. S2 E), consistent with the idea that sisR-1 is a more stable molecule. We estimated that there are ∼2 × 107 molecules of sisR-1 in 10 μg of total RNA.
sisR-1 can be processed from the rga pre-mRNA
Intronic ncRNAs can be cotranscribed and processed with the cognate pre-mRNA or independently transcribed from its own promoter (Okamura et al., 2007; Ruby et al., 2007; Brown et al., 2008; Heo and Sung, 2011; Yin et al., 2012). Recent studies have provided evidence that sisRNAs are not independently transcribed molecules in Xenopus and mammalian cell lines (Gardner et al., 2012; Yin et al., 2012; Zhang et al., 2013). We therefore asked if sisR-1 can be processed from its cognate pre-mRNA. We used a P-element insertion line EY10731 that allows Gal4-inducible expression of the rga pre-mRNA (Fig. 3 A). Overexpression of rga pre-mRNA in the ovaries by MTD-Gal4, led to a concomitant increase in expression of sisR-1 (Fig. 3 B), consistent with the idea that the rga pre-mRNA is processed into sisR-1. To ask whether the full-length intronic sequence itself is sufficient to produce sisR-1, we cloned both full length isoforms of the rga fourth intron. Long and short rga introns differ by their 5′ splice sites, with intact splice sites in between coding sequences of dsRed and myc (Fig. 3 C; Okamura et al., 2007) and generated transgenic flies. Overexpression of both intron isoforms in the ovaries by MTD-Gal4 resulted in increased expressions of sisR-1 (Fig. 3 D), indicating that the intronic sequence itself is sufficient to generate sisR-1. To test whether the rga intron contains an independent promoter, and the transgene insertion has any effect on endogenous sisR-1 expression, we examined the levels of sisR-1 in control (y w) versus UAS-rga intron-L-myc (without Gal4 induction) females. We did not detect any differences in sisR-1 levels (Fig. S2 F), therefore excluding the presence of an intronic promoter and indirect effect of transgene insertion.
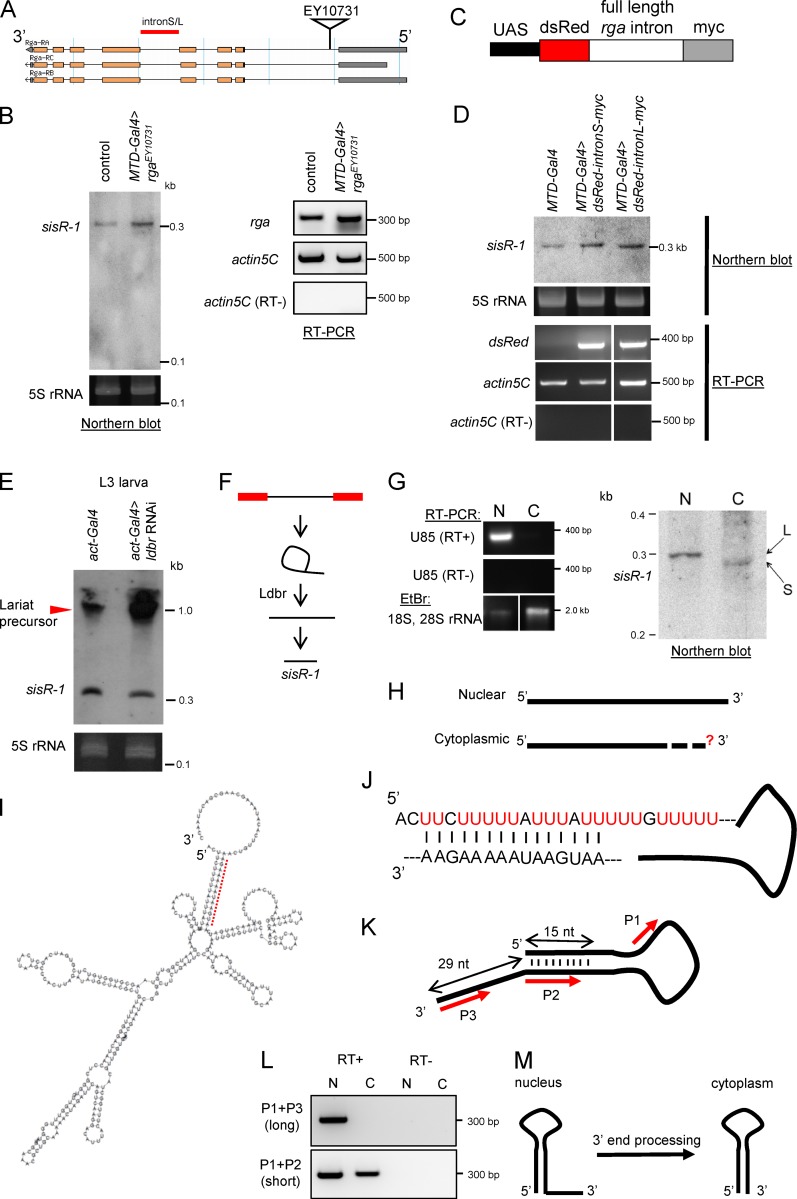
Figure 3.
