Visualization of nascent lipid droplets reveals that they form lens-like structures inside the ER membrane bilayer and that FIT proteins are necessary for lipid droplet protrusion toward the cytoplasm.
Abstract
Lipid droplets (LDs) are found in all cells and play critical roles in lipid metabolism. De novo LD biogenesis occurs in the endoplasmic reticulum (ER) but is not well understood. We imaged early stages of LD biogenesis using electron microscopy and found that nascent LDs form lens-like structures that are in the ER membrane, raising the question of how these nascent LDs bud from the ER as they grow. We found that a conserved family of proteins, fat storage-inducing transmembrane (FIT) proteins, is required for proper budding of LDs from the ER. Elimination or reduction of FIT proteins in yeast and higher eukaryotes causes LDs to remain in the ER membrane. Deletion of the single FIT protein in Caenorhabditis elegans is lethal, suggesting that LD budding is an essential process in this organism. Our findings indicated that FIT proteins are necessary to promote budding of nascent LDs from the ER.
Introduction
Lipid droplets (LDs) are ubiquitous organelles that play central roles in energy and lipid metabolism (Murphy and Vance, 1999; Murphy, 2001; Martin and Parton, 2006). Neutral lipid sequestration into LDs is crucial for cellular defense against lipotoxicity (Listenberger et al., 2003; Unger and Scherer, 2010). It is also becoming clear that LDs have additional functions, including important roles in protein degradation (Hartman et al., 2010; Olzmann et al., 2013), the ER stress response (Fei et al., 2009), and viral replication (Miyanari et al., 2007).
LDs have a unique architecture: a core of neutral lipids, triacylglycerols (TAGs), and sterol esters (SEs) surrounded by a phospholipid monolayer (Murphy, 2001; Martin and Parton, 2006; Wilfling et al., 2014). LD biogenesis is known to occur in the ER, but the mechanism is not well understood (Murphy and Vance, 1999; Wältermann et al., 2005; Andersson et al., 2006; Wolins et al., 2006; Ploegh, 2007; Ohsaki et al., 2009, 2014; Robenek et al., 2009; Skinner et al., 2009; Walther and Farese, 2009; Brasaemle and Wolins, 2012; Pol et al., 2014; Wilfling et al., 2014). The most widely held model states that as neutral lipids are synthesized in the ER, they accumulate between the two leaflets, forming lens (or blisters) that grow and bud into the cytoplasm (Fig. 1 A; Murphy and Vance, 1999; Czabany et al., 2007; Gross and Silver, 2014; Ohsaki et al., 2014; Wilfling et al., 2014). Mature LDs may completely separate from the ER, or it has been suggested that, at least in yeast, LDs remain connected to the ER by a membrane bridge (Jacquier et al., 2011).
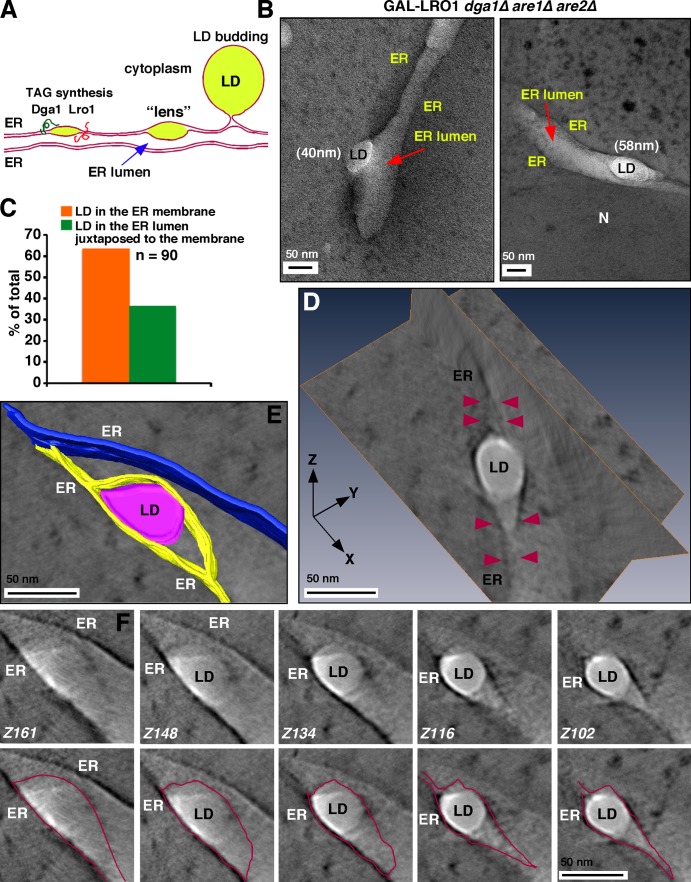
Figure 1.
Nascent LDs form lenses in the ER membrane. (A) Model of LD biogenesis. Dga1 and Lro1 are the two TAG synthases in yeast. (B) GAL-LRO1 dga1Δ are1Δ are2Δ cells were grown to mid-logarithmic growth phase in a medium with raffinose. Galactose was added to the medium to induce TAG formation, and 10 min later, the cells were harvested and visualized by cryo-EM. Two examples are shown. Arrows indicates the ER lumen. N, nucleus. (C) Quantification of location of nascent LDs. (D) Tomographic reconstruction of an early-stage LD inside the ER bilayer membrane. Orthogonal projection x/y and x/z are shown; arrowheads indicate the bifurcating ER membrane that encases the nascent LD. (E) Segmentation analysis of the tomogram. The bifurcating ER membrane encasing the LD (marked in purple) is shown in yellow. The nonbifurcating ER membrane is shown in blue. (F) Sequential tomography slices (with or without contours shown in red) of the LD shown in D. Z, slice number.
