Tracking the kinetics of equilibration of H2O2 between compartments reveals unexpected isolation of the endoplasmic reticulum and hints at a hitherto unsuspected local source of peroxide.
Abstract
The endoplasmic reticulum (ER)–localized peroxiredoxin 4 (PRDX4) supports disulfide bond formation in eukaryotic cells lacking endoplasmic reticulum oxidase 1 (ERO1). The source of peroxide that fuels PRDX4-mediated disulfide bond formation has remained a mystery, because ERO1 is believed to be a major producer of hydrogen peroxide (H2O2) in the ER lumen. We report on a simple kinetic technique to track H2O2 equilibration between cellular compartments, suggesting that the ER is relatively isolated from cytosolic or mitochondrial H2O2 pools. Furthermore, expression of an ER-adapted catalase to degrade lumenal H2O2 attenuated PRDX4-mediated disulfide bond formation in cells lacking ERO1, whereas depletion of H2O2 in the cytosol or mitochondria had no similar effect. ER catalase did not effect the slow residual disulfide bond formation in cells lacking both ERO1 and PRDX4. These observations point to exploitation of a hitherto unrecognized lumenal source of H2O2 by PRDX4 and a parallel slow H2O2-independent pathway for disulfide formation.
Introduction
Oxidative protein folding in the ER relies on protein disulfide isomerase (PDI) machinery that accepts electrons from client cysteine thiols generating native disulfides (Hudson et al., 2015). A major advance in our understanding of this machinery came with the discovery of an ER-localized PDI oxidase, endoplasmic reticulum oxidase 1 (ERO1; Frand and Kaiser, 1998; Pollard et al., 1998), which accepts electrons from reduced PDI and hands them over to molecular oxygen, catalyzing oxygen-mediated disulfide bond formation (Tsai and Rapoport, 2002; Araki et al., 2013). ERO1 is conserved in eukaryotes. The marked impairment in disulfide bond formation in yeast lacking ERO1 suggested an essential role in accelerating dithiol oxidation in the ER (Frand and Kaiser, 1998; Pollard et al., 1998). Surprisingly, targeted mutagenesis of the genes encoding animal ERO1 orthologues, ERO1L/ERO1α and ERO1LB/ERO1β, revealed a remarkably mild phenotype (Tien et al., 2008; Zito et al., 2010a).
In ERO1-deficient cells and tissues, life-sustaining rates of disulfide bond formation depend on the ER-localized enzyme peroxiredoxin 4 (PRDX4; Zito et al., 2010b, 2012). PRDX4 accepts electrons from reduced PDI and transfers them to hydrogen peroxide (H2O2; Tavender et al., 2010; Zito et al., 2010b). ERO1-mediated electron transfer from reduced PDI to oxygen reduces the latter to H2O2 (Gross et al., 2006). Thus, the sequential action of ERO1 and PRDX4 can produce two disulfides from every molecule of oxygen converted to water.
Though it is dispensableunder normal circumstances (Iuchi et al., 2009), the PRDX4-mediated reaction becomes limiting in cells lacking ERO1 (Zito et al., 2010b), but the identity of the source of H2O2 that fuels PRDX4 in cells lacking ERO1 is unknown. Resolving this mystery has been hampered by difficulties in measuring changes in H2O2 concentration in the ER of living cells. To circumvent this issue, we exploited the kinetic properties of intravital fluorescent thiol redox probes that diverge in their reactivity with the oxidized/reduced PDI couple and H2O2. Our studies point to an ER-localized, ERO1-independent source of H2O2 that fuels PRDX4-mediated disulfide bond formation in cells lacking ERO1.
Results and discussion
PRDX4-mediated ER thiol oxidation is fueled by H2O2 independently of ERO1
Combined deficiency of ERO1 and PRDX4 is detrimental to mammalian development and enhances lethality of mutant mice. Nonetheless, enfeebled, viable compound mutant cell lines can be cultured in vitro (Zito et al., 2012). Remarkably, such triple-mutant cells (referred to as triple knockout [TKO]; Ero1IGt(xst171)Byg/Gt(XST171)Byg; Ero1IbGt(P077G11)Wrst/Gt(P077G11)Wrs; Prdx4tm1.1JuFu/tm1.1JuFu) defend a steady-state PDI thiol redox couple that is indistinguishable from wild-type cells. However, the deficit in oxidative power of the TKO cells is revealed in a kinetic assay that tracks recovery of disulfide bonds after a reductive pulse of DTT (Avezov et al., 2013).
These measurements were previously performed by tracking changes in the fluorescence lifetime of an ER-localized intravital redox probe based on GFP containing E147 insertion (ERroGFPiE) that equilibrates rapidly with PDI and is specially tuned to PDI’s oxidative state found in the ER (Lohman and Remington, 2008; Avezov et al., 2013). However, this theoretical advantage of ERroGFPiE is obviated by the normal steady state of the PDI redox couple in TKO cells. Furthermore, the fluorescent lifetime imaging measurements necessary to track the redox state of the ER-tuned roGFPiE are difficult to acquire. Therefore, we sought an alternative method to detect the kinetic defect in ER thiol reoxidation observed in TKO cells after a DTT pulse.