Processing of sisR-1. (A) Diagram showing the insertion of an EP element EY10731 used to overexpress the rga pre-mRNA. L, long; S, short. (B) A Northern blot showing expression of sisR-1 in the ovaries of controls versus rga overexpression flies. RT-PCR showing expression of rga mRNA and actin5C in the ovaries of controls versus rga overexpression flies. (C) Diagram showing the construct used to overexpress rga intronic sequences. (D) Northern blot showing expression of sisR-1 in ovaries of controls versus rga intronic sequence overexpression flies. RT-PCR verifies expression of dsRed mRNA in the ovaries of dsRed-intron-myc overexpression flies but not in controls. (E) Northern blots showing expression of sisR-1 in the controls versus ldbr RNAi third instar larvae. (F) Model for processing of sisR-1 from the rga pre-mRNA. (G) Northern blot showing detection of sisR-1 in both the nuclear (N) and cytoplasmic (C) fractions of 14–24-h embryos. RT-PCR showing the expression of U85 only in the nuclear fraction. EtBr staining showing the enrichment of 18S, 28S rRNA in the cytoplasmic fraction. RNAs were run on the same gel, and the intervening lanes were removed (white vertical lines). (H) Diagram showing the 5′ and 3′ ends of the nuclear and cytoplasmic sisR-1 determined by RACE. Question mark depicts unknown 3′ end of cytoplasmic sisR-1. (I) Predicted structure of nuclear sisR-1 by Vienna RNA fold software. Red dotted region is shown in J. (J) Magnified view of the dotted region in I shows the base pairing. (K) Diagram with red arrows to indicate regions where the primers anneal to. (L) RT-PCR showing the detection of the short, but not long, isoform of sisR-1 in the cytoplasmic fraction. (M) Model showing the processing of nuclear sisR-1 to cytoplasmic sisR-1 by 3′ end processing.
Because snoRNAs and mirtrons are processed from spliced intronic transcripts and require lariat debranching (ldbr) enzyme activity for their biogenesis (Brown et al., 2008), we asked whether ldbr is required for the production of sisR-1. Knockdown of ldbr in third instar larvae using a previously published ldbr RNAi transgenic fly (Conklin et al., 2005; Okamura et al., 2007) by act-Gal4 resulted in a decrease in expression of sisR-1 (Fig. 3 E). Importantly, we also observed a concomitant accumulation of higher molecular weight precursors, which were presumably the intron lariats (Fig. 3 E, arrowhead). Collectively, our data are consistent with a model in which sisR-1 is processed from a predominant pathway that requires canonical splicing and debranching of intron lariats (Fig. 3 F).
3′ end processing of sisR-1
To determine whether sisR-1 localizes to the nucleus or cytoplasm, we collected 14–24-h embryos where sisR-1 expression is the highest during embryogenesis and performed nuclear-cytoplasmic fractionation. Relatively clean nuclear and cytoplasmic fractions were obtained as revealed by nuclear-specific expression of U85 and cytoplasmic-enrichment of 18S and 28S rRNA (Fig. 3 G). We performed Northern blotting and detected sisR-1 in both the nuclear and cytoplasmic fractions (Fig. 3 G). Curiously, cytoplasmic sisR-1 migrated faster than nuclear sisR-1, suggesting that they had different sizes (Fig. 3 G). It further implies processing of nuclear sisR-1 to produce cytoplasmic sisR-1.
Both nuclear and cytoplasmic sisR-1 were susceptible to RNase R-mediated degradation, indicating that they are not circular molecules or lariats (Fig. S2 G). We therefore mapped the ends of nuclear sisR-1 by 5′ and 3′ rapid amplification of cDNA ends (RACE) to determine its full-length sequence (Fig. S2 H). Using the same method, we found that cytoplasmic and nuclear sisR-1 have the same 5′ end (Fig. 3 H). However, despite various attempts, we were unable to map the 3′ end of cytoplasmic sisR-1, probably because of the presence of an unknown RNA modification at the 3′ end, which made it technically difficult (Raabe et al., 2014). We next used the Vienna RNA fold web server to predict the secondary structure of nuclear sisR-1. The 5′ end of nuclear sisR-1 contains a U-rich tract that was predicted to form 15 nucleotide base-pairing with a region near its 3′ end, leaving behind an exposed 3′ tail of 29 nucleotides (Fig. 3, I and J). Collectively, our findings suggest a possibility that cytoplasmic sisR-1 is formed by 3′ end processing of nuclear sisR-1. To test this idea, we designed primers that anneal to different regions of the 3′ end of nuclear sisR-1 (P2 anneals to both long and short sisR-1 and P3 anneals specifically to long sisR-1; Fig. 3 K). By performing RT-PCR using the different primer pairs, we detected the presence of short, but not long, sisR-1 in the cytoplasm (Fig. 3 L), verifying that cytoplasmic sisR-1 is formed by 3′ end processing (Fig. 3 M).