How nascent LDs form in the ER is not well understood, and what role proteins play is unclear. Three families of proteins are thought to participate in de novo LD biogenesis: seipins (Szymanski et al., 2007), lipins (Adeyo et al., 2011), and fat-storage-inducing transmembrane (FIT) proteins (Kadereit et al., 2008; Gross et al., 2011). Lipins play important roles in regulating TAG formation (Carman and Han, 2009; Kohlwein, 2010; Henry et al., 2012; Siniossoglou, 2013), and there is evidence that they promote TAG formation in regions of the ER where LD biogenesis occurs (Adeyo et al., 2011; Karanasios et al., 2013). Seipin (BSCL2 in humans and Fld1p in yeast) is an ER transmembrane protein that localizes to ER-LD junctions in yeast and modulates LD abundance and size (Szymanski et al., 2007; Fei et al., 2008). Mutations in human BSCL2/Seipin result in the most severe form of congenital generalized lipodystrophy (Magré et al., 2001; Agarwal and Garg, 2004). FIT proteins are conserved transmembrane proteins that are ER localized (Kadereit et al., 2008). Humans have two FIT proteins, FIT1 and FIT2; FIT1 is primarily found in muscle, whereas FIT2 is ubiquitously expressed. FIT proteins are not homologous with other known proteins (Kadereit et al., 2008). They bind TAG in vitro (Gross et al., 2011), and it has been found that knockdown of FIT2 causes a dramatic reduction in the number and size of lipid droplets (Kadereit et al., 2008). Knockdown of FIT2 in the adipose tissue of mice results in lipodystrophy and insulin resistance (Miranda et al., 2014). The role of FIT proteins in LD biogenesis is not well understood. It has been proposed that FIT proteins generate nascent LDs by concentrating TAG between the leaflets of the ER, thus promoting LD budding (Goh and Silver, 2013). Consistent with this proposal, it was recently found that postnatal knockdown of FIT2 causes the absence of cytosolic LDs in the intestinal cells of mice (Goh et al., 2015).
In this study, we show that FIT proteins facilitate proper LD budding from the ER. We find that nascent LDs form lens-like structures in the ER membrane and that in the absence of FIT proteins, nascent LDs fail to bud from the ER and instead grow and remain in the ER membrane. This function of FIT proteins is conserved in yeast, worms, and mammalian cells. Interestingly, worms lacking the single FIT gene in this organism are not viable, indicating that proper LD budding from the ER is essential in higher eukaryotes.
Results and discussion
To visualize early stages of LD biogenesis, we used a Saccharomyces cerevisiae strain in which de novo LD biogenesis can be controlled. Four ER-resident enzymes synthesize neutral lipids in yeast: Dga1p and Lro1p catalyze TAG formation, and Are1p and Are2p produce SEs. Yeast cells lacking all four proteins are viable but lack detectable LDs and neutral lipids (Sandager et al., 2002). In this study, we used a strain that lacks three of these enzymes, whereas expression of the fourth is under the regulatable GAL1 promoter (GAL1-LRO1, dga1Δ are1Δ are2Δ; Jacquier et al., 2011). This promoter is off in media containing raffinose but is activated when galactose is added to the medium. Thus, in media containing raffinose, GAL1-LRO1 dga1Δ are1Δ are2Δ cells lack LDs but begin to produce TAG and form LDs when galactose is added to the medium.
Nascent LDs form lenses in the ER membrane
To visualize nascent LD formation, GAL-LRO1 dga1Δ are1Δ are2Δ cells growing in a raffinose-containing medium were supplemented with galactose and grown for 10 min, which is sufficient to induce TAG production (Jacquier et al., 2011). The cells were then subjected to high-pressure freezing (HPF), automatic freeze substitution, and visualized by EM and EM tomography. Small, white, lens-like, electron-transparent structures were observed (labeled LD in Fig. 1 B). Typically, one or two of these structures were observed per cell. These structures are probably nascent LDs for three reasons. First, they were never observed before TAG production was induced (Jacquier et al., 2011). Second, it is well established that LDs are electron-transparent structures in EM of the ultrathin sections of cells (Binns et al., 2006; Adeyo et al., 2011; Jacquier et al., 2011, 2013; Cartwright et al., 2015). Third, the electron-transparent structures were only found in the ER, where LD biogenesis is known to occur; all nascent LDs were found either in the ER membrane or in the ER lumen closely apposed to the ER membrane (Fig. 1 C). The nascent LDs had diameters that ranged from 30 to 60 nm. Lens formation upon TAG production was not unique to Lro1p, which unlike Dga1p has an active site facing the ER lumen (Choudhary et al., 2011). Similar lens-like structures were observed when TAG was produced by Dga1p instead of Lro1p (Fig. S1 A).
To determine if the nascent LDs are in or next to the ER membrane, we performed EM tomography and 3D reconstruction. This analysis revealed that the nascent LDs are encased in the ER membrane, which appears to split where the nascent LDs are present (Fig. 1, D–F). Thus, nascent LDs form lens-like structures that are probably between the two leaflets of the ER membrane (Fig. 1, D and E), as has been predicted by some models of LD biogenesis (Murphy and Vance, 1999; Khandelia et al., 2010; Ohsaki et al., 2014; Wilfling et al., 2014).
Nascent LD formation is abnormal in cells lacking FIT proteins
As nascent LD lens grow larger, they are thought to protrude and bud from the ER toward the cytoplasm to form mature LDs (Fig. 1 A). We examined the role of FIT proteins in LD growth and maturation. S. cerevisiae has two homologues of FIT2, called Scs3p and Yft2p (Kadereit et al., 2008; Moir et al., 2012), and no FIT1 homologues. We verified that both FIT proteins in yeast reside in the ER, as has been shown for the mammalian proteins (Kadereit et al., 2008; Fig. S1 B).