ER-localized roGFP2 (ERroGFP2) is recognized as a substrate by PDI, and kinetic parameters deduced from the recovery of its oxidized state after a DTT pulse and washout thus reflect the activity of the enzymatic machinery for PDI reoxidation in vivo (Tsunoda et al., 2014). Its enhanced brightness enables measuring its redox state ratiometrically, by comparing the emission intensity of the probe at 530 nm when excited at 405 and 488 nm (Hanson et al., 2004), and it does not require the more exacting measurements of fluorescent lifetime.
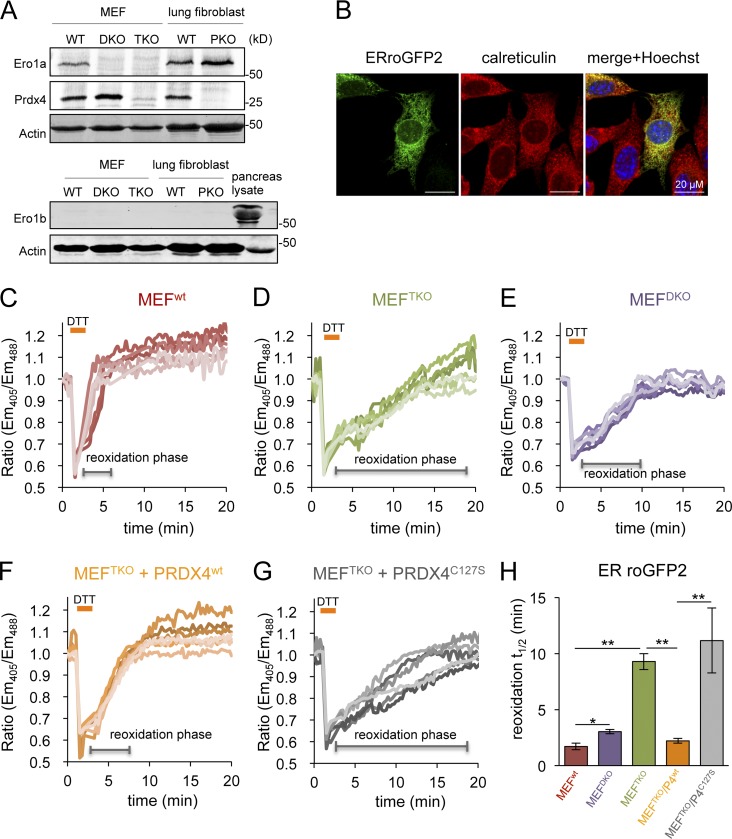
The genetic defect in mouse fibroblasts lacking key ER redox enzymes was confirmed: ERO1α, normally present in fibroblasts, was undetectable in homozygous double knockout (DKO) mutant Ero1IGt(xst171)Byg/Gt(XST171)Byg, Ero1IbGt(P077G11)Wrst/Gt(P077G11)Wrst mouse embryonic fibroblasts (MEFs) and in TKO mutants that are also homozygous for a null allele of Prdx4tm1.1JuFu/tm1.1JuFu. PRDX4 was readily detected in fibroblasts and was eliminated by mutation of its encoding gene, whereas the pancreatic-specific isoform of ERO1, ERO1β, was undetectable in fibroblasts of all genotypes tested (Fig. 1 A).
Figure 1.
PRDX4 supports a near normal rate of ER thiol oxidation in the absence of ERO1. (A) Immunoblot of endogenous Ero1α, Ero1β, and Prdx4 in lysates of MEFs with the indicated genotypes: wild-type (WT), Ero1α; Ero1β double mutant (DKO), Ero1α; Ero1β; Prdx4 triple mutant (TKO), and Prdx4 single mutant (PKO). An anti-actin blot serves as the loading control. (B) Fluorescent photomicrographs of MEF cells transiently expressing ERroGFP2, immunostained for calreticulin as an ER marker. The merge panels show an overlap of the GFP signal with calreticulin and the karyophilic dye Hoechst 33258 (to reveal the nuclei). (C–G) Traces of time-dependent changes in the fluorescence excitation ratio, reflecting the alterations in the oxidation state of roGFP2 expressed in the ER of cells with the indicated genotypes: WT, DKO, TKO, and TKO expressing an active, PRDX4WT, or inactive, PRDX4C127S, enzyme. Cells were exposed to a brief (1 min) reductive pulse of DTT (2 mM), followed by a washout. Each line traces the fluorescence profile of an individual cell. (H) Bar diagram of the t1/2 to recovery of the oxidized form of ERroGFP2 after the reductive DTT pulse, calculated from fitting the data in C–G. Shown are means ± SEM (n > 20). *, P < 0.05; **, P < 0.01.
As expected, ERroGFP2 was localized to the ER of transfected mouse fibroblasts (Fig.1 B) and was rapidly reoxidized after a DTT reductive pulse and washout of the reductant (Fig.1 C). The conversion of the reduced probe to its oxidized, pretreatment steady state (reoxidation phase) occurs with a t1/2 of 1.7 ± 0.3 min in wild-type MEFs (Fig. 1, C and H), whereas the reoxidation was approximately fivefold slower in TKO cells (t1/2 of 9.28 ± 0.8 min; Fig.1, D and H). These values, obtained for ERroGFP2, are in agreement with earlier measurements performed with the ER-tuned probes ERroGFPiL in HT1080 cells and ERroGFPiE in wild-type and TKO MEF cells (van Lith et al., 2011; Avezov et al., 2013).
