In C. elegans, a meiotic checkpoint monitors synapsis between homologous chromosomes. Mad1, Mad2, and Bub3 are required for this checkpoint and negatively regulate synapsis, suggesting a possible role for these proteins in monitoring the stability of homologue pairing.
Abstract
Homologue synapsis is required for meiotic chromosome segregation, but how synapsis is initiated between chromosomes is poorly understood. In Caenorhabditis elegans, synapsis and a checkpoint that monitors synapsis depend on pairing centers (PCs), cis-acting loci that interact with nuclear envelope proteins, such as SUN-1, to access cytoplasmic microtubules. Here, we report that spindle assembly checkpoint (SAC) components MAD-1, MAD-2, and BUB-3 are required to negatively regulate synapsis and promote the synapsis checkpoint response. Both of these roles are independent of a conserved component of the anaphase-promoting complex, indicating a unique role for these proteins in meiotic prophase. MAD-1 and MAD-2 localize to the periphery of meiotic nuclei and interact with SUN-1, suggesting a role at PCs. Consistent with this idea, MAD-1 and BUB-3 require full PC function to inhibit synapsis. We propose that SAC proteins monitor the stability of pairing, or tension, between homologues to regulate synapsis and elicit a checkpoint response.
Introduction
Cell cycle checkpoints ensure accurate chromosome segregation by monitoring the progression of critical events (Murray, 1992). When errors occur, checkpoints prevent the production of aneuploid daughter cells by either arresting the cell cycle to promote error correction or targeting the cell for apoptosis. Aneuploidy is a hallmark of tumor cells undergoing mitosis (Kops et al., 2005) and is associated with birth defects and infertility during sexual reproduction (Hassold and Hunt, 2001).
Sexual reproduction requires meiosis, a specialized cell division that produces gametes such as eggs and sperm. During meiotic prophase, homologous chromosomes pair and synapse to undergo crossover recombination, a prerequisite for proper meiotic chromosome segregation (Bhalla and Dernburg, 2008). In Caenorhabditis elegans, the synapsis checkpoint induces apoptosis to remove nuclei with unsynapsed chromosomes (Bhalla and Dernburg, 2005). This checkpoint depends on cis-acting sites near one end of each chromosome termed pairing centers (PCs; Bhalla and Dernburg, 2005), which are also essential for pairing and synapsis (MacQueen et al., 2005). Early in meiotic prophase PCs recruit factors, such as HIM-8, ZIM-1, ZIM-2, and ZIM-3 (Phillips et al., 2005; Phillips and Dernburg, 2006), to assemble a transient regulatory platform that interacts with the conserved nuclear envelope proteins SUN-1 and ZYG-12. This interaction allows PCs access to the cytoplasmic microtubule network and the microtubule-associated motor, dynein (Penkner et al., 2007; Sato et al., 2009; Labrador et al., 2013). The mobilization of chromosomes by cytoskeletal forces is a conserved feature of meiotic prophase (Bhalla and Dernburg, 2008) that facilitates homologue pairing and synapsis (Sato et al., 2009; Labrador et al., 2013).
When dynein function is abolished, chromosomes pair but fail to synapse (Sato et al., 2009). These data have led to a working model in which dynein is dispensable for homologue pairing but licenses synapsis through a tension-sensing mechanism (Sato et al., 2009; Wynne et al., 2012). This model proposes that when a chromosome identifies its homologue and remains stably paired, it resists the pulling forces of dynein. This resistance, or tension, is thought to initiate synapsis at PCs. However, if nonhomologous chromosomes interact, they cannot resist dynein’s pulling forces and restart the homology search. How tension between PCs could be monitored to regulate synapsis is unknown.
Despite their functional differences, PCs have been compared with centromeres (Dernburg, 2001; Labella et al., 2011). Both are cis-acting chromosomal sites that nucleate transient structures to mediate microtubule binding, promote specific chromosome behavior, and generate a checkpoint response. In addition, centromeres can act as sites for meiotic synapsis initiation in budding yeast (Tsubouchi and Roeder, 2005; Tsubouchi et al., 2008) and Drosophila (Takeo et al., 2011; Tanneti et al., 2011). Centromeres assemble kinetochores to orchestrate chromosome segregation (Cheeseman and Desai, 2008). Kinetochores also provide a platform for the spindle assembly checkpoint (SAC), which inhibits the anaphase-promoting complex (APC) and halts cell cycle progression until all kinetochores are successfully bioriented (Foley and Kapoor, 2013).
Because of the similarities between PCs and centromeres, we hypothesized that components of the SAC might act at PCs during meiotic prophase. We report that MAD-1, MAD-2, and BUB-3 are required for the synapsis checkpoint and negatively regulate synapsis in C. elegans. Mutation of mad-1, mad-2, or bub-3 suppresses synapsis defects in dynein mutants, implicating SAC components in the tension-sensing mechanism that is thought to license meiotic synapsis. These roles in monitoring and regulating synapsis are independent of a conserved APC component, indicating MAD-1, MAD-2, and BUB-3 are performing a role aside from inhibiting the APC. MAD-1 and MAD-2 localize to the nuclear periphery and coimmunoprecipitate with SUN-1. Furthermore, MAD-1 and BUB-3 require full PC function to inhibit synapsis. Altogether, these data strongly suggest that SAC proteins function at PCs. Therefore, we propose that the ability of some SAC components to monitor tension is conserved and may have been coopted in a variety of biological contexts to maintain genomic integrity.
Results and discussion
MAD-1, MAD-2, and BUB-3 are required for the synapsis checkpoint
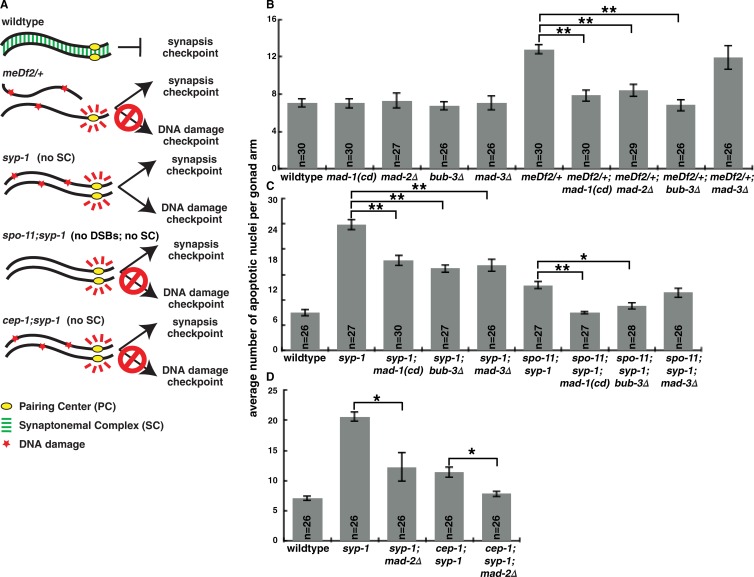
To determine whether SAC proteins are required for the synapsis checkpoint, we used a hypomorphic allele of mad-1 (mdf-1[av19]) defective in SAC function (Stein et al., 2007; Yamamoto et al., 2008) and null mutations of three core but nonessential SAC components, mad-2Δ, mad-3Δ (known as mdf-2 and mdf-3/san-1, respectively, in C. elegans), and bub-3Δ (Kitagawa and Rose, 1999; Essex et al., 2009). We refer to the mad-1(av19) allele as mad-1(cd) for checkpoint deficient. meDf2 is a deficiency that removes the X chromosome PC (Villeneuve, 1994). Animals heterozygous for meDf2 (meDf2/+) have unsynapsed X chromosomes in a subset of meiotic nuclei because synapsis cannot initiate efficiently (MacQueen et al., 2005). The synapsis checkpoint responds to these unsynapsed chromosomes by elevating germline apoptosis above wild-type physiological levels (Fig. 1, A and B; Bhalla and Dernburg, 2005). We introduced SAC mutations into meDf2/+ and found that loss of mad-1, mad-2, or bub-3, but not mad-3, decreased apoptosis in meDf2/+ to wild-type levels (Fig. 1 B), illustrating that MAD-1, MAD-2, and BUB-3 are required for the synapsis checkpoint when X chromosomes are unsynapsed.