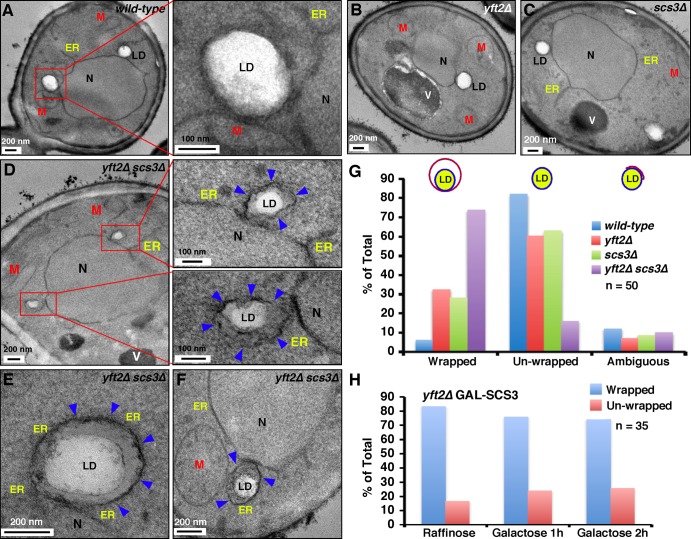
To determine whether FIT proteins play a role in nascent LD protrusion and budding from the ER toward the cytoplasm, we deleted SCS3 and YFT2 in GAL-LRO1, dga1Δ are1Δ are2Δ cells. The resulting strain was grown in a raffinose-containing medium. TAG production was induced by adding galactose to the medium and the cells were grown for 45 min, chemically fixed, and visualized by EM. Surprisingly, an electron-dense membrane was found surrounding most LDs (Fig. S1 C). This finding prompted us to examine mature LDs in cells lacking only the FIT proteins and not lacking neutral lipid synthesizing enzymes. Wild-type cells and yeast mutants lacking Scs3p, Yft2p, or both proteins were chemically fixed and visualized by EM. In cells missing either or both FIT proteins, some LDs were wrapped by an additional membrane that was not usually found around LDs in wild-type cells (Fig. 2, A–F). Quantification revealed that ∼30% of LDs were membrane wrapped in cells lacking either of the FIT proteins and ∼70% of LDs were wrapped in cells that lacked both proteins (Fig. 2 G).
Figure 2.
Most LDs in yft2Δ scs3Δ cells are surrounded by a membrane. (A–F) Cells were grown in YPD medium and visualized by EM after chemical fixation. Arrowheads indicate membranes wrapping the LDs. In A and D, the red boxes indicate regions shown at higher magnification. M, mitochondria; N, nucleus; V, vacuole. (G and H) Quantification of percentage of LDs that are wrapped in the indicated strains (n = 50 in G, n = 35 in H).
Yeast lacking Scs3p grow very poorly in media lacking inositol (Hosaka et al., 1994). However, we have no evidence that the inositol auxotrophy of cells lacking Scs3p is related to the abnormal LD biogenesis in cells lacking FIT proteins. EM analysis of LDs in scs3Δ yft2Δ cells grown in media with or without inositol revealed no morphological difference (unpublished data).
The membrane surrounding LDs in FIT mutants is not related to autophagy
We investigated the origin of the membrane that wraps most LDs in cells lacking FIT proteins. The membrane is probably not related to autophagosomal degradation of LDs in cells lacking FIT proteins. LDs in yeast can be degraded by macroautophagy and microlipophagy, the direct engulfment of LDs by the vacuole (van Zutphen et al., 2014).
We found that membrane wrapping of LDs still occurs in cells lacking both FIT proteins and Atg proteins needed for both macroautophagy and microlipophagy (Fig. S2 A). We also did not find evidence that LDs are undergoing degradation in cells lacking FIT proteins. If lipophagy were occurring, LDs would be seen inside the vacuolar lumen and fluorescently tagged proteins on the surface of lipid droplets, such as Erg6p, would be in vacuoles (van Zutphen et al., 2014). However, we did not see LDs or Erg6-GFP in the vacuoles of cells lacking FIT proteins (Fig. S2 B). Together, these observations suggest that the membranes surrounding LDs in cells lacking FIT proteins are unrelated to autophagy of LDs.
The membrane surrounding LDs in FIT mutants is the ER
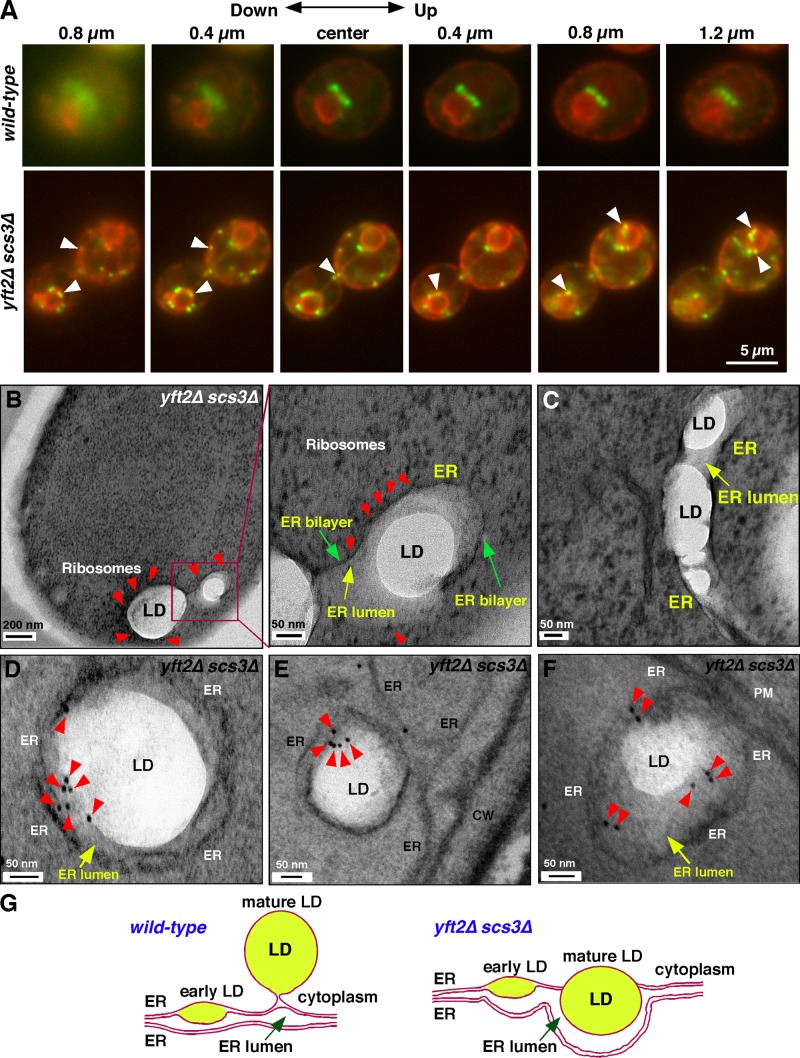
Analysis of cells lacking FIT proteins by fluorescence microscopy suggested that the membrane wrapping LDs is part of the ER. We expressed the ER luminal marker RFP-HDEL in cells and visualized LDs using the lipophilic dye BODIPY 493/503, which is known to stain LDs (Guo et al., 2008; Adeyo et al., 2011; Jacquier et al., 2011; Cartwright et al., 2015). In wild-type cells, LDs appeared to be adjacent to the ER but usually did not overlap with it (Fig. 3 A), suggesting that the ER does not surround LDs. In contrast, most LDs in cells lacking Scs3p and Yft2p remain closely associated with or colocalized with the ER (Fig. 3 A), suggesting that the membrane found surrounding many LDs in cells lacking FIT proteins is the ER.