In cells with wild-type ERO1 activity, PRDX4 has no measureable effect on the rate of ER thiol reoxidation after a DTT pulse (Fig. S1). Although PRDX4 inactivation by excess ERO1-driven H2O2 production in DTT pulsed cells may underestimate PRDX4’s normal role in thiol oxidation (Tavender and Bulleid, 2010), our observations are consistent with the inconspicuous phenotype of Prdx4tm1.1JuFu/tm1.1JuFu (peroxiredoxin 4 knockout [PKO]) mice (Iuchi et al., 2009). Conversely, lack of ERO1 measurably delayed oxidation kinetics because a consistently longer t1/2 was measured in ERO1-deficient DKO cells (which contain PRDX4; Fig. 1 A, lane 2) than in their isogenic control (Fig. 1, E and H). Nevertheless, the importance of PRDX4 to the kinetics of disulfide bond formation in cells lacking ERO1 was highlighted by the restoration of the t1/2 of recovery of oxidized ERroGFP2 in TKO cells transduced with wild-type PRDX4 and by the inactivity of the enzymatically inactive PRDX4C127S mutant lacking the peroxidiac cysteine (Fig. 1, F–H). These observations confirm that PRDX4 assumes an important role in the kinetics of disulfide bond formation when ERO1 activity is limiting, and they correlate with the previous phenotypic analysis of compound mutant cells (Zito et al., 2010b).
Compartment-specific responsiveness of the H2O2-sensitive probe HyPer reveals a barrier to oxidant diffusion across the ER membrane
The above observations raised the question of the source of the oxidative process that fuels PRDX4 recycling from its reduced to its oxidized state in ERO1-deficient cells lacking the major known source of H2O2 production in the ER lumen. To address this question, we sought tools to estimate changes in lumenal H2O2 concentration.
HyPer is a genetically encoded in vivo sensor for fluctuations in H2O2 concentration. It is based on direct H2O2-mediated formation of a disulfide bond between peroxidiac cysteine199 that reacts with H2O2 to form a sulfenic acid (and water) and the resolving cysteine208 of the Escherichia coli peroxide sensor OxyR (Zheng et al., 1998). The intramolecular C199-C208 disulfide is coupled to changes in the probes’ fluorescent properties by incorporating the OxyR sensor into a circularly permuted YFP (Belousov et al., 2006; Markvicheva et al., 2011). In normally reduced cellular compartments, such as the cytosol and mitochondrial matrix, reduced thioredoxin maintains the OxyR cysteines in their reduced state, ready to respond to H2O2 (Belousov et al., 2006). In the ER, however, HyPer is severely compromised in its ability to sense H2O2, likely by a competing H2O2-independent, disulfide exchange–mediated formation of a C199-C208 disulfide (Malinouski et al., 2011; Mehmeti et al., 2012).
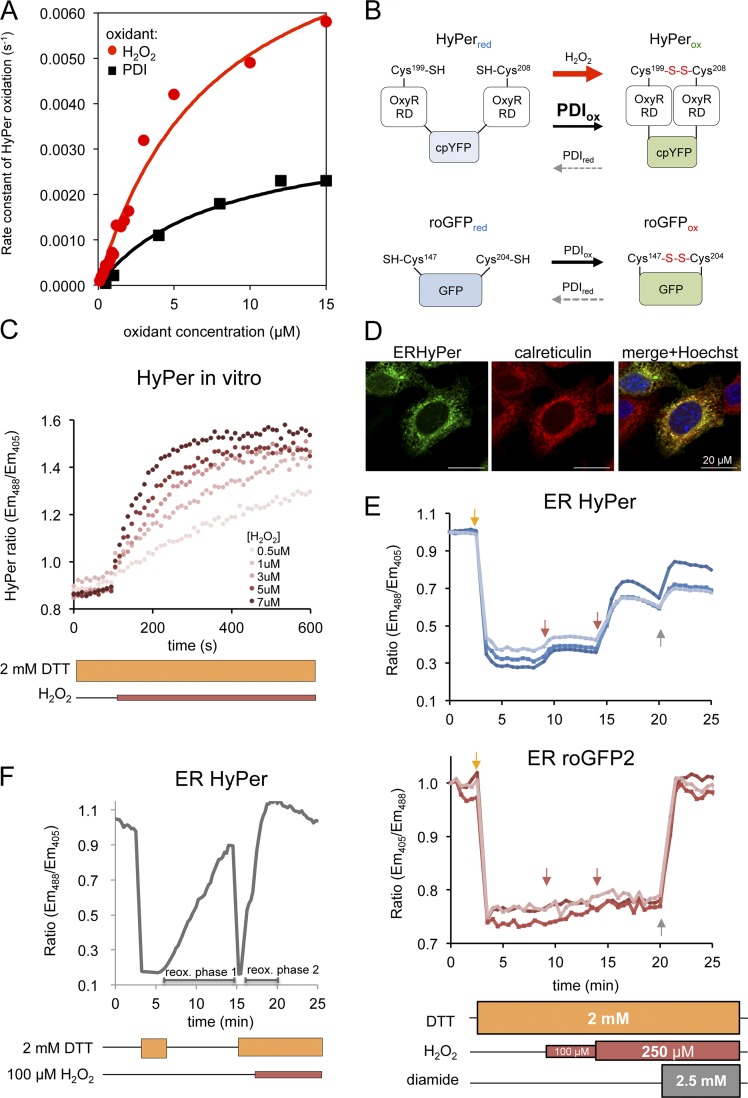
Inactivation of ER-localized HyPer fits well with our observation that HyPer readily served as a substrate for oxidized PDI (Fig. 2 A and Fig. S2). However, these same in vitro experiments revealed an important kinetic advantage to H2O2 over oxidized PDI in converting HyPer from its reduced to its oxidized form (Fig. 2 A; and Fig. S2, A and B). To determine whether this kinetic advantage could be exploited to sense H2O2 in the ER, we compared the effect of exogenous H2O2 on the rate of reoxidation of ERHyPer with that of ERroGFP2, which is indifferent to H2O2 (Gutscher et al., 2009), in a DTT washout experiment in TKO cells. H2O2 enhanced the typically sluggish reoxidation of ERHyPer but had no effect on ERroGFP2 (Fig. S2, C and D).