ASTR promotes the expression of rga pre-mRNA
To search for potential molecular functions of sisR-1, we examined the genome organization of the rga locus. There is an annotated long ncRNA CR43628 transcribed in the antisense orientation with respect to rga (Fig. 4 A). We named this cis-NAT ASTR. The 3′ end of ASTR is predicted to form a 76-nucleotide base pairing with the 3′ end of nuclear sisR-1 (Fig. 4 A), suggesting that sisR-1 may bind to and regulate ASTR. Furthermore, the predicted secondary structure of sisR-1 suggests that the 3′ end of nuclear sisR-1 has an exposed 29 nt tail, further raising the possibility that it may bind to ASTR in the nucleus. By performing Northern blotting using a probe that is specific to ASTR (Fig. 4 A), we found that ASTR was highly expressed in 2–14-h embryos (Fig. 4 B). ASTR was also found to be predominantly in the nucleus as assayed by strand-specific RT-PCR (Fig. 4 C).
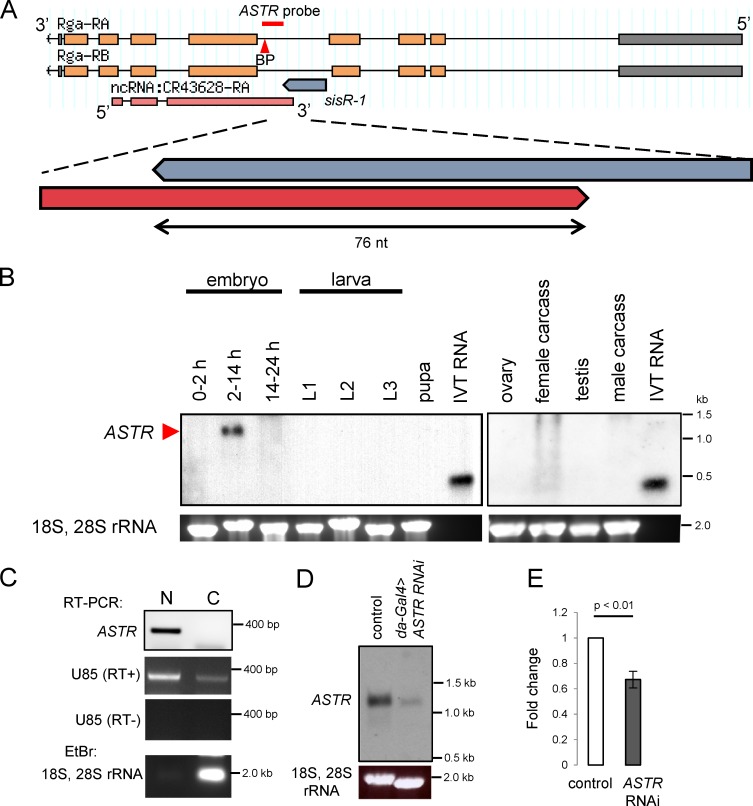
Figure 4.
ASTRpromotes robust expression of rga pre-mRNA. (A) Genomic locus showing regions of rga, sisR-1, and ASTR. Red bar indicates the region where the ASTR probe binds. Red arrowhead points to a putative branch point (BP) CTAAT. (B) Northern blots showing expression of ASTR in 2–14-h embryos. In vitro transcribed (IVT) RNA was used as positive controls. The gels were stained with EtBr to visualize 18S, 28S rRNA as a loading control. (C) Strand-specific RT-PCR detecting the presence of ASTR in the nuclear but not the cytoplasmic fraction of 2–14-h embryos. RT-PCR showing the high expression of U85 in the nuclear fraction. EtBr staining showing the enrichment of 18S, 28S rRNA in the cytoplasmic fraction. (D) Northern blot showing the expression of ASTR in controls versus da-Gal4>ASTR RNAi 2–14-h embryos. The gel was stained with EtBr to visualize 18S, 28S rRNA as a loading control. (E) qPCR data showing relative expression of rga pre-mRNA normalized to actin5C in the controls versus da-Gal4>ASTR RNAi 2–14-h embryos. Error bars represent SD. n = 3. P-value represents t test.