Figure 3.
The membrane surrounding LDs in yft2Δ scs3Δ cells is the ER. (A) Wild-type and yft2Δ scs3Δ cells expressing ER luminal marker RFP-HDEL were grown to mid-logarithmic phase and stained with BODIPY 493/503 to visualize LDs. Optical sections showing consecutive 0.4-µm z-section images are shown. Arrowheads indicate close association of LDs with the ER. (B and C) Visualization of LDs in scs3Δ yft2Δ cells using cryo-EM. Red arrowheads indicate the ribosomes. Yellow arrows indicate the ER lumen, and green arrows denote the ER membrane. (D–F) Immunogold EM images of scs3Δ yft2Δ cells expressing ss-GFP-HDEL using anti-GFP antibodies. Arrowheads indicate the gold particles. CW, cell wall; PM, plasma membrane. (G) Model of LD growth in wild-type and scs3Δ yft2Δ cells.
Further evidence that it is the ER membrane that surrounds LDs in cells lacking FIT proteins was obtained when they were imaged by EM after HPF and automatic freeze substitution. This analysis revealed that the membrane surrounding LDs in the cells is decorated with ribosomes and is thus part of the ER (Fig. 3, B and C). Notably, ribosomes were excluded from the space between LDs and the surrounding membrane.
LDs fail to emerge from the ER in cells lacking FIT proteins
There are two reasons why ribosomes might be excluded from the regions between LDs and the ER membrane that surrounds them in cells lacking FIT proteins. Either the ER becomes so closely associated with LDs that ribosomes are excluded or LDs remain in the ER membrane and therefore the region between LDs and the surrounding membrane is the ER lumen (Fig. 3 G). We found that the second possibility is correct by using immunogold labeling to determine whether an ER luminal protein was present in region between LDs and the surrounding ER membrane. We have previously shown that GFP with an N-terminal signal sequence and the C-terminal ER retention signal HDEL (ss-GFP-HDEL) resides in the ER lumen (Prinz et al., 2000). Immunogold labeling of cells lacking FIT proteins and expressing ss-GFP-HDEL revealed that it is present in the area between LDs and the surrounding membrane (Fig. 3, D–F), indicating that this region is part of the ER lumen. It should be noted that the majority of the gold particles were on the ER (∼75% of 800 gold particles observed). Therefore, most LDs in cells lacking FIT proteins fail to emerge from the ER and remain in the ER membrane (Fig. 3 G). Consistent with this, it has recently been shown that mouse intestinal cells lacking FIT2 are devoid of cytoplasmic LDs (Goh et al., 2015).
We wondered whether the LDs that fail to emerge from the ER membrane in cells lacking FIT proteins could bud normally from the ER when FIT proteins are reexpressed in these cells. To address this, expression of Scs3p was placed under the GAL1 promoter in cells lacking Yft2p. In media with raffinose, this strain lacks FIT proteins and ∼80% of LDs are wrapped by the ER membrane (Fig. 2 H). When galactose was added to the medium to induce Scs3p production, there was almost no reduction in the percent of ER-wrapped LDs (Fig. 2 H), suggesting that once LDs fail to emerge from the ER membrane, they cannot be rescued by FIT proteins.
LDs in cells lacking FIT proteins remain accessible to cytoplasmic proteins
We wondered whether some LDs in cell lacking FIT proteins completely detach from the ER membrane into the ER lumen and would therefore no longer be accessible to lipases, which probably have active sites facing the cytoplasm and not the ER lumen (Athenstaedt and Daum, 2003, 2005; Köffel et al., 2005). If this were correct, neutral lipid mobilization from LDs might be impaired. However, we found no difference in the rates at which neutral lipids were mobilized from wild-type cells and those lacking FIT proteins. TAGs and SEs are mobilized from LDs when yeast cells are treated with the fatty acid synthesis inhibitor cerulenin and terbinafine, which inhibits sterol biosynthesis. We treated cultures of wild-type and scs3Δ yft2Δ cells with these drugs and found no significant difference in the rates of neutral lipid mobilization (Fig. S3 A). This indicates that LDs in scs3Δ yft2Δ cells remain accessible to lipases in the cytoplasm and thus probably do not bud completely into the ER lumen. Consistent with this, we showed that Erg6p, an LD protein that binds to the cytosolic surface of LDs (Yang et al., 2012), was found on all LDs in scs3Δ yft2Δ cells (Fig. S3 B). Together, these findings indicated that LDs in cells lacking FIT proteins are accessible to proteins in the cytoplasm and do not bud completely into the ER lumen.