Figure 2.
ERHyPer responds to exogenous H2O2 in a chemically imposed reducing environment. (A) Plot of the rate of in vitro oxidation of HyPer (1 µM) as a function of H2O2 or oxidized PDI (PDIox) concentration, calculated from the linear phase of the initial oxidation reaction traced ratiometrically (Fig. S2). (B) Schema representing response modes of ERroGFP2 and ERHyper. Both probes undergo oxidation by PDI. Unlike roGFP2, reduced HyPer also undergoes rapid oxidation by H2O2. Thus, HyPer’s responsiveness to H2O2 can be unmasked in settings with a low concentration of oxidized PDI. (C) Traces of time-dependent changes to the redox-sensitive excitation ratio of HyPer exposed to various concentrations of H2O2 in vitro, in the continued presence of DTT. (D) Fluorescent photomicrographs of MEFs transiently transfected with ERHyPer encoding vector, immunostained for calreticulin, as an ER marker. (E) Trace of time-dependent changes to the oxidation state of ERHyPer (top plot) or ERroGFP2 (bottom plot) expressed in MEF cells challenged repeatedly with H2O2 (arrows) in the continuous presence of DTT. The general oxidant, diamide, was added in excess, to reveal the responsiveness of both probes at the end of the experiment. (F) Trace of time-dependent changes to the redox state of ERHyPer expressed in MEF cells, first briefly exposed to DTT, followed by wash out, and then exposed to DTT, followed by introduction of H2O2 to the reducing medium.
The reactivity of HyPer with PDI observed in vitro (Fig. 2 A) explains the inactivity of the probe in the ER under baseline conditions (Fig. 2 B). Furthermore, in wild-type cells, with a normal complement of ERO1, PDI-driven reoxidation of HyPer dominates, precluding detection of H2O2. However, if H2O2 oxidation of HyPer were to exceed the rate of its reduction by a counteracting reductant (e.g., DTT), changes in H2O2 could be detected in the face of the continued presence of a reductant, neutralizing the contribution of PDI. Therefore, we tested the ability of HyPer to respond to H2O2 in vitro in the presence of DTT.
Fig. 2 C indicates that in vitro HyPer retains sensitivity to low concentrations of H2O2 (0–7 µM) in the presence of higher concentration of DTT (2 mM). These features are also observed in vivo, as in wild-type cells exposed continuously to 2 mM DTT (a concentration sufficient to fully reduce PDI thiols), addition of H2O2 led to a rapid oxidation of ERHyPer but not ERroGFP2 (Fig. 2, D and E). Reoxidation of HyPer by H2O2 in the presence of DTT was faster than that afforded by the core machinery after DTT washout (Fig. 2 F, compare the first and second oxidation phases). Moreover, the presence of DTT had no observable effect on the response of cytosolically located HyPer (cytoHyPer) exposed to a gradient of H2O2 (Fig. 3 A). These observations are consistent with rapid formation of the OxyR disulfide by reaction with H2O2 and its slow reduction by DTT, paralleling the hierarchy observed with PDI (Fig. 2, A and B).