cis-NATs have been shown to regulate the expression of pre-mRNAs of their cognate genes by various mechanisms in different contexts (Wight and Werner, 2013; Herzog et al., 2014; Xue et al., 2014). ASTR and rga show similar expression patterns during embryogenesis, where both are highly expressed in the 2–14-h embryos and expression levels drop in the 14–24-h embryos and later developmental stages (Figs. 4 B and 5 D; and Fig. S2, A and B). This suggests a possibility that ASTR promotes rga pre-mRNA expression. We therefore tested whether ASTR has a role in regulating the rga pre-mRNA expression. We designed a shRNA that specifically targets ASTR under the UASp promoter and drove the expression in the embryos using da-Gal4. Expression of ASTR shRNA resulted in a robust knockdown of ASTR in 2–14-h embryos as assayed by Northern blotting (Fig. 4 D). By performing quantitative PCR (qPCR), we observed a significant decrease in the expression of rga pre-mRNA in ASTR RNAi 2–14-h embryos compared with controls (Fig. 4 E). This result could not have been the result of an indirect knockdown of rga pre-mRNA by the passenger strand because we and others had shown that shRNAs do not target pre-mRNAs (Fig. S3, A and B; discussed in the following section; Yin et al., 2012; Zhang et al., 2013). Therefore, our data suggest that ASTR plays a role in promoting robust rga pre-mRNA expression during embryogenesis.
Figure 5.
sisR-1promotes robust repression of ASTR. (A) Northern blot showing the expression of ASTR in controls versus da-Gal4>sisR-1 2–14-h embryos. (B) qPCR data showing relative expression of rga pre-mRNA normalized to actin5C in the controls versus da-Gal4>sisR-1 2–14-h embryos. Error bars represent SD. n = 3. P-value represents t test. (C) Northern blots showing expression of ASTR in controls versus embryos expressing sisR-1 shRNA. The gels were stained with EtBr to visualize 18S, 28S rRNA as a loading control. (D) qPCR data showing fold change of rga pre-mRNA expression (normalized to actin5C) from 2–14-h to 14–24-h embryos in controls versus sisR-1 RNAi versus sisR-1 RNAi + ASTR RNAi embryos. Error bars represent SD. n = 3. P-value represents t test. (E) Model for the regulatory relationship between sisR-1, ASTR, and rga pre-mRNA during embryogenesis.
sisR-1 represses the expression of ASTR
The expression pattern of ASTR during a specific window of 2–14-h embryogenesis suggests a mechanism that facilitates robust downregulation of ASTR during late embryogenesis (14–24 h). Interestingly, expression of sisR-1 increased gradually during embryogenesis and peaked only at 14–24 h (Fig. 2 D). These reciprocal expression patterns between sisR-1 and ASTR during embryogenesis suggest that sisR-1 may facilitate the robust downregulation of ASTR during late embryogenesis.
We overexpressed sisR-1 in 2–14-h embryos by driving UAS-dsRed-rga intronL-myc with da-Gal4, and we observed a robust downregulation of ASTR (Fig. 5 A). This result indicated that sisR-1 acts in trans to repress ASTR. Further supporting a role for ASTR in promoting rga pre-mRNA expression, we also observed a concomitant drop in rga pre-mRNA expression in sisR-1–overexpressing 2–14-h embryos (Fig. 5 B).
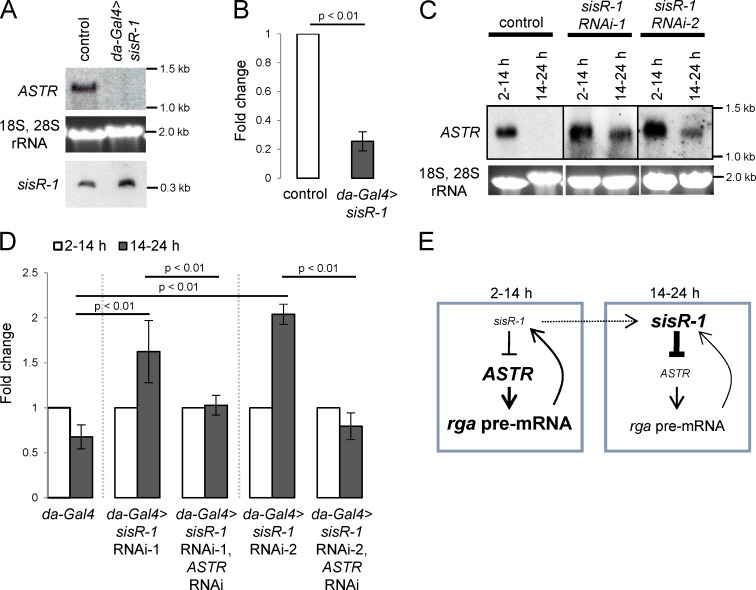
We generated transgenic flies expressing two independent shRNAs that target specifically to two regions of sisR-1 under the UASp promoter. Expression of sisR-1 shRNAs driven by da-Gal4 in ovaries resulted in four- to fivefold decrease in sisR-1, but not the rga pre-mRNA levels (Fig. S3, A and B), consistent with published studies that shRNAs specifically target sisRNAs but not pre-mRNAs (Yin et al., 2012; Zhang et al., 2013). This result indicates that the knockdown was specific to sisR-1 but not the rga pre-mRNA. Next, we tested the idea that sisR-1 may facilitate robust downregulation of ASTR during late embryogenesis. In support of this idea, we observed a delay in the downregulation of ASTR during 14–24-h embryogenesis in sisR-1 RNAi embryos, but not in controls (Fig. 5 C). The hatching rates of sisR-1 RNAi embryos were similar to that of the controls (Fig. S3 C), indicating that the delay in downregulation of ASTR was not the result of a delay in developmental progression.