Knockdown of FIT proteins in mammalian cells results in LD wrapping
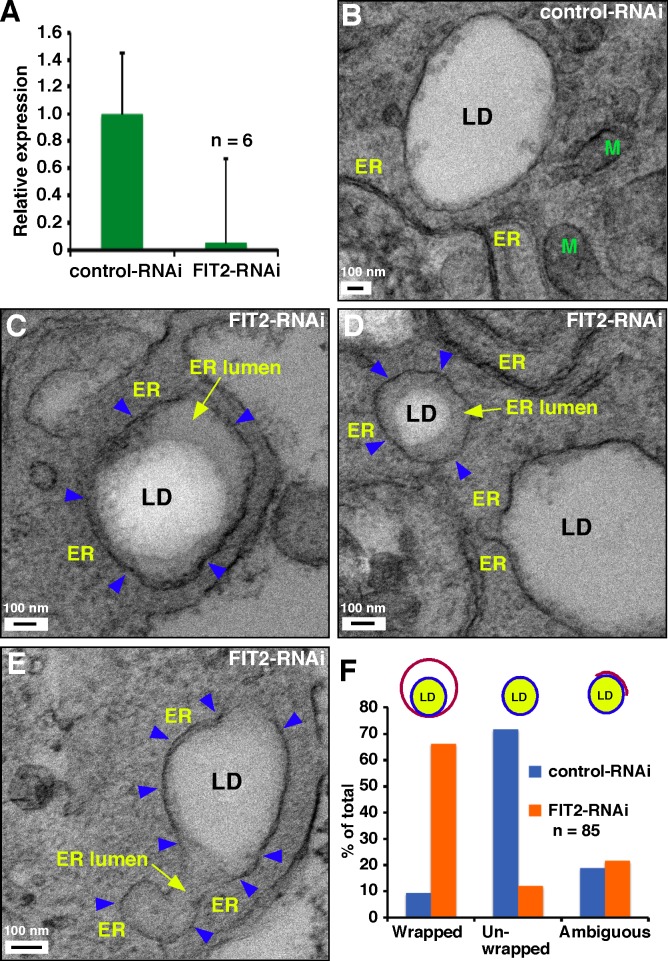
We next asked if FIT proteins are necessary for proper LD budding from the ER in higher eukaryotes. Mouse 3T3-L1 fibroblasts express only FIT2 (Kadereit et al., 2008), and we were able to efficiently knock down expression of the FIT2 gene in these cells using siRNA (Fig. 4 A). 3T3-L1 cells treated with control RNAi and FIT2-RNAi were chemically fixed and processed for EM. Although few LDs were wrapped with a membrane in cells treated with control RNAi (Fig. 4 B), most LDs in FIT2-RNAi–treated cells were membrane wrapped (Fig. 4, C–E); 66% of LDs were membrane wrapped in cells depleted of FIT2, and only 9% of LDs were membrane wrapped in control cells (Fig. 4 F). Analysis of the EM images suggested that the membranes wrapping LDs in FIT2-RNAi–treated cells are continuous with the ER. Thus, in both a mouse cell line and in yeast in which FIT proteins have been eliminated or reduced, the majority of LDs become wrapped by the ER, probably because LDs fail to emerge from the ER membrane.
Figure 4.
FIT2 knockdown in 3T3-L1 cells causes LDs to become surrounded by a membrane. (A) Knockdown of expression of FIT2 by RNAi in cells quantitated by RT-PCR (mean ± SD, n = 6 from two independent experiments). (B–E) LDs from cells treated with control RNAi (B) or FIT2 RNAi (C–E). M, mitochondria. Blue arrowheads indicate ER membrane wrapping LDs and yellow arrows point to ER lumen. (F) Quantification of LD wrapping (n = 85).
The sole FIT protein in Caenorhabditis elegans is essential and plays a role in LD biogenesis
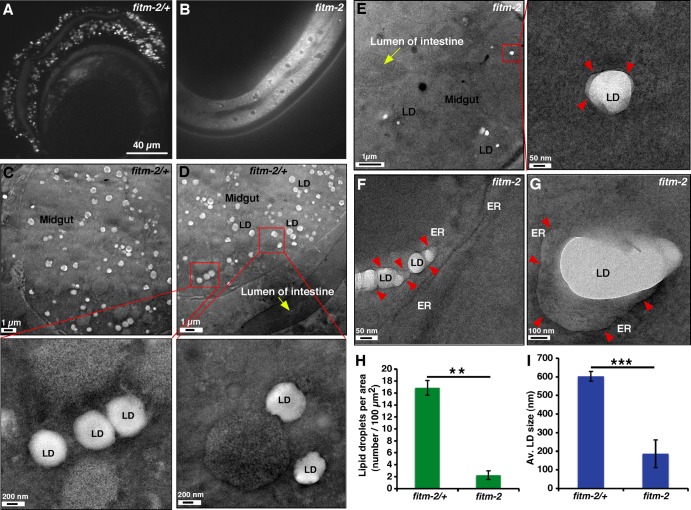
To determine if loss of FIT proteins has adverse effects on higher eukaryotes we used C. elegans, which has only one homologue of FIT2 and no homologue of FIT1 (Kadereit et al., 2008). The C. elegans ORF ZK265.9 encodes a protein that is 29% identical to human FIT2. We named this gene fitm-2. Two mutant alleles of fitm-2 were characterized: a deletion (tm3632Δ) generated by the National Bioresource Project (Japan) and an allele with early nonsense mutations (av41) that we generated using the CRISPR/Cas9 coconversion protocol (Arribere et al., 2014). Animals with the tm3632Δ or the av41 alleles had identical phenotypes; homozygous animals segregating from fitm-2 heterozygous mothers hatched and developed normally through the larval stages but died as young adults. Homozygous animals had very small broods, most of which hatched and arrested as L1 and L2 larvae (mean brood size was 28 with a range of 0–128; 36% of mothers burst). Development of nematodes involves embryogenesis, followed by four larval stages (L1, L2, L3, and L4) and then adulthood. Survival to the adult stage could be due to maternal rescue or could indicate that the fitm-2 gene product is not required until late in development. Homozygous fitm-2(av41) animals had dramatically fewer and smaller LDs than heterozygous animals (Fig. 5, A and B). These results are consistent with an earlier study that found that depletion of FIT2 in 3T3-L1 cells or in zebrafish caused a reduction in the number and size of LDs (Kadereit et al., 2008). In addition, EM analysis revealed that a membrane wrapped most LDs in the intestinal cells of homozygous fitm-2(av41) animals, but not in heterozygous animals (Fig. 5, C–G), suggesting that aberrant LD budding occurs in the intestinal cells of C. elegans lacking FITM-2 as it does in yeast cells lacking FIT proteins. We used EM to quantitate the dramatic decrease in number and size of LDs in homozygous worms compared with heterozygous worms. There was a significant eightfold decrease in the number of LDs per area in fitm-2 worms compared with fitm-2/+ worms (fitm-2 ≈ 2 LDs, fitm-2/+ ≈ 17 LDs per 100 µm2; n = 5 worms; Fig. 5 H). Similarly, homozygous worms had a significant threefold decrease in mean LD size compared with heterozygous worms (mean LD size in fitm-2 ≈ 200 nm, and in fitm-2/+ ≈ 600 nm; n = 50; Fig. 5 I). These findings suggest that homozygous fitm-2(av41) animals are dying because of a severe LD biogenesis defect caused by aberrant LD budding. It may be that the presence of LDs in the ER membrane somehow disrupts lipoprotein production in the ER. This could explain why FIT proteins are essential in C. elegans but not in yeast; worms produce lipoproteins in the ER lumen (Branicky et al., 2010; Zhang et al., 2013), but yeast do not.