Figure 1.
MAD-1, MAD-2, and BUB-3 are required for the synapsis checkpoint. (A) Meiotic checkpoints in C. elegans. (B) Mutation of mad-1, mad-2, or bub-3, but not mad-3, reduces germline apoptosis in meDf2/+. (C) Mutation of mad-1 or bub-3 reduces germline apoptosis in syp-1 and spo-11;syp-1 mutants, whereas mutation of mad-3 reduces apoptosis in syp-1 but not in spo-11;syp-1 mutants. (D) Mutation of mad-2 reduces germline apoptosis in syp-1 and cep-1;syp-1 mutants. Error bars represent ±SEM. *, P < 0.01; **, P < 0.0001 in all graphs.
meDf2 homozygotes exhibit asynapsis in almost all meiotic nuclei (MacQueen et al., 2005). However, these mutant worms exhibit elevated germline apoptosis as the result of the DNA damage checkpoint (Fig. S1, A and B) because functional PCs are required for the synapsis checkpoint (Bhalla and Dernburg, 2005). Mutation of mad-1, mad-2, mad-3, or bub-3 did not reduce apoptosis in meDf2 homozygotes (Fig. S1 B). Therefore, MAD-1, MAD-2, and BUB-3 are specifically required to induce germline apoptosis in response to the synapsis checkpoint.
Next, we tested the requirement for SAC components in the synapsis checkpoint when all chromosomes are unsynapsed. Synapsis requires the assembly of the synaptonemal complex (SC) between homologous chromosomes (Bhalla and Dernburg, 2008). syp-1 mutants do not load SCs between homologues (MacQueen et al., 2002), leading to high levels of checkpoint-induced germline apoptosis as a result of both the synapsis and DNA damage checkpoints (Fig. 1, A, C, and D; Bhalla and Dernburg, 2005). Mutation of mad-1, mad-2, mad-3, or bub-3 in the syp-1 mutant background reduced apoptosis to intermediate levels, indicating loss of one checkpoint but not both (Fig. 1, C and D).
To determine which checkpoint these genes are required for, we prevented the DNA damage checkpoint response in syp-1 mutants by mutating either spo-11 or cep-1 (Fig. 1 A; Bhalla and Dernburg, 2005). SPO-11 generates double-strand breaks that initiate meiotic recombination (Dernburg et al., 1998), and CEP-1 (the C. elegans p53 orthologue) promotes germline apoptosis in response to DNA damage (Derry et al., 2001; Schumacher et al., 2001). Therefore, the elevated apoptosis in spo-11;syp-1 and cep-1;syp-1 double mutants is solely a consequence of the synapsis checkpoint (Fig. 1 A; Bhalla and Dernburg, 2005). Mutation of mad-1 or bub-3 in the spo-11;syp-1 background or mad-2 in the cep-1;syp-1 background produced wild-type levels of apoptosis (Fig. 1, C and D). However, mutation of mad-3 in spo-11;syp-1 mutants did not further decrease apoptosis (Fig. 1 C), suggesting that MAD-3 is required for the DNA damage checkpoint in syp-1 mutants. Because this differs from our results with meDf2;mad-3Δ double mutants (Fig. S1 B), we infer that the DNA damage checkpoint responds differently if all chromosomes are unsynapsed (syp-1 mutants) versus if one pair of chromosomes are unsynapsed (meDf2 mutants). More importantly, these data establish that MAD-1, MAD-2, and BUB-3, but not MAD-3, are required for the synapsis checkpoint when all chromosomes are unsynapsed.
Mad2, Bub3, and Mad3 form the mitotic checkpoint complex (MCC), which inhibits the APC activator Cdc20 and entry into anaphase (Sudakin et al., 2001). MAD-3’s primary role during the SAC response may be inhibition of the APC (Shonn et al., 2003), suggesting that the APC might also not be involved in the synapsis checkpoint. To test this, we used a temperature-sensitive allele of mat-3, the orthologue of Cdc23/Apc8 and an essential subunit of the APC (Golden et al., 2000). We predicted that if SAC components were acting through the APC, loss of APC activity would elevate germline apoptosis as the result of an inappropriate checkpoint response (Fig. S1 C). However, germline apoptosis in mat-3, syp-1;mat-3;mad-1(cd), or syp-1;mat-3;bub-3Δ mutants was unaffected in comparison with wild-type, syp-1;mad-1(cd), and syp-1;bub-3Δ backgrounds, respectively (Fig. S1 D). We also evaluated whether Cdc20 (FZY-1 in C. elegans) was involved in the synapsis checkpoint using a loss of function allele (Kitagawa et al., 2002) but detected no change in apoptosis in fzy-1 or syp-1;fzy-1;mad-1(cd) mutants when compared with wild-type or syp-1;mad-1(cd) worms, respectively (Fig. S1 E). Together these data indicate that the APC is unlikely to be the target of SAC components in the synapsis checkpoint. Intriguingly, orthologues of some SAC components, but not APC components, have been identified in the Giardia genome (Gourguechon et al., 2013). Loss of these conserved SAC components produces chromosome segregation errors (Vicente and Cande, 2014), suggesting that they may have additional roles in regulating chromosome behavior.
In addition to the synapsis checkpoint, asynapsis is associated with a delay in meiotic progression in which chromosomes remain asymmetrically localized (clustered) in meiotic nuclei (Fig. S2 A; MacQueen et al., 2002). To evaluate whether SAC proteins affect meiotic progression, we quantified the percentage of nuclei in which chromosomes appeared clustered (Fig. S2, B and C). mad-1(cd) and bub-3Δ mutants had slightly more nuclei with clustered chromosomes than wild-type germlines, suggesting defects in synapsis or recombination (Fig. S2, B and C). However, neither of the single mutants exhibited any achiasmate chromosomes (not depicted), indicating that crossover recombination is not disrupted. More importantly, we did not detect any difference in the percentage of nuclei with clustered chromosomes between syp-1;mad-1(cd) or syp-1;bub-3Δ double mutants and syp-1 single mutants (Fig. S2, B and C). Therefore, MAD-1 and BUB-3 are specifically required for the synapsis checkpoint and not for the delay in meiotic progression that accompanies asynapsis.