In control embryos, the expression of rga pre-mRNA decreases from 2–14 h to 14–24 h (Fig. 5 D), which correlates with a decrease in the expression of ASTR (Fig. 4 B). Because we observed a less robust downregulation of ASTR in sisR-1 RNAi embryos, and ASTR promotes rga pre-mRNA expression, we asked if the downregulation of rga pre-mRNA was also perturbed. As assayed by qPCR, the downregulation of rga pre-mRNA from 2–14-h to 14–24-h embryos was significantly perturbed in sisR-1 RNAi embryos (Fig. 5 D). Importantly, the failure to downregulate rga pre-mRNA in sisR-1 RNAi embryos was rescued by simultaneously knocking down ASTR, indicating that the effect was mediated by ASTR (Fig. 5 D). Therefore, our data suggest that sisR-1 represses ASTR to promote robust downregulation of rga pre-mRNA from 2–14-h to 14–24-h embryogenesis (Fig. 5 E).
In this study, we report a sisRNA (sisR-1) produced from the rga locus, likely processed from the rga pre-mRNA. sisR-1 functions to repress the expression of ASTR, which is a positive regulator of the rga pre-mRNA. Collectively, our findings suggest a model whereby sisR-1 modulates its host gene expression by repressing ASTR during embryogenesis in Drosophila (Fig. 5 E). Our results provide an explanation for the wild-type expression patterns of sisR-1, ASTR, and rga pre-mRNA from 2–14-h to 14–24-h embryos. During 2–14-h embryogenesis, a high level of ASTR promotes robust expression of rga pre-mRNA. A high level of rga pre-mRNA in turn contributes to production of more sisR-1. Over time, sisR-1 accumulates to a level that is sufficient to repress ASTR during 14–24-h embryogenesis. Downregulation of ASTR then results in a lower expression of rga pre-mRNA. Therefore, sisR-1 appears to modulate a negative feedback loop in promoting robust downregulation of its host gene during development. ncRNAs, such as miRNAs and long ncRNAs, have been shown to modulate gene expression via feedback loops (Herranz and Cohen, 2010; Peláez and Carthew, 2012; Xue et al., 2014). Feedback loops play important roles in biological systems to maintain a robustness network of gene expression. We propose that sisR-1 is present in the nucleus to repress any uncontrolled accumulation of ASTR, which may have deleterious effects on rga expression.
Among the remaining sisRNA loci that we identified, only the Decondensation factor 31 (Df31) locus have an annotated cis-NAT. It is possible that Df31 sisRNAs regulate the cis-NAT in the same locus. Because sisR-1 can act in trans, it is possible that other sisRNAs may have targets in trans that remain to be discovered. In general, sisRNAs may be present as a quality control and surveillance mechanism to target any antisense transcripts, which may affect gene expression.
In conclusion, we have shown that sisRNAs are present in D. melanogaster and characterized a novel sisRNA (sisR-1) that functions to modulate its host gene expression during embryogenesis by repressing a cis-NAT.
Materials and methods
Fly strains
The y w strain was used as controls unless otherwise stated. Fly strains used were rgaEY10731 (a P-element insertion in the rga locus that allows Gal4-inducible expression; Bloomington 20207), MTD-Gal4 (a driver that expresses in the germline; Petrella et al., 2007), da-Gal4 (a driver that expresses ubiquitously), actin-Gal4 (a driver that expresses ubiquitously), and UAS-ldbr RNAi (an RNAi line that knocks down the expression of ldbr; Conklin et al., 2005). For embryo collection, females were fed with wet yeast for 1–2 d before mating. They were then mated to males in bottles at 25°C. For generation of dsRed-intron-myc transgenic flies, PCR of full-length intronic sequences was performed using primers that contain the AscI–NotI restriction sites. The PCR product was purified and cut using AscI and NotI, and ligated into AscI–NotI digested UAS-dsRed-intron-myc plasmid (Okamura et al., 2007). Transgenic flies were generated by Genetics Services using P-element–mediated insertion (Rubin and Spradling, 1982). For generation of sisR-1 and ASTR shRNA transgenic flies, design of shRNAs targeting sisR-1 and ASTR and cloning into Valium22 plasmid were performed as previously described (Ni et al., 2011). Sequences were chosen to avoid off-target effects. Transgenic flies were generated by Genetics Services using phiC31 integrase-mediated insertion into 25C7 landing site (Bischof et al., 2007). Oligo sequences are available in Table S2.