Figure 5.
Deletion of fitm-2 in C. elegans causes a profound defect in LD biogenesis and results in most LDs being wrapped by a membrane. Adult animals were stained with BODIPY to visualize LDs and imaged live. A single focal plane focusing on the intestine in heterozygous (A) and homozygous (B) fitm-2 animals is shown. (C–G) EM images of LDs from intestinal cells in adult heterozygous (C and D) and homozygous (E–G) fitm-2 worms. Boxed regions are shown in higher magnification. Red arrowheads indicate the membrane that is wrapping the LD in the homozygous fitm-2 animals. (H) Quantification of LDs per area. Bar graph showing average number of LDs per 100 µm2 area in homozygous and heterozygous fitm-2 worms (mean ± SD, n = 5 individual worms). (I) Quantification of average size of LD. Histogram showing average size of LDs in homozygous and heterozygous fitm-2 worms (mean ± SD, n = 70 for fitm-2/+, n = 50 for fitm-2 worms). **, P < 0.01; ***, P < 0.001, independent two-tailed t test.
In summary, we have found that nascent LDs form lens-like structures in the ER membrane and that FIT proteins are necessary for LD budding from the ER; in the absence of FIT proteins, LDs fail to leave the ER membrane, causing them to frequently become wrapped by the ER membrane. How FIT proteins facilitate LD emergence toward the cytoplasm is a fascinating question. One possibility is that FIT proteins directly facilitate bending of the ER membrane at sites of LD biogenesis. However, FIT proteins are not homologous to proteins known to facilitate membrane deformation. Our findings suggest that FITs play a role in LD biogenesis after the initial formation of neutral lipid lenses in the ER membrane, which may not require any proteins. Indeed, we found that the formation of nascent LD lenses is normal in cells lacking FIT proteins (Fig. S1 D). We propose that FIT proteins incorporate TAG into nascent LDs lenses in a way that promotes LD protrusion toward the cytoplasm, perhaps by adding TAG to the cytosolic side of nascent LD lenses in the ER membrane. Understanding how FIT proteins regulate the growth and budding of nascent LDs in the ER membrane will be critical for understanding the earliest stages of de novo LD biogenesis.
Materials and methods
Yeast strains and growth conditions
Yeast strains and plasmids used in this study are listed in Table S1. Cells were grown in YPD medium (1% Bacto yeast extract, 2% Bacto Peptone, and 2% glucose) unless otherwise indicated. In some cases, the glucose in YPD was replaced with 2% raffinose. Galactose was added to 2% to media containing raffinose. Cells containing plasmids were grown in synthetic complete media containing 6.7 g/l yeast nitrogen base without amino acids (USBiological), an amino acid mix (USBiological), containing either 2% glucose or raffinose.
Fluorescence microscopy and BODIPY staining of yeast
Cells were washed once with PBS and imaged live using an Olympus BX61 microscope, a UPlanApox100/1.35 lens, and a QImaging Retiga EX camera, and processed using iVision software (version 4.0.5). Images being directly compared were obtained using identical microscope settings. Brightness and contrast were adjusted using Photoshop CS2. To visualize LDs in yeast, cells growing in synthetic complete media were stained with BODIPY 493/503 (1 µg/ml final concentration; Invitrogen) for 15 min at RT. After washing twice with PBS, cells were visualized live using the GFP filter.
Mobilization of neutral lipids
Mobilization of neutral lipids was performed as described previously (Jacquier et al., 2011). Cells were grown to stationary growth phase in YPD medium (20–24 h) and diluted to an OD600 of 2.5 in fresh YPD containing cerulenin (10 µg/ml) and terbinafine (30 µg/ml). At the indicated times, 5 ml of the cultures was collected and lipids were extracted as described (Parks et al., 1985). To quantitate TAG and SE, the lipids were spotted onto silica gel 60 TLC plates (Merck) and developed with hexane-diethylether-acetic acid (70:30:1). Glycerolphospholipids were separated by TLC as described (Vaden et al., 2005). Lipids on TLC plates were quantified on a RITA Star Thin Layer Analyzer (Raytest).
Electron microscopy of yeast cells
Sample preparation for EM was done as described previously (Bernales et al., 2006; Jacquier et al., 2011; Lahiri et al., 2014). Yeast cells were grown to mid-logarithmic growth phase, and 10 OD600 units of cells were harvested and fixed in 1 ml fixative media (1% glutaraldehyde, 0.2% paraformaldehyde, and 40 mM potassium phosphate, pH 7.0) for 10 min at RT. Cells were harvested and again resuspended in 1 ml fresh fixative media and incubated on ice for 50 min. Cells were centrifuged and washed twice with 0.9% NaCl and once with water. Cells were incubated with 2% solution of KMnO4 for 5 min at RT, centrifuged, and again resuspended with fresh solution of 2% KMmO4 for 45 min at RT for en bloc staining. The cells were then dehydrated using graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%) for 10 min each, with two more incubations in 100% ethanol from a freshly opened bottle. The samples were subsequently embedded stepwise using Spurr’s low-viscosity resin (EMS). Samples were infiltrated for 2 h each with a 3:1, 1:1, 1:3 dehydrating agent/embedding media mixture. Cells were incubated overnight with 100% fresh resin. The next day, cells were again resuspended in fresh 100% resin for 2–3 h, transferred into BEEM capsules (EMS), and polymerized at 70°C for 72 h. Semi- and ultrathin sections were produced with a diamond knife (Diatome) on an ultramicrotome (Ultracut UCT; Leica Microsystems), collected on 200 mesh copper grids (EMS), poststained with uranyl acetate and lead citrate, and visualized with a Tecnai T12 transmission electron microscope (TEM; FEI), operating at 120 kV. Pictures were recorded on a below-mounted Gatan 2k × 2k charge coupled device (CCD) camera.