MAD-1 and MAD-2 interact with SUN-1 and localize to the periphery of meiotic nuclei
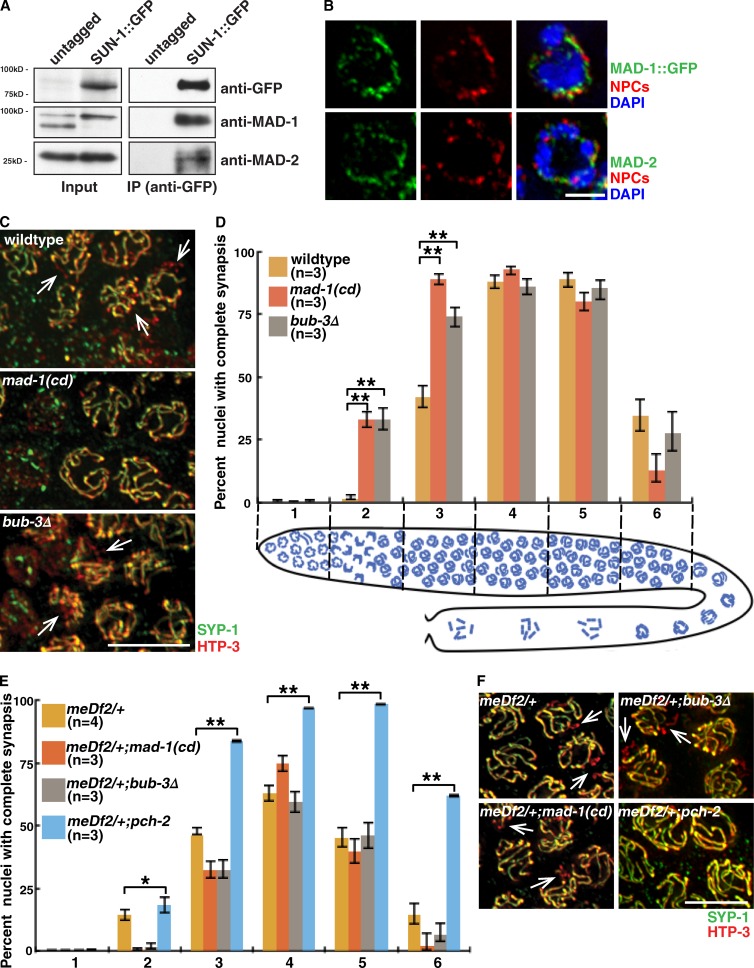
SAC components localize to unattached kinetochores to initiate checkpoint signaling (Foley and Kapoor, 2013). In meiotic prophase, SUN-1 is present at the nuclear periphery and colocalizes with PCs during pairing and synapsis (Penkner et al., 2007; Sato et al., 2009). To test whether SAC proteins also interact with PC-associated proteins to promote synapsis checkpoint signaling, we performed coimmunoprecipitations (co-IPs) of SUN-1::GFP and probed the immunoprecipitates with antibodies against MAD-1 and MAD-2 (Fig. 2 A). Both MAD-1 and MAD-2 coimmunoprecipitated with SUN-1::GFP but not with our untagged control (Fig. 2 A). We also assessed whether BUB-3 coimmunoprecipitated with SUN-1::GFP but were unable to detect an interaction (not depicted). Therefore, both MAD-1 and MAD-2 interact with the PC-associated protein SUN-1.
Figure 2.
SAC proteins interact with PC-associated protein SUN-1, localize to the periphery of meiotic nuclei, and inhibit synapsis in a PC-dependent manner. (A) MAD-1 and MAD-2 coimmunoprecipitate with SUN-1::GFP. Lysates and IPs from untagged and tagged worm strains blotted with antibodies against GFP, MAD-1, and MAD-2. (B) MAD-1::GFP and MAD-2 are at the nuclear periphery marked with NPCs. Images of partial projections of meiotic nuclei stained to visualize DNA (blue), MAD-1::GFP or MAD-2, and NPCs. (C) Images of nuclei during synapsis initiation in wild-type worms and mad-1(cd) and bub-3Δ mutants stained to visualize SYP-1 and HTP-3. (D) mad-1(cd) and bub-3Δ mutants accelerate synapsis. Cartoon depicts worm germline. Meiotic progression is from left to right. (E) Mutation of mad-1 or bub-3 does not accelerate synapsis in meDf2/+. (F) Mutation of mad-1 or bub-3 does not rescue the synapsis defect in meDf2/+. Images of nuclei in meDf2/+, meDf2/+;mad-1(cd), meDf2/+;bub-3Δ, and meDf2/+;pch-2 mutants stained to visualize SYP-1 and HTP-3. (C and F) Arrows indicate unsynapsed chromosomes. Error bars represent 95% confidence intervals. *, P < 0.01; **, P < 0.0001 in all graphs. Bars: (B) 2 µm; (C and F) 5 µm.
We evaluated whether SAC proteins are at the nuclear periphery by staining germlines with antibodies against nuclear pore complexes (NPCs) and MAD-1::GFP or MAD-2. MAD-1::GFP and MAD-2 localized to the nuclear periphery in a punctate pattern (Fig. 2 B), and colocalization with SUN-1::GFP confirmed that both proteins were inside the nucleus (not depicted). MAD-1 localization to the nuclear periphery in embryos is dependent on the nonessential NPC component NPP-5 (Ródenas et al., 2012). However, in meiotic nuclei, MAD-1 or MAD-2 localization is not disrupted in npp-5Δ, sun-1Δ, or npp-5Δ;sun-1Δ mutants (not depicted). We stained for BUB-3 but were unable to localize it in meiotic nuclei (not depicted).
We attempted to localize SAC proteins with PCs. However, despite the biochemical interaction with SUN-1, neither MAD-1 nor MAD-2 colocalized with PC proteins in wild-type, syp-1, or meDf2/+ germlines (not depicted). We provide two potential explanations for the inconsistency between our biochemical and cytological experiments: (1) the interaction of SAC proteins at PCs is transient, and/or (2) the pool of MAD-1 and MAD-2 that interacts with PCs is a small fraction of the total protein present in meiotic nuclei. Similar explanations have been made to argue that Mad1 and Mad2 sense tension during mitosis despite an inability to localize these proteins to tensionless kinetochores (Maresca and Salmon, 2010).
MAD-1 and BUB-3 inhibit synapsis in a PC-dependent manner
Given that the synapsis checkpoint component PCH-2 inhibits synapsis (Deshong et al., 2014), we hypothesized that SAC proteins might also regulate this process. We were concerned that defects in germline organization might complicate our analysis (Kitagawa and Rose, 1999; Stein et al., 2007), so we first analyzed meiotic entry in SAC mutants. In C. elegans, premeiotic nuclei undergo mitotic divisions at the distal end of the germline until they enter meiotic prophase (Fig. S2 D). Meiotic entry is accompanied by the temporary clustering of chromosomes within nuclei and the appearance of phosphorylated SUN-1 (SUN-1pSer8) at the nuclear envelope (Penkner et al., 2009; Woglar et al., 2013). We assessed whether meiotic entry was affected in SAC mutants by quantifying the number of rows of mitotic germline nuclei from the distal tip to the appearance of SUN-1pSer8 in nuclei with clustered chromosomes (Fig. S3 E). Although mad-2Δ mutants delayed the onset of meiosis, as indicated by an increased number of rows of mitotic germline nuclei compared with wild-type worms, we did not detect any significant difference in the number of rows of mitotic germline nuclei in wild-type, mad-1(cd), and bub-3Δ mutants (Fig. S2, E and F). For this reason, additional analysis of meiotic prophase events was performed in mad-1(cd) and bub-3Δ mutants.