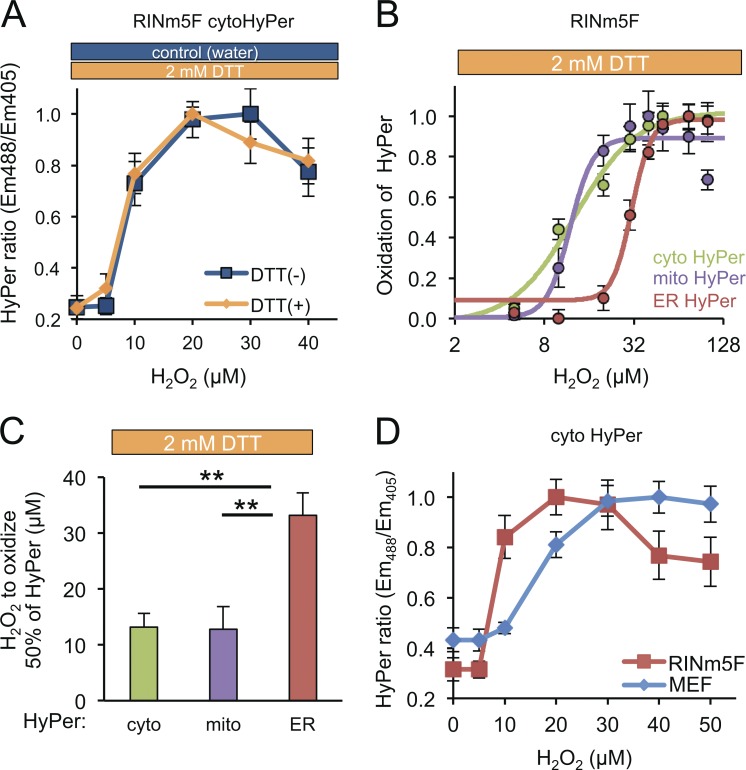
Figure 3.
Sluggish transit of H2O2 into the ER lumen. (A) Plot showing the dependence of the redox state of cytoHyPer-expressed in RINm5F cells (measured by excitation ratio) on concentration of H2O2, introduced to the culture medium in the continued presence or absence of DTT (2 mM). (B) Plot of the relationship between HyPer oxidation state (linearly normalized, by setting the maximal observed value to 1 and the minimal observed value to 0) in the different compartments and concentration of H2O2 in the media in RINm5F cells exposed continuously to DTT (as in A). (C) Bar diagram of the mean H2O2 concentration in the culture media required to effect 50% of maximal oxidation of cytosolic, mitochondrial, or ER-localized HyPer (the midpoint in the traces shown in B; mean ± SEM, n = 4). **, P < 0.01. (D) Titration of H2O2 as described in C, performed in the presence of DTT in RINm5F or MEF cells. Note the enhanced sensitivity of RINm5F cells compared with the MEFs.
Having developed a measurement method responsive to exogenously imposed changes in H2O2 concentration in the ER, we next set out to compare the responsiveness of cytoHyPerand mitochondrial HyPer and ERHyPer to mounting concentrations of exogenous H2O2. Remarkably, reoxidation of ERHyPer by exogenous H2O2 was considerably less efficient than that of the cytosolic or mitochondrially localized probe (midpoint of 33.2, 13.2, and 12.8 µM of extracellular H2O2, respectively; Fig. 3, B and C). To minimize the potential impact of ROS quenching enzymes in the various cellular compartments, we chose to perform these experiments in catalase/peroxidase-deficient RINm5F pancreatic cells (Tiedge et al., 1997). In our system, this deficiency is manifested by higher sensitivity of cytoHyPer to a gradient of H2O2 (Fig. 3 D).
The H2O2-mediated ER thiol oxidation pathway relies on an internal ER source of the oxidant
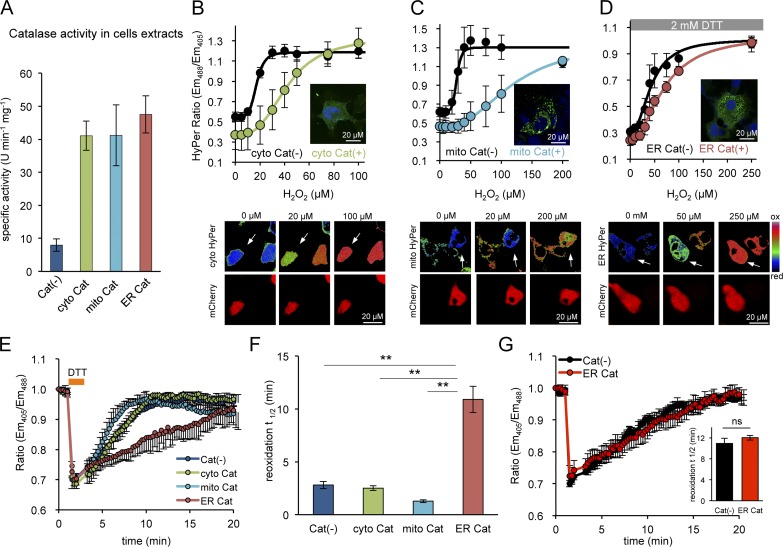
The aforementioned observations suggest the existence of a functional barrier to the rapid diffusion of H2O2 to the ER and therefore an ER source of H2O2 to fuel PRDX4-mediated disulfide bond formation, even in cells lacking ERO1. To further explore this possibility, we sought to selectively eliminate H2O2 from the different compartments by localized expression of catalase and then monitor the effects on ER thiol oxidation rate in DKO cells, a system in which this parameter is dependent on H2O2-fueled PRDX4 activity.
Targeting catalase to the cytosol, mitochondria, or ER markedly elevated the total catalase activity in lysates of transfected cells, indicating that the enzyme was active in all three compartments (Fig. 4 A). Activity of catalase was further confirmed by the observation that its presence in any of the three compartments attenuated the response of a colocalized HyPer to exogenous H2O2 (Fig. 4, B–D). The (obligatory) presence of DTT in the assay of ER catalase (Fig. 4 D) and differences in compartment size, obscure the correlation between catalase expression level and the magnitude of the attenuating effect on colocalized HyPer. Nonetheless, our observations point to the activity of catalase in all three compartments.