Immunogold EM of yeast cells
Sample preparation for immunogold EM was performed as described previously (Wright, 2000). Cells were fixed in Pipes buffer (0.2 M Pipes, pH 6.8, 0.2 M Sorbitol, 2 mM MgCl2, 2 mM CaCl2, 1% glutaraldehyde, and 4% formaldehyde) for 1 h on ice. Cells were harvested and washed twice with water. Thereafter, cells were treated with 1% sodium metaperiodate (15 min) and 1% NH4Cl (15 min) and washed twice with water. Samples were dehydrated in graded ethanol series (50%, 70%, 80%, 90%, 95%, and 100%) for 10 min each, with two more incubations in 100% ethanol from a freshly opened bottle. Samples were infiltrated and embedded using London resin white resin (Polysciences). Samples were infiltrated for 2 h each with a 3:1, 1:1, 1:3 dehydrating agent/embedding media mixture. Cells were incubated overnight with 100% fresh London resin white resin. Next day cells were again resuspended in fresh 100% resin for 2–3 h, transferred into gelatin (size 00) capsules (EMS), and polymerized at 45°C for 24–48 h. Blocks were sectioned, and ultrathin sections (80–100 nm) were collected onto 200 mesh nickel grids (EMS). Sections were blocked and immunogold-labeled using a mouse anti–GFP antiserum (Roche) and 10 nm protein A-gold (EMS) as per the manufacturer’s instructions, poststained with uranyl acetate and lead citrate, and visualized with a Tecnai T12 TEM, operating at 120 kV. Pictures were recorded on a below-mounted 2k × 2k CCD camera. For quantification, a gold particle was assigned to a compartment if it was within 25 nm of the limiting membrane or cytoplasm. The relative distribution of the immunogold labeling was determined by randomly analyzing 800 gold particles in yft2Δ scs3Δ cells.
EM of mammalian cells
3T3-L1 fibroblasts treated with control RNAi or FIT2-RNAi were collected after trypsin digestion and fixed in a mixture of 2.5% glutaraldehyde, 1.25% paraformaldehyde, and 0.1 M sodium cacodylate buffer, pH 7.4, for 1 h at RT. Cells were then washed in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h, and washed twice in distilled water. To enhance contrast, fixed cells were stained with 1% aqueous uranyl acetate for 1 h. Samples were then dehydrated in increasing concentration of ethanol (50%, 70%, 80%, 90%, 95%, and 100%) for 10 min each, with two more incubations in 100% ethanol from a freshly opened bottle. Samples were rinsed twice in acetone (EM grade), infiltrated and embedded in Embed 812 resin (EMS) for 2 h each with a 3:1, 1:1, 1:3 acetone/embedding media mixture. Cells were incubated overnight with 100% fresh resin. The next day, cells were again resuspended in fresh 100% resin for 2–3 h, transferred into BEEM capsules (EMS), and polymerized at 65°C for 48 h. Semi and ultrathin sections were produced with a diamond knife (Diatome) on an ultramicrotome (Ultracut UCT; Leica Microsystems), collected on 200 mesh copper grids (EMS), poststained with uranyl acetate and lead citrate, and examined with a Tecnai T12 TEM, operating at 120 kV. Pictures were recorded on a below-mounted Gatan 2k × 2k CCD camera.
EM of C. elegans
EM fixation on worms was adapted from Zhou et al. (2014), with some modifications. Homozygous and heterozygous fitm-2 worms were fixed in a mixture of 2.5% glutaraldehyde, 1% osmium tetroxide, and 0.1 M sodium cacodylate buffer, pH 7.4, overnight at 4°C. The next morning, animals were rinsed several times in 0.1 M sodium cacodylate buffer and then restained in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h at RT. Samples were washed twice in distilled water. Later, one or two worms were aligned next to each other on an agar pad (2% agarose in dH2O) under dissection microscope and overlaid with molten agar, and the sandwich was allowed to solidify. Agar block was trimmed under dissection microscope so as to have worms sandwiched in a small area of agarose. Trimmed blocks of agarose were placed in glass vials with caps or scintillation vials. To enhance the contrast, blocks were stained en bloc with 2% aqueous uranyl acetate for 1 h at RT. Samples were dehydrated in increasing concentration of ethanol, embedded in Embed 812, and polymerized as described above. Ultrathin sections were produced using a diamond knife, collected onto formvar-coated copper grids, poststained, and visualized with a Tecnai T12 TEM, operating at 120 kV as described in the previous section.
HPF and automatic freeze substitution
HPF and automatic freeze substitution was performed as previously described (Walther and Ziegler, 2002). HPF allows for rapid cryo-immobilization of nonpretreated samples up to 200-µm thick with minimal or no ice crystal damage (Studer et al., 2008). The HPF method was used in Fig. 1, Fig. 3, and Fig. S1. Yeast cells were centrifuged, decanted, and residual media aspirated to have a thick pellet. 0.5–1 µl of cells was loaded into carriers and cryofixed on a Leica Microsystems EMPACT-2 at ∼2,050 bar and automatically cooled into liquid nitrogen. After HPF, the samples were transferred under liquid nitrogen to a Leica Microsystems AFS-2 unit for freeze substitution using 2% (wt/vol) osmium tetroxide, 0.1% (wt/vol) uranyl acetate, and 3% (vol/vol) water in acetone (water free). The automatic freeze substitution unit was programmed to keep samples frozen at −90°C for 22 h. The temperature was increased by 2°C/h for 15 h and then was held at −60°C for 8 h before increasing again at a rate of 2°C/h for 15 h and was held at −30°C for 8 h. Samples were held at 0°C for 2 h, followed by three washes in acetone. Samples were embedded stepwise in increasing concentration of epon (3:1, 1:1, 1:3) for 2 h each. Cells were incubated overnight with 100% fresh resin. The next day, cells were resuspended in fresh 100% resin for 2–3 h, transferred into BEEM capsules, and polymerized at 70°C for 72 h. Semi- and ultrathin sections were produced with a diamond knife (Diatome) on an ultramicrotome (Ultracut UCT; Leica Microsystems), collected on 200 mesh copper grids (EMS), poststained with uranyl acetate and lead citrate, and visualized with a Tecnai T12 TEM, operating at 120 kV. Pictures were recorded on a below-mounted Gatan 2k × 2k CCD camera.
EM tomography
Electron tomography reconstructions were performed on 200 nm–thick cryosections. Single-axis tilt series were recorded using a Tecnai T12 TEM, operating at 120 kV, with an angular range of −60° to +60° with 1° increments. The acquired tilt series were aligned and 3D reconstruction analysis were performed using INSPECT 3D software (FEI). Orthogonal projection and segmentation analyses of 3D tomograms were performed using AMIRA software (FEI).