To test whether MAD-1 and BUB-3 negatively regulate synapsis, we assayed SC assembly by staining for HTP-3, an axial element protein that assembles on chromosomes before synapsis (MacQueen et al., 2005), and SYP-1, a central element component whose addition to the SC is concomitant with synapsis (MacQueen et al., 2002). Extensive stretches of HTP-3 without SYP-1 indicate the presence of unsynapsed chromosomes (arrows in Fig. 2 C and Fig. S4 A), and colocalization of HTP-3 with SYP-1 indicates complete synapsis (Fig. 2 C and Fig. S4 A). Because nuclei in the germline are arrayed in a spatiotemporal gradient, we divided germlines into six equivalent zones and calculated the percentage of nuclei that had completed synapsis (Fig. 2 D). mad-1(cd) and bub-3Δ mutants accelerated synapsis, exhibiting significantly more nuclei with complete synapsis in zones 2 and 3 than wild-type germlines (Fig. 2, C and D; and Fig. S4 A). Null mad-1 (mad-1Δ) mutants also exhibited normal meiotic entry (Fig. S2, E and F) and similar acceleration of synapsis as bub-3Δ and mad-1(cd) mutants (not depicted). We tested whether accelerated synapsis in mad-1(cd) and bub-3Δ mutants produced nonhomologous synapsis by monitoring pairing at a PC locus (HIM-8; Phillips et al., 2005) and a non-PC locus (5S rDNA; Fig. S3, C and D). We did not detect any defects in pairing in these mutant backgrounds. These data indicate that MAD-1 and BUB-3 normally restrain synapsis but are not involved in homology assessment.
SAC proteins depend on functional kinetochores to elicit a checkpoint response (Essex et al., 2009). Therefore, we evaluated whether the effect of MAD-1 and BUB-3 on synapsis relied on functional PCs. Because PCs are essential for synapsis, we tested this in meDf2/+, in which the loss of a single PC on one of the two X chromosomes results in a fraction of nuclei exhibiting asynapsis of the X chromosomes (MacQueen et al., 2005). PCH-2’s regulation of synapsis does not depend on full PC function because loss of pch-2 accelerates synapsis even in meDf2/+, completely suppressing its synapsis defect (Deshong et al., 2014). Therefore, if MAD-1 or BUB-3’s ability to inhibit synapsis depends on PCs, mutation of either of these genes should not affect the rate or extent of synapsis in meDf2/+. Unlike meDf2/+;pch-2 double mutants, meDf2/+;mad-1(cd) and meDf2/+;bub-3Δ mutants did not accelerate synapsis when compared with meDf2/+ single mutants (Fig. 2 E, zones 2 and 3) and had meiotic nuclei with unsynapsed chromosomes (arrows in Fig. 2 F and Fig. S4 B). Therefore, SAC proteins negatively regulate synapsis in a PC-dependent manner.
MAD-1, MAD-2, and BUB-3 enforce the reliance on dynein to promote synapsis
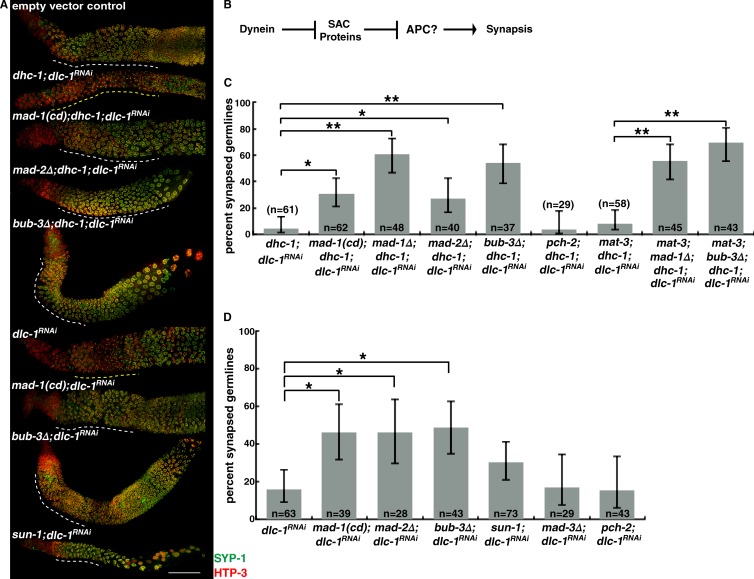
If SAC proteins inhibit synapsis until homologous chromosomes have generated the appropriate amount of dynein-dependent tension, loss of SAC components should abrogate the requirement for dynein in licensing synapsis (Fig. 3 B). To test whether mutations in SAC components suppress synapsis defects in dynein mutants, we used a temperature-sensitive mutation of dynein heavy chain, dhc-1(or195) (Hamill et al., 2002), which produces defects in both germline mitosis and meiosis. We specifically affected meiotic nuclei by inactivating dynein light chain (dlc-1) by RNAi, which partially suppresses the mitotic defects of dhc-1 mutants (O’Rourke et al., 2007). dhc-1;dlc-1RNAi mutants exhibited extensive asynapsis in 95% of germlines, as illustrated by the inability to load SYP-1 onto meiotic chromosomes that have already localized HTP-3 and SYP-1’s aggregation into polycomplexes (Fig. 3 A; Sato et al., 2009). When we combined mad-1(cd), mad-1Δ, mad-2Δ, or bub-3Δ mutations with dhc-1;dlc-1RNAi mutants, we observed robust synapsis (Fig. 3 A). We quantified the level of suppression of the asynapsis phenotype and found that 31% of mad-1(cd);dhc-1;dlc-1RNAi, 60% of mad-1Δ;dhc-1;dlc-1RNAi, 28% of mad-2Δ;dhc-1;dlc-1RNAi, and 54% of bub-3Δ;dhc-1;dlc-1RNAi germlines exhibited synapsed chromosomes (Fig. 3 C).
Figure 3.
Loss of MAD-1, MAD-2, or BUB-3 suppresses the synapsis defects in dynein mutants. (A) Images of germlines from wild-type, dhc-1;dlc-1RNAi, mad-1(cd);dhc-1;dlc-1RNAi, mad-2Δ;dhc-1;dlc-1RNAi, bub-3Δ;dhc-1;dlc-1RNAi;dlc-1RNAi, mad-1(cd);dlc-1RNAi, mad-2Δ;dlc-1RNAi, bub-3Δ;dlc-1RNAi, and sun-1;dlc-1RNAi mutants stained to visualize SYP-1 and HTP-3. Regions of asynapsis are indicated by yellow dashed lines, and regions of normal synapsis are indicated by white dashed lines. Bar, 30 µm. (B) Schematic of the possible role of the APC in regulating synapsis. (C) mad-1(cd), mad-1Δ, mad-2Δ, or bub-3Δ suppresses the synapsis defect in dhc-1;dlc-1RNAi germlines. Mutation of mat-3 does not affect synapsis in mad-1Δ;dhc-1;dlc-1RNAi or bub-3Δ;dhc-1;dlc-1RNAi mutants. (D) mad-1(cd), mad-2Δ, or bub-3Δ suppresses the synapsis defect in dlc-1RNAi germlines. Error bars represent 95% confidence intervals. *, P < 0.01; **, P < 0.0001 in all graphs.
The synapsis in mad-1(cd);dhc-1;dlc-1RNAi, mad-2Δ;dhc-1;dlc-1RNAi, and bub-3Δ;dhc-1;dlc-1RNAi mutants was homologous, as assayed by staining for the PC proteins ZIM-2 and HIM-8 (Fig. S3 E; Phillips et al., 2005; Phillips and Dernburg, 2006). In addition, mad-1(cd);bub-3Δ;dhc-1;dlc-1RNAi mutants resembled bub-3Δ;dhc-1;dlc-1RNAi mutants with regard to percentage of synapsed germlines (not depicted), indicating that MAD-1 and BUB-3 act in the same pathway. Only 5% of pch-2;dhc-1;dlc-1RNAi triple mutants exhibited normal synapsis, similar to dhc-1;dlc-1RNAi double mutants (Fig. 3 B), suggesting that PCH-2’s effect on regulating synapsis is independent of the role that dynein and SAC components play in this process.