Figure 4.
Purging the ER of its H2O2 content selectively retards PRDX4-mediated ER oxidation. (A) Bar diagram showing specific activities of catalase in extracts of untransfected MEFDKO cells and cells expressing catalase variants targeted to the indicated compartments. (B–D) Plot of the relationship between H2O2 concentration in the media and the excitation ratio of cytoHyper, mitochondrially located HyPer (mitoHyPer), or ERHyPer, expressed alone or alongside catalase in MEFDKO cells (with insets of fixed anti-catalase immunostained cells showing the localization of the protein). The titration measuring the ER-localized probe was performed in presence of 2 mM DTT. The plots are presented along their corresponding ratiometric photomicrographs (B–D, bottom panels). The catalase-expressing cells are marked by the coexpression of mCherry (encoded on the same plasmid, denoted by white arrows). Note, the ratio indicates a more reduced state (color coded in blue-green) at the intermediate H2O2 concentration in catalase-positive cells compared with the mostly oxidized state (color coded yellow-red) in catalase-negative cells, as the latter are desensitized to H2O2 by catalase overexpression. (E) Traces of oxidation recovery of ERroGFP2 after a DTT pulse (2 mM, 1 min) in MEFDKO cells with catalase expressed in the indicated compartments. (F) Bar diagram of the t1/2 to recovery of the oxidized form of ERroGFP2 after the reductive DTT pulse, calculated from fitting the data in E. Shown are mean t1/2 values ± SEM (n > 10; **, P < 0.01). (G) Traces of oxidation recovery of ERroGFP2 after a DTT (2 mM, 1 min) pulse in parental TKO cells [Cat(−)] and TKO cells expressing catalase in their ER [ER Cat], identified by the presence of the coexpressed mCherry marker, as in E–G]. Inset shows a bar diagram of the corresponding ERroGFP2 reoxidation t1/2 values. Shown are means ± SEM (n > 10).
Despite their ability to degrade cytosolic and mitochondrially localized H2O2, neither cytosolic nor mitochondrially localized catalase affected the rate of reformation of disulfide bonds in the ER: the t1/2 to recovery of ERroGFP2 in DKO cells after a DTT pulse was similar in untransfected cells and cells transfected with cytosolic or mitochondrially localized catalase (Fig. 4, E and F). In contrast, ER-localized catalase markedly attenuated the reoxidation of the H2O2 inert ERroGFP2 in DKO cells, increasing its t1/2 to recovery from 2.8 ± 0.6 to 10.9 ± 1.24 min (compare the red and purple traces in Fig. 4 E). Thus, ER catalase eliminates the effect of PRDX4, converting DKO cells to functionally TKO cells.
In contrast, ER catalase had no effect on the rate of ERroGFP2 reoxidation in TKO cells, lacking the H2O2-using enzyme PRDX4 (Fig. 4 G). Together, these observations support the conclusion that the attenuated reoxidation of ERroGFP2 in the DKO cells reflects the depletion of H2O2, an essential substrate of PRDX4. Moreover, the lack of an effect of ER catalase on the rate of disulfide bond formation in cells lacking PRDX4 (TKO cells; Fig. 4 G) suggests a nonredundant role for PRDX4 in using ER H2O2 as an oxidant to drive ER thiol oxidation in the absence of ERO1 and indicates the existence of an additional, slow, residual H2O2-independent mechanism for disulfide formation in TKO cells.
Our findings are consistent with the evolution of an ER-localized ERO1-independent mechanism for disulfide bond formation consisting of PRDX4 and an alternative lumenal source of H2O2. For now, the physiological significance of this backup mechanism remains obscure, because the bland phenotype of the PRDX4-deficent mouse provides no clues. Nonetheless, the finding that the ER membrane poses a barrier to mobility of H2O2 is consistent with the need to protect the cytosol and nucleus from lumenally generated H2O2, which has at least two sources: ERO1 and yet-to-be-identified parallel processes uncovered here. The cost of sluggish transport across the ER membrane is reflected in a diminished contribution of mitochondrially generated H2O2 to ER disulfide bond formation. This fits previous observations whereby inhibition of mitochondrial respiration in ERO1-deficient yeast cells did not affect the ability of PRDX4 to rescue disulfide bond formation (Zito et al., 2010b).
Materials and methods
Plasmid construction
Table S1 lists the plasmids used, their laboratory names, description, published references, and a notation of their appearance in the figures.
Protein purification and in vitro enzymatic assays
Human PDI (PDIA1 18-508) and HyPer were expressed in the E. coli BL21 (DE3) strain and purified with Ni-NTA affinity chromatography as previously described (Avezov et al., 2015). Time-dependent changes in redox of HyPer, in the presence of PDI or H2O2, were measured as described (Avezov et al., 2013; Tsunoda et al., 2014). In brief, the ratio of fluorescence emission at 535 nm of samples sequentially excited at 405 nm and 488 nm was measured using a Tecan 500 microplate reader (Tecan Group). For measurements of catalase activity in vitro, cells were homogenized in PBS by sonication; decomposition of substrate (H2O2) was traced by ultraviolet spectroscopy monitoring the absorbance at 240 nm, as described (Tiedge et al., 1998). Specific activity was calculated using the following equation: U/mg = ΔA × min−1 × 1,000 × ml Reaction Mix/43.6 × mg protein, where U is activity units in micromoles per minute.