RNA extraction
Tissues were homogenized with a plastic pestle in 1.5 ml plastic Eppendorf tubes, and RNA was extracted using the TRIzol extraction protocol (Ambion; Thermo Fisher Scientific), Direct-zol RNA miniprep kit (Zymo Research), or RNeasy plus mini kit (QIAGEN). For deep sequencing, rRNA depletion was performed using the Ribo-Zero magnetic kit (Epicentre). RNA was quantified with the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and further characterized with a Bioanalyzer 2100 (Agilent Technologies).
Sequencing and sequence analysis
Construction of cDNA libraries, sequencing, and analysis were performed as previously described (Gardner et al., 2012). RNA was fragmented, and cDNA library was constructed using random hexamers as primers following the procedure in the Illumina TruSeq RNA sample preparation guide. 100-bp sequencing was performed using the Illumina HiSeq 2000 sequencer. Reads were aligned to the D. melanogaster genome (dm3 genome release) using TopHat version 1.4.0 and Bowtie version 0.12.9 sequence alignment programs (Langmead et al., 2009; Trapnell et al., 2009).
Identification of candidate sisRNAs
Sequenced and aligned reads were passed through a Perl-filtering algorithm to determine if they were internal to an intron or overlapped with the 5′ or 3′ junction of each intron in the D. melanogaster (dm3) genome. Each intron was indexed, and reads that aligned to the index were parsed and counted to determine molecule size. Sequence information was then extracted for each putative molecule and pairwise aligned using Blast (Altschul et al., 1997). Positive Blast hits were then aligned with Clustalw2 to determine actual sequence alignment for further downstream analysis (Larkin et al., 2007). Candidates with ≤100 reads were first selected because they are less likely to be abundant alternatively spliced mRNAs, snoRNAs, or other annotated ncRNAs. Candidates were manually checked on the Integrative Genomics Viewer browser (Nicol et al., 2009) with cross-reference to FlyBase to eliminate candidates that might be alternatively spliced mRNAs, intronic or overlapping protein-coding genes, or ncRNAs (including mirtrons, snoRNAs, and long ncRNAs). Furthermore, on visual examination, the reads should cluster at a particular section of the intron to form a discernible peak and not be scattered along the entire intron.
RT-PCR
For standard RT-PCR, total RNA was reverse transcribed for 1 h using AMV-RT (New England BioLabs) or M-MLV RT (Promega) with random hexamers. The cDNA was then used for PCR. For strand-specific RT-PCR, the one-step RT-PCR kit (QIAGEN) was used. RT was performed with a single primer, followed by PCR with both primers at 40 cycles. PCR products were visualized on 1%–2% agarose gels. For qPCR, SYBR Fast qPCR kit master mix (2×) universal (Kapa Biosystems) with addition of ROX reference dye high was used with the Applied Biosystems 7900HT Fast Real-Time PCR system.
5′ RACE and 3′ RACE
For 5′ RACE, a 5′ adaptor was ligated to RNA using T4 RNA ligase (Roche). RT was performed using sisR-1–specific reverse primer. For 3′ RACE, poly(U) tailing was performed to RNA using poly(U) polymerase (New England BioLabs). RT was performed using an adaptor containing poly(A). Nested PCR was performed using sisR-1– and adaptor-specific primers. PCR products were run on an agarose gel, purified and cloned into pGEM-T-Easy vector (Promega), and sequenced.
Nuclear-cytoplasmic fractionation
Nuclear-cytoplasmic fraction was performed as described previously (Liu et al., 2011). Embryos were dechorionated in 50% bleach for 2 min and washed extensively before homogenization in cold lysis buffer (100 mM potassium acetate, 0.1% Triton X-100, 50 mM Hepes, pH 7.4, 2 mM magnesium acetate, 10% glycerol, 1 mM DTT, and 1× complete mini EDTA-free protease inhibitor cocktail; Roche). Lysate was spun at 1,300 g for 10 min at 4°C. The supernatant was collected as the cytoplasmic fraction. The crude nuclear pellet was homogenized in cold lysis buffer and spun at 1,300 g for 10 min at 4°C. The resultant pellet was collected as the nuclear fraction. After nuclear-cytoplasmic fractionation, RNA was extracted and dissolved in equal volumes of water. Equal volumes equivalent of RNA were used for Northern blot and RT-PCR analyses.
RNase R treatment
RNA was incubated with 10X RNase R reaction buffer (Epicentre) at 65°C for 5 min to denature the RNA. Samples were then cooled on ice for 2 min before adding 1 µl RNasin (40 U/µl; Promega) and 1 µl RNase R (20 U/µl; Epicentre) and incubated at 37°C overnight.