Similar to a meiosis-specific mutation of sun-1, sun-1(jf18) (Sato et al., 2009), mad-1(cd), mad-2Δ, and bub-3Δ mutations suppressed defects in synapsis when only dlc-1 was knocked down by RNAi (Fig. 3, A and D). Loss of pch-2 also did not suppress the asynapsis phenotype of dlc-1RNAi (Fig. 3 D). Furthermore, mad-3Δ;dlc-1RNAi worms had extensive asynapsis (Fig. 3 D), consistent with our finding that mad-3 is not required for the synapsis checkpoint (Fig. 1, B and C).
To further test the involvement of the APC in synapsis, we assessed synapsis in mat-3;dhc-1;dlc-1RNAi mutants, as well as mat-3;mad-1Δ;dhc-1;dlc-1RNAi and mat-3;bub-3Δ;dhc-1;dlc-1RNAi mutants. Mutation of mat-3 had no effect on the percentage of germlines with synapsed chromosomes (Fig. 3 C). These data indicate that when specific SAC proteins are absent, the reliance on dynein to license synapsis is lost and this is independent of the APC.
MAD-1 and BUB-3 regulate synapsis by a mechanism redundant with PCH-2
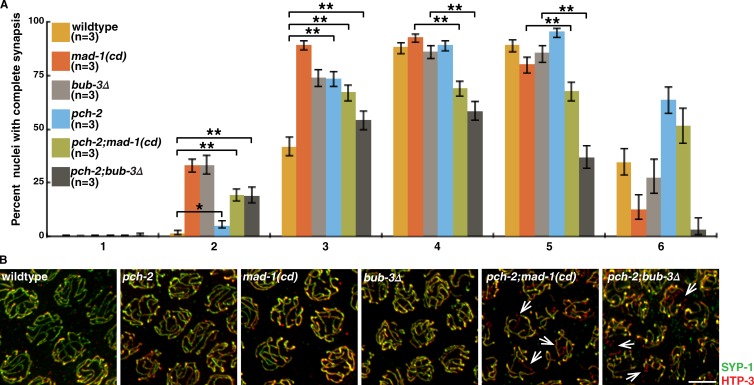
Our experiments in meDf2/+ (Fig. 2, E and F) and dynein mutants (Fig. 3, C and D) suggest that SAC components and PCH-2 regulate synapsis by independent mechanisms. If so, loss of both of these mechanisms should affect synapsis more severely than loss of only one. First, we verified that meiotic entry in pch-2;mad-1(cd) and pch-2;bub-3Δ double mutants was unaffected (Fig. S2 F). We then assessed synapsis in pch-2;mad-1(cd) and pch-2;bub-3Δ double mutants and detected a similar acceleration of synapsis as single mutants, indicating that loss of both of these mechanisms does not further hasten synapsis (Fig. 4 A, zones 2 and 3). However, these double mutants, particularly pch-2;bub-3Δ, had significantly more nuclei with unsynapsed chromosomes throughout the germline (Fig. 4, A [zones 4 and 5] and B; and Fig. S4 C). Therefore, loss of both PCH-2 and SAC components produces defects in synapsis that are more severe than any of the single mutants, indicating that the regulation of synapsis by SAC components and PCH-2 are distinct, biologically parallel processes.
Figure 4.
Loss of both PCH-2 and MAD-1 or BUB-3 results in more severe defects in synapsis. (A) pch-2;mad-1(cd) and pch-2;bub-3Δ double mutants exhibit more severe synapsis defects than pch-2, mad-1(cd), and bub-3Δ single mutants (mad-1(cd) and bub-3Δ data from Fig. 2 D). Error bars represent 95% confidence intervals. *, P < 0.01; **, P < 0.0001. (B) Images of nuclei in wild-type worms and pch-2, mad-1(cd), bub-3Δ, pch-2;mad-1(cd), and pch-2;bub-3Δ mutants stained to visualize SYP-1 and HTP-3. Arrows indicate unsynapsed chromosomes. Bar, 5 µm.
The loss of both SAC components and PCH-2 does not result in nonhomologous synapsis (not depicted), suggesting that the mechanisms controlling synapsis are distinct from those that assess homology between chromosomes. Given that synapsis initiation, not homologous interactions, is the rate-limiting step for synapsis (Rog and Dernburg, 2015) and SC assembly on meiotic chromosomes is highly processive, even when confronted with extensive regions of nonhomologous sequence (MacQueen et al., 2005), it seems likely that synapsis between homologous PCs must overcome multiple barriers, such as those enforced by PCH-2 and SAC components. Why these multiple barriers exist and how they contribute to accurate meiotic chromosome segregation are currently unknown.
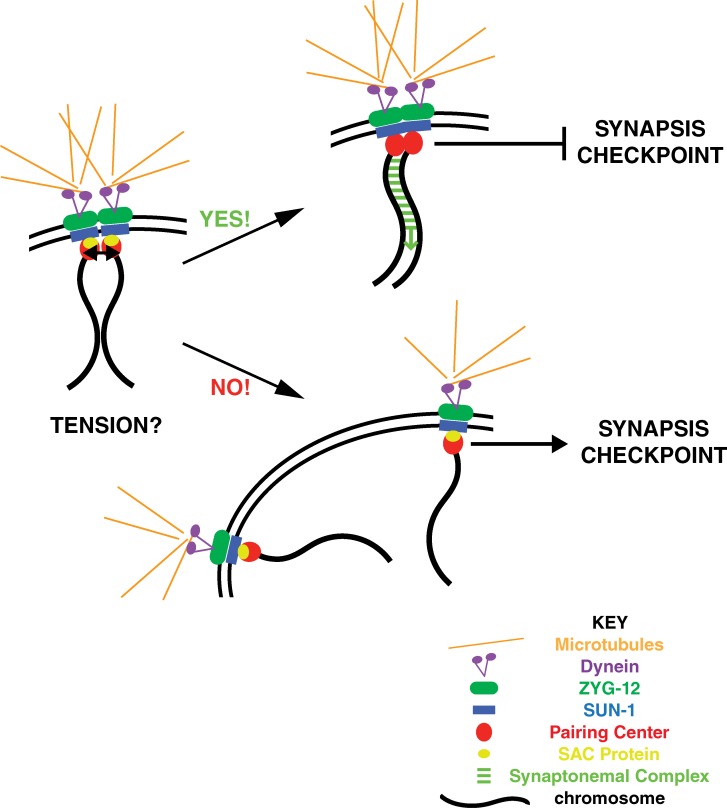
We previously proposed that highly stable PC pairing, whether accomplished normally through synapsis or via the inappropriate stabilization of paired PCs, as in pch-2 mutants, satisfies the synapsis checkpoint (Deshong et al., 2014). Our analysis of mad-1, mad-2, and bub-3 mutants introduces another layer of complexity to the mechanisms that control synapsis: tension, potentially generated by stably paired PCs, may be translated into a molecular signal that silences the checkpoint and initiates synapsis. Because we cannot cytologically detect SAC components at PCs (not depicted), an alternate interpretation is that SAC components perform some other role at the nuclear envelope that affects the checkpoint and synapsis. However, our data support a model in which once stable PC pairing has generated sufficient tension to resist the pulling forces of the microtubule motor dynein, SAC proteins are either inactivated or removed from PCs to initiate synapsis and the synapsis checkpoint is silenced (Fig. 5).
Figure 5.