Transfections, immunoblotting, immunofluorescence, and cell culture
Mouse fibroblasts deficient in both ERO1a and ERO1b (DKO cells, genotype: Ero1IGt(xst171)Byg/Gt(XST171)Byg; Ero1IbGt(P077G11)Wrst/Gt(P077G11)Wrs; Zito et al., 2010a) and ERO1a- and ERO1b-deficient cells compounded further by deletion of PRDX4 (TKO cells, genotype: Ero1IGt(xst171)Byg/Gt(XST171)Byg; Ero1IbGt(P077G11)Wrst/Gt(P077G11)Wrs; Prdx4tm1.1JuFu/tm1.1JuFu; Zito et al., 2012), as well as single deficiency of PRDX4 (PKO, genotype Prdx4tm1.1JuFu/tm1.1JuFu; Iuchi et al., 2009; a gift from R. Edgar and A. Reddy, University of Cambridge Institute of Metabolic Science, Cambridge, England, UK) and wild-type counterpart cells were cultured in DMEM; RINm5F cells (ATCC) were cultured in RPMI (Sigma-Aldrich), both supplemented with 10% fetal calf serum.
Transfections were performed using the Neon Transfection System (Invitrogen) applying 3 µg of ERroGFP2 or ERHyPer DNA, 6 µg of PRDX4, and 10 µg of catalase DNA/106 cells.
For immunoblotting, cells from confluent 100-mm dishes were washed in PBS and lysed in 0.5% Triton X-100, 150 mM NaCl, 20 mM Hepes, pH 7.4, and protease inhibitors. Proteins were resolved by 12% SDS-PAGE and blotted with rabbit ERO1α, ERO1β, or PRDX4 antisera (Zito et al., 2010b) or with mouse monoclonal anti–actin IgG (Abcam).
Before immunofluorescence staining, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100/PBS, and blocked with 10% goat serum/PBS. To visualize the ER, rabbit anti–calreticulin IgG (Pierce) and goat anti–rabbit IgG conjugated to DyLight 543 (Jackson ImmunoResearch Laboratories) were used as the primary and secondary antibodies, respectively. Catalase was detected using goat IgG (Abcam) and donkey anti–goat IgG conjugated to Alexa Fluor 488 (Jackson ImmunoResearch Laboratories) as primary and secondary antibodies, respectively. ERroGFP2 and ERHyPer were detected by their fluorescence. Nuclei were counterstained with Hoechst 33342 (2 µg/ml in PBS).
Confocal microscopy and image analysis
Cells transfected with the redox reporters (roGFP2 or HyPer) were acquired and analyzed by a laser scanning confocal microscopy system running Zen Blue software (LSM 510 Meta; Carl Zeiss) with a Plan-Apo Chromat 60× oil immersion lens (NA 1.4), coupled to a microscope incubator, maintaining standard tissue culture conditions (37°C, 5% CO2; Okolab), in complete DMEM culture medium. Fluorescence ratiometric intensity images (512 × 512 points, 16-bit) of live cells were acquired. A 405-nm diode and 488-nm Argon lasers (2% and 0.5% output, respectively) were used for excitation of the ratiometric probes in multitrack mode with an HFT 488/405 beam splitter. The signal was detected with 518- to 550-nm filters, and the detector gain was arbitrarily adjusted to yield an intensity ratio of the two channels approximating one.
The recovery t1/2 was extracted from fitting the intensity ratio changes over time to an exponential equation: I (t) = A (1 − e−τt), where I is intensity, t is time, and τ is the fitted parameter. After obtaining τ from the fitting curve, the recovery t1/2 was calculated using the formula t1/2 = ln(0.5)/−τ. Images were analyzed using ImageJ (National Institutes of Health) and Zen (Carl Zeiss) software.
Online supplemental material
Fig. S1 shows measurements of ERroGFP2 reoxidation kinetics in PRDX4 KO cells. Fig. S2 shows HyPer oxidation rates driven by H2O2 or PDI, in vitro and in vivo. Table S1 lists the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506123/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201506123.dv.
Supplementary Material
Acknowledgments
We are grateful to Joseph E. Chambers for critical comments on the manuscript.