Northern blotting
For sisRNA analysis, 10–30 µg of total RNA was separated on an 8% denaturing polyacrylamide gel (8 M urea, 1× TBE buffer). RNA was transferred by electrophoresis onto a nylon membrane (Zeta-Probe GT membrane; Bio-Rad Laboratories). For ASTR analysis, 8–10 µg of total RNA was separated on a 0.8% agarose/formaldehyde gel. RNA was transferred by capillary action onto a nylon membrane (Zeta-Probe GT membrane; Bio-Rad Laboratories). RNA was then UV cross-linked to the membranes, prehybridized with salmon sperm DNA, and hybridized overnight (14–16 h) with probes in DIG Easy Hyb Granules (Roche) at 51°C (for mbt sisRNA) or 42°C (for all other sisRNAs). DIG-labeled DNA probes were synthesized by PCR with genomic DNA as the template and purified before using. After hybridization, the membranes were rinsed once with 2× SSC, followed by one wash with 2× SSC and 0.1% SDS, and two washes with 0.1× SSC and 0.1% SDS. Detection was performed using the CDP-Star chemiluminescent substrate (Roche) and exposed on x-ray films.
Actinomycin D treatment
Ovaries were incubated with 20 µg/ml actinomycin D in Grace’s medium with constant rocking at room temperature.
5-ethynyluridine labeling
Ovaries were incubated with 5-ethynyluridine in Grace’s medium (1:50) at room temperature. Samples were fixed in 4% paraformaldehyde in Grace’s medium for 10 min, rinsed twice in PBST (0.2% Triton X-100), and blocked for 30 min. Detection using Alexa Fluor 488 was performed in accordance with the manufacturer’s protocol (Life Technologies). Ovaries were stained with DAPI (4′,6-diamidino-2-phenylindole), mounted in Vectashield (Vector Laboratories), and examined under a fluorescence microscope (Olympus BX61) using 10× dry NA 0.2 and 40× dry NA 0.75 objectives at room temperature. Images were taken using a digital charge couple device camera (C4742-95; Hamamatsu) and acquired using Slidebook 4.1 software and processed (adjustment of brightness and contrast) using ImageJ and Photoshop software.
Accession number
Expression data were deposited in the NCBI Gene Expression Omnibus under accession no. GSE69212.
Online supplemental material
Fig. S1 shows the identification of sisRNAs by RT-PCR and Northern blotting. Fig. S2 shows the characterization of sisR-1 and rga expression. Fig. S3 shows the verification of sisR-1 knockdown by shRNAs. Table S1 shows a list of candidate sisRNAs in 0–2 h embryos. Table S2 shows a list of oligonucleotides used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507065/DC1.
Supplementary Material
Acknowledgments
We thank Joseph Gall in whose laboratory these studies were started. We thank Megan Kutzer, Allison Pinder, Nicholas Ingolia, Eugene Gardner, and Fred Tan for help in deep sequencing and bioinformatics, Allan Spradling and members of the Gall, Spradling, Kai, and Pek laboratories for technical support and discussion, Katsutomo Okamura for sharing fly strains, plasmids, and critical reading of the manuscript, and the Bloomington Stock Center. We acknowledge the core facilities at the Carnegie Institution (Department of Embryology) and Temasek Life Sciences Laboratory for their support over the course of this study.
J.W. Pek is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. The authors are supported by the Temasek Life Sciences Laboratory.
The authors declare no further competing financial interests.
Footnotes
Abbreviations used in this paper:
- cis-NAT
- cis-natural antisense transcript
- ncRNA
- noncoding RNA
- qPCR
- quantitative PCR
- pre-mRNA
- precursor mRNA
- rRNA
- ribosomal RNA
- sisRNA
- stable intronic sequence RNA
- snoRNA
- small nucleolar RNA
- RACE
- rapid amplification of cDNA ends
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J.. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Chung W.J., Willis J., Cuppen E., and Lai E.C.. 2007. Mammalian mirtron genes. Mol. Cell. 28:328–336. 10.1016/j.molcel.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W., Marshall D.F., and Echeverria M.. 2008. Intronic noncoding RNAs and splicing. Trends Plant Sci. 13:335–342. 10.1016/j.tplants.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Bushati N., and Cohen S.M.. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23:175–205. 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Cech T.R., and Steitz J.A.. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 157:77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Clement J.Q., Qian L., Kaplinsky N., and Wilkinson M.F.. 1999. The stability and fate of a spliced intron from vertebrate cells. RNA. 5:206–220. 10.1017/S1355838299981190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Goldman A., and Lopez A.J.. 2005. Stabilization and analysis of intron lariats in vivo. Methods. 37:368–375. 10.1016/j.ymeth.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Frolov M.V., Benevolenskaya E.V., and Birchler J.A.. 1998. Regena (Rga), a Drosophila homolog of the global negative transcriptional regulator CDC36 (NOT2) from yeast, modifies gene expression and suppresses position effect variegation. Genetics. 148:317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner E.J., Nizami Z.F., Talbot C.C. Jr., and Gall J.G.. 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 26:2550–2559. 10.1101/gad.202184.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Soler M., Portela A., Carrère J., Fonalleras E., Gómez A., Villanueva A., and Esteller M.. 2012. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 19:664–670. 10.1038/nsmb.2315 [DOI] [PubMed] [Google Scholar]