Model for synapsis initiation and checkpoint satisfaction in C. elegans. A pair of chromosomes with PCs interact with proteins at the nuclear envelope, including SUN-1 and ZYG-12, to gain access to the cytoplasmic microtubule network and dynein. SAC components are presumed to function at PCs despite our inability to colocalize them. When a chromosome encounters another chromosome, homology is assessed by unknown mechanisms. If chromosomes are homologous and remain stably paired, they resist the pulling forces of the microtubule motor dynein, generating tension (black arrows between PCs) that is monitored by SAC components. Once sufficient tension has been generated (YES!), SAC components are removed, synapsis is initiated, and the checkpoint is silenced. If chromosomes are not homologous, they cannot resist the pulling forces of dynein, are pulled apart, and do not generate tension (NO!). Unsynapsed PCs initiate the synapsis checkpoint response. If unsynapsed chromosomes persist, these nuclei are removed by apoptosis.
Materials and methods
Genetics and worm strains
The wild-type C. elegans strain background was Bristol N2 (Brenner, 1974). All experiments were performed on adult hermaphrodites at 20°C under standard conditions unless otherwise stated. Mutations and rearrangements used were as follows: LG I: mnDp66, dhc-1(or195), san-1/mdf-3(ok1580), cep-1(gk138); LG II: fzy-1(h1983), pch-2(tm1458), npp-5(tm3039), bub-3(ok3437), jfSi1[Psun-1::GFP::sun-1::cb-unc-119(+)]), mIn1 [mIs14 dpy-10(e128)]; LG III: mat-3(or180), jzIs1[pRK139; Ppie-1::GFP::mdf-1::unc-119(+)], unc-119(ed3); LG IV: mdf-2(tm2190), spo-11(ok79), nT1[unc-?(n754) let-?(m435)], nTI [qIs51]; LG V: dpy-11(e224), mdf-1(av19), mdf-1(gk2), syp-1(me17), sun-1(jf18), sun-1(ok1282), bcIs39(Pim::ced-1::GFP); and LG X: meDf2.
meDf2 is a terminal deficiency of the left end of the X chromosome that removes the X chromosome PC as well as numerous essential genes (Villeneuve, 1994). For this reason, homo- and hemizygous meDf2 animals also carry a duplication (mnDp66) that includes these essential genes but does not interfere with normal X chromosome segregation (Herman and Kari, 1989) or the synapsis checkpoint (Bhalla and Dernburg, 2005). For clarity, it has been omitted from the text. Because the mad-2 gene is closely linked to spo-11, we used cep-1 to prevent DNA damage checkpoint–induced apoptosis in mad2Δ mutants.
Quantification of germline apoptosis
Scoring of germline apoptosis was performed as previously descried in Bhalla and Dernburg (2005). L4 hermaphrodites were allowed to age for 22 h at 20°C, except for the mat-3 temperature-sensitive mutation, which was aged for 18 h at the restrictive temperature of 25°C. We verified that MAT-3 function had been reduced by the increase in the number of germline nuclei positive for phosphorylation of histone H3 serine 10 (Golden et al., 2000). Because nuclei progress from mitosis to meiosis as they travel through the germline, this incubation period guaranteed that early meiotic prophase nuclei, but not mitotic nuclei at the start of the germline, had sufficient time to progress to where checkpoint-induced apoptosis occurs in late meiotic prophase (Jaramillo-Lambert et al., 2007). Live worms were mounted under coverslips on 1.5% agarose pads containing 0.2 mM levamisole for wild-type moving strains or 0.1 mM levamisole for dpy-11 strains. A minimum of 25 germlines were analyzed for each genotype by performing live fluorescence microscopy and counting the number of cells fully surrounded by CED-1::GFP. Significance was assessed using a paired t test. All experiments were performed at least twice.
Antibodies, immunostaining, fluorescence in situ hybridization, and microscopy
Immunostaining was performed on worms 20–24 h after L4 stage. Gonad dissections were performed in 1× EBT (250 mM Hepes-Cl, pH 7.4, 1.18 M NaCl, 480 mM KCl, 20 mM EDTA, and 5 mM EGTA) + 0.1% Tween 20 and 20 mM sodium azide. An equal volume of 2% formaldehyde in EBT (final concentration was 1% formaldehyde) was added and allowed to incubate under a coverslip for 5 min. The sample was mounted on HistoBond slides (75 × 25 × 1 mm from Lamb), freeze-cracked, and incubated in methanol at −20°C for slightly more than 1 min and transferred to PBST (PBS with Tween 20). After several washes of PBST, the samples were incubated for 30 min in 1% bovine serum albumin diluted in PBST. A hand-cut paraffin square was used to cover the tissue with 50 µl of antibody solution. Incubation was conducted in a humid chamber overnight at 4°C. Slides were rinsed in PBST and then incubated for 2 h at room temperature with fluorophore-conjugated secondary antibody at a dilution of 1:500. Samples were rinsed several times and DAPI stained in PBST, then mounted in 13 µl of mounting media (20 M N-propyl gallate [Sigma-Aldrich] and 0.14 M Tris in glycerol) with a no. 1 1/2 (22 mm2) coverslip, and sealed with nail polish.
Primary antibodies were as follows (dilutions are indicated in parentheses): rabbit anti–SYP-1 (1:500; MacQueen et al., 2002), chicken anti–HTP-3 (1:250; MacQueen et al., 2005), rabbit anti–MAD-2 (1:10,000; Essex et al., 2009), mouse anti-NPC MAb414 (1:5,000; Covance; Davis and Blobel, 1986), guinea pig anti–HIM-8 (1:250; Phillips et al., 2005), rat anti–HIM-8, guinea pig anti–ZIM-2 (1:2,500; Phillips and Dernburg, 2006), guinea pig anti–SUN-1pSer8 (1:500; Penkner et al., 2009), and goat anti-GFP (1:10,000; Hua et al., 2009). Secondary antibodies were Cy3 anti–mouse, anti–rabbit, anti–guinea pig, anti–rat, and anti–chicken (Jackson ImmunoResearch Laboratories, Inc.) and Alexa Fluor 488 anti–goat, anti–guinea pig, and anti–rabbit (Invitrogen). Antibodies against SYP-1 were provided by A. Villeneuve (Stanford University, Palo Alto, CA). Antibodies against HTP-3, HIM-8, and ZIM-2 were provided by A. Dernburg (University of California, Berkeley/E.O. Lawrence Berkeley National Lab, Berkeley, CA). Antibodies against MAD-2 were provided by A. Desai (Ludwig Institute/University of California, San Diego, La Jolla, CA). Antibodies against SUN-1pSer8 were provided by V. Jantsch (Max F. Perutz Laboratories, Vienna, Austria). Antibodies against GFP were provided by S. Strome (University of California, Santa Cruz, Santa Cruz, CA).
Fluorescence in situ hybridization was performed as described in Phillips et al. (2005). 5S rDNA probe was generated using genomic DNA as a template by PCR and gel purified. The PCR product was digested with the TasI restriction enzyme and ethanol precipitated. 10 µg of digested DNA was diluted into 50 µl water, denatured for 2 min at 95°C, chilled on ice, and spun briefly. At room temperature, 20 µl Roche 5× TdT reaction buffer (Tris-HCl, pH 7.2, potassium cacodylate, and BSA), 20 µl of 25 mM CoCl2 solution, 3.3 ml of 1 mM aa-dUTP, 6.6 ml of 1 mM unlabelled dTTP, and 2 µl (800 U) recombinant terminal deoxynucleotidyl transferase were added. This solution was incubated for 1 h at 37°C. EDTA was added to 5 mM, and the DNA was ethanol precipitated. The probe was conjugated with Cy3 dye (Life Technologies) by adding 5 µl of 1 mg/ml resuspended probe and 3 µl of 1 M bicarbonate buffer to one aliquot of dry dye. The reaction was mixed, shielded from light, and incubated for 1 h at room temperature and ethanol precipitated.