This study was supported by grants from the Wellcome Trust (Wellcome 084812), the European Commission (EU FP7 Beta-Bat 277713), and Fundação para a Ciência e Tecnologia (PTDC/QUI-BIQ/119677/2010), as well as a Wellcome Trust Strategic Award for core facilities to the Cambridge Institute for Medical Research (Wellcome 100140). D. Ron is a Wellcome Trust Principal Research Fellow. T. Konno was supported by the Japan Society for the Promotion of Science Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- DKO
- double knockout
- MEF
- mouse embryonic fibroblast
- PDI
- protein disulfide isomerase
- PKO
- peroxiredoxin 4 knockout
- roGFP
- redox-sensitive GFP
- TKO
- triple knockout
References
- Araki K., Iemura S., Kamiya Y., Ron D., Kato K., Natsume T., and Nagata K.. 2013. Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 202:861–874. 10.1083/jcb.201303027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E., Cross B.C., Kaminski Schierle G.S., Winters M., Harding H.P., Melo E.P., Kaminski C.F., and Ron D.. 2013. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J. Cell Biol. 201:337–349. 10.1083/jcb.201211155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E., Konno T., Zyryanova A., Chen W., Laine R., Crespillo-Casado A., Melo E.P., Ushioda R., Nagata K., Kaminski C.F., et al. . 2015. Retarded PDI diffusion and a reductive shift in poise of the calcium depleted endoplasmic reticulum. BMC Biol. 13:2 10.1186/s12915-014-0112-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., and Lukyanov S.. 2006. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 3:281–286. 10.1038/nmeth866 [DOI] [PubMed] [Google Scholar]
- Frand A.R., and Kaiser C.A.. 1998. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1:161–170. 10.1016/S1097-2765(00)80017-9 [DOI] [PubMed] [Google Scholar]
- Gross E., Sevier C.S., Heldman N., Vitu E., Bentzur M., Kaiser C.A., Thorpe C., and Fass D.. 2006. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA. 103:299–304. 10.1073/pnas.0506448103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., and Dick T.P.. 2009. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 284:31532–31540. 10.1074/jbc.M109.059246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., and Remington S.J.. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044–13053. 10.1074/jbc.M312846200 [DOI] [PubMed] [Google Scholar]
- Hudson D.A., Gannon S.A., and Thorpe C.. 2015. Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic. Biol. Med. 80:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi Y., Okada F., Tsunoda S., Kibe N., Shirasawa N., Ikawa M., Okabe M., Ikeda Y., and Fujii J.. 2009. Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem. J. 419:149–158. 10.1042/BJ20081526 [DOI] [PubMed] [Google Scholar]
- Lohman J.R., and Remington S.J.. 2008. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 47:8678–8688. 10.1021/bi800498g [DOI] [PubMed] [Google Scholar]
- Malinouski M., Zhou Y., Belousov V.V., Hatfield D.L., and Gladyshev V.N.. 2011. Hydrogen peroxide probes directed to different cellular compartments. PLoS ONE. 6:e14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markvicheva K.N., Bilan D.S., Mishina N.M., Gorokhovatsky A.Y., Vinokurov L.M., Lukyanov S., and Belousov V.V.. 2011. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg. Med. Chem. 19:1079–1084. 10.1016/j.bmc.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Mehmeti I., Lortz S., and Lenzen S.. 2012. The H2O2-sensitive HyPer protein targeted to the endoplasmic reticulum as a mirror of the oxidizing thiol-disulfide milieu. Free Radic. Biol. Med. 53:1451–1458. 10.1016/j.freeradbiomed.2012.08.010 [DOI] [PubMed] [Google Scholar]
- Pollard M.G., Travers K.J., and Weissman J.S.. 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 1:171–182. 10.1016/S1097-2765(00)80018-0 [DOI] [PubMed] [Google Scholar]
- Tavender T.J., and Bulleid N.J.. 2010. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J. Cell Sci. 123:2672–2679. 10.1242/jcs.067843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender T.J., Springate J.J., and Bulleid N.J.. 2010. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29:4185–4197. 10.1038/emboj.2010.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge M., Lortz S., Drinkgern J., and Lenzen S.. 1997. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 46:1733–1742. 10.2337/diab.46.11.1733 [DOI] [PubMed] [Google Scholar]
- Tiedge M., Lortz S., Munday R., and Lenzen S.. 1998. Complementary action of antioxidant enzymes in the protection of bioengineered insulin-producing RINm5F cells against the toxicity of reactive oxygen species. Diabetes. 47:1578–1585. 10.2337/diabetes.47.10.1578 [DOI] [PubMed] [Google Scholar]
- Tien A.C., Rajan A., Schulze K.L., Ryoo H.D., Acar M., Steller H., and Bellen H.J.. 2008. Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J. Cell Biol. 182:1113–1125. 10.1083/jcb.200805001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., and Rapoport T.A.. 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol. 159:207–216. 10.1083/jcb.200207120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S., Avezov E., Zyryanova A., Konno T., Mendes-Silva L., Pinho Melo E., Harding H.P., and Ron D.. 2014. Intact protein folding in the glutathione-depleted endoplasmic reticulum implicates alternative protein thiol reductants. eLife. 3:e03421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lith M., Tiwari S., Pediani J., Milligan G., and Bulleid N.J.. 2011. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J. Cell Sci. 124:2349–2356. 10.1242/jcs.085530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Aslund F., and Storz G.. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 279:1718–1722. 10.1126/science.279.5357.1718 [DOI] [PubMed] [Google Scholar]
- Zito E., Chin K.T., Blais J., Harding H.P., and Ron D.. 2010a ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 188:821–832. 10.1083/jcb.200911086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Melo E.P., Yang Y., Wahlander Å., Neubert T.A., and Ron D.. 2010b Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell. 40:787–797. 10.1016/j.molcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Hansen H.G., Yeo G.S., Fujii J., and Ron D.. 2012. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol. Cell. 48:39–51. 10.1016/j.molcel.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.