- Heo J.B., and Sung S.. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 331:76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Herranz H., and Cohen S.M.. 2010. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 24:1339–1344. 10.1101/gad.1937010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog V.A., Lempradl A., Trupke J., Okulski H., Altmutter C., Ruge F., Boidol B., Kubicek S., Schmauss G., Aumayr K., et al. . 2014. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat. Genet. 46:973–981. 10.1038/ng.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins A.A., and Moore M.J.. 2012. The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem. Sci. 37:179–188. 10.1016/j.tibs.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.Y., and Salic A.. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA. 105:15779–15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., and Kim V.N.. 2007. Processing of intronic microRNAs. EMBO J. 26:775–783. 10.1038/sj.emboj.7601512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T., Colognori D., and Lee J.T.. 2013. Long noncoding RNAs: Past, present, and future. Genetics. 193:651–669. 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. . 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lasko P. 2012. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4:a012294 10.1101/cshperspect.a012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T. 2012. Epigenetic regulation by long noncoding RNAs. Science. 338:1435–1439. 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- Leverette R.D., Andrews M.T., and Maxwell E.S.. 1992. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 71:1215–1221. 10.1016/S0092-8674(05)80069-8 [DOI] [PubMed] [Google Scholar]
- Liu J., and Maxwell E.S.. 1990. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 18:6565–6571. 10.1093/nar/18.22.6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Qi H., Wang J., and Lin H.. 2011. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 138:1863–1873. 10.1242/dev.059287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., and Terns M.P.. 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220. 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- Moss W.N., and Steitz J.A.. 2013. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics. 14:543 10.1186/1471-2164-14-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Q., Zhou R., Czech B., Liu L.P., Holderbaum L., Yang-Zhou D., Shim H.S., Tao R., Handler D., Karpowicz P., et al. . 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 8:405–407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G. Jr., Raja A., and Loraine A.E.. 2009. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 25:2730–2731. 10.1093/bioinformatics/btp472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J.W., Duan H., Tyler D.M., and Lai E.C.. 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 130:89–100. 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez N., and Carthew R.W.. 2012. Biological robustness and the role of microRNAs: A network perspective. Curr. Top. Dev. Biol. 99:237–255. 10.1016/B978-0-12-387038-4.00009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L.N., Smith-Leiker T., and Cooley L.. 2007. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 134:703–712. 10.1242/dev.02766 [DOI] [PubMed] [Google Scholar]
- Raabe C.A., Tang T.H., Brosius J., and Rozhdestvensky T.S.. 2014. Biases in small RNA deep sequencing data. Nucleic Acids Res. 42:1414–1426. 10.1093/nar/gkt1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., and Chang H.Y.. 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81:145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M., and Spradling A.C.. 1982. Genetic transformation of Drosophila with transposable element vectors. Science. 218:348–353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., and Bartel D.P.. 2007. Intronic microRNA precursors that bypass Drosha processing. Nature. 448:83–86. 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E., Payen-Groschêne G., Bucheton A., and Pélisson A.. 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 166:1313–1321. 10.1534/genetics.166.3.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A., Konarksa M.M., Grabowski P.J., Lamond A.I., Marciniak R., and Seiler S.R.. 1987. Splicing of messenger RNA precursors. Cold Spring Harb. Symp. Quant. Biol. 52:277–285. 10.1101/SQB.1987.052.01.033 [DOI] [PubMed] [Google Scholar]
- Spradling A.C. 1993. Developmental Genetics of Oogenesis. In The Development of Drosophila melanogaster. Bate A.M., editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 1–70. [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L.. 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Shu M.D., and Steitz J.A.. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7:1176–1190. 10.1101/gad.7.7a.1176 [DOI] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., and Lührmann R.. 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell. 136:701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wight M., and Werner A.. 2013. The functions of natural antisense transcripts. Essays Biochem. 54:91–101. 10.1042/bse0540091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Ye Q., Anson S.R., Yang J., Xiao G., Kowbel D., Glass N.L., Crosthwaite S.K., and Liu Y.. 2014. Transcriptional interference by antisense RNA is required for circadian clock function. Nature. 514:650–653. 10.1038/nature13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q.F., Yang L., Zhang Y., Xiang J.F., Wu Y.W., Carmichael G.G., and Chen L.L.. 2012. Long noncoding RNAs with snoRNA ends. Mol. Cell. 48:219–230. 10.1016/j.molcel.2012.07.033 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., and Chen L.L.. 2013. Circular intronic long noncoding RNAs. Mol. Cell. 51:792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.