Worms were dissected 24 h after L4 stage in 30 µl EBT (1× egg buffer, 0.1% Tween 20, and 20 mM sodium azide). We added 30 µl of 1× egg buffer and 0.5% EGS (ethylene glycol bis[succinimidylsuccinate] in dimethyl formamide) and pipetted to extrude gonads. 30 µl of this liquid was removed, and the sample was mounted on HistoBond slides (75 × 25 × 1 mm from Lamb) and allowed to incubate in a humid chamber for 30 min at room temperature. Samples were freeze-cracked and incubated in methanol at −20°C for slightly more than 1 min and transferred to 2× SSCT (2× SSC and 0.1× Tween 20) at room temperature. Samples were placed in 3.7% formaldehyde in 1× egg buffer for 5 min, rinsed briefly in 2× SSCT, and washed twice in 2× SSCT for 5 min. The samples were incubated in 50% formamide in 2× SCCT for 5 min, transferred to fresh 50% formamide in 2× SCCT, and incubated at 37°C overnight. Samples were cooled to room temperature, and 20 ng of 5S rDNA probe in hybridization solution (50% formamide, 3% SSC, 10% dextran sulphate) was added and sealed onto the sample with a coverslip and nail polish. Slides were denatured on a hot block at 95°C for 3 min and placed in a humid chamber at 37°C overnight. Coverslips were removed, and the samples were washed twice in 50% formamide in 2× SSCT for 30 min each. Samples were rinsed several times and DAPI stained in 2× SSCT. Samples were mounted in 13 µl of mounting media (20 M N-propyl gallate [Sigma-Aldrich] and 0.14 M Tris in glycerol) with a no. 1 1/2 (22 mm2) coverslip and sealed with nail polish.
Quantification of synapsis and pairing was performed with a minimum of three whole germlines per genotype as in Phillips et al. (2005) on animals 24 h after L4 stage. The gonads were divided into six equal-sized regions, beginning at the distal tip of the gonad and progressing through the end of pachytene. Significance was assessed by performing Fisher’s exact test.
Quantification of rows of mitotic nuclei was performed as in Stevens et al. (2013), and a minimum of 18 germlines were analyzed on animals 24 h after L4 stage. Significance was assessed by performing a paired t test.
Quantification of meiotic progression was performed with a minimum of three whole germlines per genotype on animals 24 h after L4 stage by quantifying the percentage of nuclei with clustered chromosomes. Significance was assessed by performing Fisher’s exact test.
All images were acquired at room temperature using a DeltaVision Personal DV system (GE Healthcare) equipped with a 100× NA 1.40 oil immersion objective (Olympus), resulting in an effective xy pixel spacing of 0.064 or 0.040 µm. Images were captured using a charge-coupled device camera (Cool-SNAP HQ; Photometrics). Three-dimensional image stacks were collected at 0.2-µm z-spacing and processed by constrained, iterative deconvolution. Imaging, image scaling, and analysis were performed using functions in the softWoRx software package. Projections were calculated by a maximum intensity algorithm. Composite images were assembled, and some false coloring was performed with Photoshop software (Adobe).
IPs
Asynchronous liquid worm cultures were grown at 20°C for 4 d in S medium supplemented with concentrated HB101 bacteria, and embryos were extracted in a sodium hypochlorite solution (25% [vol/vol] NaClO and 0.25% [vol/vol] 10N NaOH) and allowed to hatch overnight on unseeded NGM plates. Hatched L1s were washed off NGM plates and grown at 19°C for 66–68 h or until the majority of animals reached adulthood. Adult worms were harvested, washed twice in sterile water and once in buffer H0.15 (50 mM Hepes, pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, pH 8.0, 0.5 mM EGTA-KOH, pH 8.0, 15% glycerol, 0.1% NP-40, and 150 mM KCl), and frozen into “popcorn” by dripping into liquid nitrogen. Popcorn was then pulverized three times for 2 min at 25 Hz in a MM-400 mixer mill (Retsch Technology) with liquid nitrogen immersion between milling sessions. Worms were lysed by adding 5 ml ice-cold buffer H0.15 supplemented with protease and phosphatase inhibitors (0.1 mM AEBSF, 5 mM benzamidine, 1:200 aprotinin, Roche Complete Mini tablets w/o EGTA, 1 mM Na4P2O7, 2 mM Na-β-glycerophosphate, 0.1 mM Na3VO4, and 5 mM NaF) to 2 g of worm powder. Lysis was continued by rotating at 4°C, followed by sonicating twice for 30 s at 40% amplitude on ice (Braun). Lysate was then spun at 48,000 g for 20 min at 4°C in a JA-20 rotor (Beckman Coulter). IPs were performed as in Akiyoshi et al. (2009) with 50 µl protein G Dynabeads (Invitrogen) cross-linked to 12.5 µg mouse GFP antibody (Roche).
For immunoblotting, samples were run on SDS-PAGE gels, transferred to nitrocellulose, blocked in a PBST + 5% (wt/vol) nonfat milk solution, and then probed with mouse anti-GFP (1:1,000; Roche), rabbit anti–MAD-1 (1:2,000; Yamamoto et al., 2008), or rabbit anti–MAD-2 (1:5,000; Essex et al., 2009) overnight at 4°C. Blots were washed three times for 10 min in PBST, probed for 1 h using an HRP-conjugated secondary antibody (rabbit or mouse; GE Healthcare), washed three times for 10 min in PBST, and then analyzed using a chemiluminescent substrate (Thermo Fisher Scientific). 0.1% of starting material is shown for all input samples, 10% of the IP elution is shown for anti-GFP Western blots, and 30% of IPs are shown for anti–MAD-1 and anti–MAD-2 Western blots. IP samples were analyzed with Pico chemiluminescent substrate (Thermo Fisher Scientific), and input samples were analyzed using Dura enhanced chemiluminescent substrate (Thermo Fisher Scientific). We estimate IP to have purified between 20 and 30% of SUN-1::GFP present in the input.
Feeding RNAi
For RNAi, dlc-1RNAi and empty vector (L4440) clones from the Ahringer laboratory (Fraser et al., 2000) were used. Bacteria strains containing dlc-1RNAi and empty vector controls were cultured overnight in 10 ml Luria broth + 50 µg/µl carbenicillin, centrifuged, and resuspended in 0.5 ml Luria broth + 50 µg/µl carbenicillin. 60 µl of the RNAi bacteria was spotted onto NGM plates containing 1 mM IPTG + 50 µg/µl carbenicillin and allowed to grow at room temperature overnight. L4 hermaphrodite worms were picked into M9, transferred to these plates, allowed to incubate for 2–3 h, and then transferred to fresh RNAi plates to be dissected 48 h after L4. Strains with the dhc-1 temperature-sensitive mutation were rinsed in M9, plated on dlc-1RNAi plates, incubated at 15°C for 24 h, and then shifted to the restrictive temperature of 25°C for 24 h and dissected 48 h after L4 as previously described (Sato et al., 2009). A minimum of 28 germlines were scored for each genotype. Significance was assessed by performing Fisher’s exact test.
Online supplemental material
Fig. S1 illustrates that MAD-2, MAD-2, and BUB-3 are not required for the DNA damage checkpoint in meDf2 homozygotes and that MAT-3 and FZY-1 are not required for the synapsis checkpoint. Fig. S2 presents data that loss of MAD-1 or BUB-3 does not affect mitotic or meiotic progression. Fig. S3 demonstrates that homologue pairing is unaffected in mad-1(cd) and bub-3Δ mutants. Fig. S4 includes grayscale images of Fig. 2 (C and F) and Fig. 4 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409035/DC1.
Supplementary Material
Acknowledgements
We would like to thank Arshad Desai, Abby Dernburg, Susan Strome, Anne Villeneuve, and Verena Jantsch for valuable strains and reagents.
This work was supported by the National Institutes of Health (NIH; grant numbers T32GM008646 [to T. Bohr and C.R. Nelson] and R01GM097144 [to N. Bhalla]). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC
- anaphase-promoting complex
- IP
- immunoprecipitation
- NPC
- nuclear pore complex
- PC
- pairing center
- SAC
- spindle assembly checkpoint
- SC
- synaptonemal complex
References
- Akiyoshi B., Nelson C.R., Ranish J.A., and Biggins S.. 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23:2887–2899. 10.1101/gad.1865909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., and Dernburg A.F.. 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 310:1683–1686. 10.1126/science.1117468 [DOI] [PubMed] [Google Scholar]
- Bhalla N., and Dernburg A.F.. 2008. Prelude to a division. Annu. Rev. Cell Dev. Biol. 24:397–424. 10.1146/annurev.cellbio.23.090506.123245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore–microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Davis L.I., and Blobel G.. 1986. Identification and characterization of a nuclear pore complex protein. Cell. 45:699–709. 10.1016/0092-8674(86)90784-1 [DOI] [PubMed] [Google Scholar]
- Dernburg A.F. 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153:F33–F38. 10.1083/jcb.153.6.F33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald K., Moulder G., Barstead R., Dresser M., and Villeneuve A.M.. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 94:387–398. 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Derry W.B., Putzke A.P., and Rothman J.H.. 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 294:591–595. 10.1126/science.1065486 [DOI] [PubMed] [Google Scholar]
- Deshong A.J., Ye A.L., Lamelza P., and Bhalla N.. 2014. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 10:e1004291 10.1371/journal.pgen.1004291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A., Dammermann A., Lewellyn L., Oegema K., and Desai A.. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 20:1252–1267. 10.1091/mbc.E08-10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., and Kapoor T.M.. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., and Ahringer J.. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330. 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P.L., Wallenfang M.R., Schumacher J.M., Hamill D.R., Bates G., Bowerman B., Seydoux G., and Shakes D.C.. 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151:1469–1482. 10.1083/jcb.151.7.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourguechon S., Holt L.J., and Cande W.Z.. 2013. The Giardia cell cycle progresses independently of the anaphase-promoting complex. J. Cell Sci. 126:2246–2255. 10.1242/jcs.121632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill D.R., Severson A.F., Carter J.C., and Bowerman B.. 2002. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell. 3:673–684. 10.1016/S1534-5807(02)00327-1 [DOI] [PubMed] [Google Scholar]
- Hassold T., and Hunt P.. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2:280–291. 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- Herman R.K., and Kari C.K.. 1989. Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics. 121:723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Kittler R., and White K.P.. 2009. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 137:1259–1271. 10.1016/j.cell.2009.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A.M., and Engebrecht J.. 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308:206–221. 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Kitagawa R., and Rose A.M.. 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1:514–521. 10.1038/70309 [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Law E., Tang L., and Rose A.M.. 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12:2118–2123. 10.1016/S0960-9822(02)01392-1 [DOI] [PubMed] [Google Scholar]
- Kops G.J., Weaver B.A., and Cleveland D.W.. 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 5:773–785. 10.1038/nrc1714 [DOI] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., and Zetka M.. 2011. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell. 21:948–958. 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Labrador L., Barroso C., Lightfoot J., Müller-Reichert T., Flibotte S., Taylor J., Moerman D.G., Villeneuve A.M., and Martinez-Perez E.. 2013. Chromosome movements promoted by the mitochondrial protein SPD-3 are required for homology search during Caenorhabditis elegans meiosis. PLoS Genet. 9:e1003497 10.1371/journal.pgen.1003497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiácovo M.P., McDonald K., and Villeneuve A.M.. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16:2428–2442. 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips C.M., Bhalla N., Weiser P., Villeneuve A.M., and Dernburg A.F.. 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 123:1037–1050. 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., and Salmon E.D.. 2010. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123:825–835. 10.1242/jcs.064790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W. 1992. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 359:599–604. 10.1038/359599a0 [DOI] [PubMed] [Google Scholar]
- O’Rourke S.M., Dorfman M.D., Carter J.C., and Bowerman B.. 2007. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3:e128 10.1371/journal.pgen.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., and Jantsch V.. 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 12:873–885. 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., and Jantsch V.. 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 139:920–933. 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., and Dernburg A.F.. 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 11:817–829. 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., and Dernburg A.F.. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 123:1051–1063. 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ródenas E., González-Aguilera C., Ayuso C., and Askjaer P.. 2012. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in Caenorhabditis elegans. Mol. Biol. Cell. 23:930–944. 10.1091/mbc.E11-11-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O., and Dernburg A.F.. 2015. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Reports. 10:1639–1645. 10.1016/j.celrep.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., and Dernburg A.F.. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919. 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Hofmann K., Boulton S., and Gartner A.. 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11:1722–1727. 10.1016/S0960-9822(01)00534-6 [DOI] [PubMed] [Google Scholar]
- Shonn M.A., Murray A.L., and Murray A.W.. 2003. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr. Biol. 13:1979–1984. 10.1016/j.cub.2003.10.057 [DOI] [PubMed] [Google Scholar]
- Stein K.K., Davis E.S., Hays T., and Golden A.. 2007. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 175:107–123. 10.1534/genetics.106.059105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Oegema K., and Desai A.. 2013. Meiotic double-strand breaks uncover and protect against mitotic errors in the C. elegans germline. Curr. Biol. 23:2400–2406. 10.1016/j.cub.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., and Yen T.J.. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S., Lake C.M., Morais-de-Sá E., Sunkel C.E., and Hawley R.S.. 2011. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21:1845–1851. 10.1016/j.cub.2011.09.044 [DOI] [PubMed] [Google Scholar]
- Tanneti N.S., Landy K., Joyce E.F., and McKim K.S.. 2011. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr. Biol. 21:1852–1857. 10.1016/j.cub.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., and Roeder G.S.. 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 308:870–873. 10.1126/science.1108283 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Macqueen A.J., and Roeder G.S.. 2008. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 22:3217–3226. 10.1101/gad.1709408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J.J., and Cande W.Z.. 2014. Mad2, Bub3, and Mps1 regulate chromosome segregation and mitotic synchrony in Giardia intestinalis, a binucleate protist lacking an anaphase-promoting complex. Mol. Biol. Cell. 25:2774–2787. 10.1091/mbc.E14-05-0975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve A.M. 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 136:887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., Daryabeigi A., Adamo A., Habacher C., Machacek T., La Volpe A., and Jantsch V.. 2013. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 9:e1003335 10.1371/journal.pgen.1003335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne D.J., Rog O., Carlton P.M., and Dernburg A.F.. 2012. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J. Cell Biol. 196:47–64. 10.1083/jcb.201106022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., and Kitagawa R.. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194. 10.1083/jcb.200805185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.