Cell culture
3T3-L1 preadipocyte cells (ATCC) were maintained in DMEM/Nutrient Mixture F-12 supplemented with 10% FBS (Gibco) and 1% streptavidin and penicillin. 3T3-L1 preadipocyte cells were used for siRNA transfections to silence FIT2. siRNA-transfected cells were later used for real-time quantitative PCR and to analyze lipid droplets using EM.
siRNA treatment
Specific siRNAs (21-mers) to silence FIT2 were purchased from Ambion as double-stranded DNA. Selected sequences of siRNA specific for mouse FIT2 are shown in Table S2. The control siRNA oligonucleotide was also obtained from Ambion. For siRNA transfections, 3T3-L1 preadipocyte cells were plated at 50% confluency in six-well plates and left overnight at 37°C in DMEM/Nutrient Mixture F-12 along with 10% FBS. The next day, cells were transfected with 25 pmol control and FIT2 siRNA oligonucleotides using Lipofectamine RNAiMax (Life Technologies) according to the manufacturer's instructions. 48 h posttransfection, cells were scraped for real-time quantitative PCR and EM analysis.
Fit2 mRNA expression and real-time quantitative PCR
Total RNA was isolated from FIT2- and control siRNA–treated 3T3 L1 preadipocyte cells using PureLink RNA Mini kit (Ambion) according to the manufacturer's instructions. cDNA was synthesized from 1 µg total RNA with qScript cDNA SuperMix (Quanta Biosciences) in 20 µl reaction volume according to the enzyme supplier's instructions. Quantitative real-time PCR was performed in a 20 µl reaction mixture containing 2 µl cDNA, primers, and SYBR Green mix (Applied Biosystems). The sequences of primers used for FIT2 amplification are shown in Table S2. The enzyme was activated at 95°C for 20 s. After activation, the reaction mixture was amplified for 40 cycles under the following conditions: denaturing for 1 s at 95°C and annealing and extension for 20 s at 60°C. Real-time PCR analysis was done on 7900HT FAST Real-Time PCR system (Applied Biosystems). Gene expression was normalized to that of actin and data are presented as the “-fold change” relative to the corresponding siRNA for control and FIT2 according to the 2-ΔΔCT (change in cycling threshold) method.
Characterization of fitm-2(tm3632Δ) C. elegans
A deletion allele was generated by the National BioResource Project of Japan; this deletion deletes most of exons 5 and 6. We obtained this deletion allele (tm3632Δ) as a heterozygote from S. Matani (Tokyo Women's Medical University School of Medicine, Tokyo, Japan), outcrossed it eight times against unc-13(e450), and maintained it as fitm-2(tm3632Δ)/unc-13(e450). To better balance this deletion, we then crossed in the translocation balancer hT2 [bli-4(e937) let-?(q782) qIs48] (I;III), which carries an insertion of myo-2::GFP. Heterozygous animals are scored as having a green pharynx. Nongreen animals were isolated for the characterization of tm3632Δ homozygotes. The deletion was followed throughout the outcrosses by PCR with the following primers: F1, 5-′CAACCAATCGTTGATGTGTGC-3′; R3, 5-′ATGCGGTAGGTCACAAACC-3′. The deletion was confirmed by sequencing and deletes 278 bp of the gene (47 of which is an intron). This deletion is predicted to delete 77 amino acids but remain in frame.
Generation of fitm-2(av41) C. elegans
To characterize the null phenotype of C. elegans lacking fitm-2, A. Fabritius (National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD) generated an early nonsense mutation in the fitm-2 gene using the CRISPR/Cas9 coconversion protocol (Arribere et al., 2014). Codons 17 and 20 were replaced with stop codons and verified by sequencing. Two guide RNAs (gRNAs) were designed (Table S2) to target two overlapping fitm-2 Cas9 sites. gRNAs were cloned into plasmid pDD162 (a gift from D.J. Dickinson and B. Goldstein, University of North Carolina, Chapel Hill, NC; Dickinson et al., 2013) using a Q5 Site-Directed Mutagenesis kit (New England Biolabs) along with gRNA primers (FITM-2 gRNA-1 and FITM-2 gRNA-2; Table S2), reverse primer 5′-CAAGACATCTCGCAATAGG-3′, and plasmid. A repair oligonucleotide was synthesized bearing mutations to convert both targeted PAM sites into stop codons (codons 17 and 20) and to introduce a HindIII restriction site for screening purposes (Table S2). The PAM site mutations were designed to prevent the gRNAs from targeting the repair template (Paix et al., 2014). Repair oligonucleotide encompassed 50 nt of flanking homology on either side of the targeted region of interest.
The edited allele, fitm-2(av41), was sequence confirmed from three independent lines. All three lines were balanced over the hT2 translocation balancer. For characterization of the mutant phenotype, non-GFP animals were picked and observed. For the tm3632Δ allele and the av41 edited nonsense allele, the same “plate” phenotype was observed.
Microinjection and screening of C. elegans
Plasmid DNA was prepared using QIAprep Spin Miniprep kit (Qiagen) and quantified using Nanodrop. 19 N2 animals were injected with DNA mixture consisting of fitm-2 gRNAs cloned in pDD162, repair oligo for fitm-2, pSS4 (the dpy-10 sgRNA in pDD162 (a gift from S. Shrestha, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD), and its repair oligo to create the dominant dpy-10(cn64) allele. Injected animals were rehydrated in M9 buffer before being placed on a small nematode growth media plate seeded with OP50. F1 progeny were collected from the animals that survived microinjection and were further screened for Roller progeny.
The F1 progeny were screened for Roller phenotypes 1–3 d after injection. 60 Roller animals were picked, pooling two Rollers each into 4 µl lysis buffer (10 mM Tris, pH 8.2, 50 mM KCl, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, and 0.01% gelatin) with Proteinase K (60 µg/ml). The worms were frozen overnight at −80°C and then incubated for 1 h at 60°C followed by 15 min at 95°C to make a crude worm genomic DNA preparation. The lysates were then used for worm PCR.
Worm PCR and sequencing
For screening of the F1 Rollers, worm lysates were PCR-amplified using thermo Taq polymerase (New England Biolabs) along with fitm-2 primers (FITM-2 PCR forward and reverse; Table S2). The enzyme was activated at 95°C for 2 min. After activation, the reaction mixture was amplified for 35 cycles under the following conditions: denaturing for 30 s at 95°C, annealing for 30 s at 48°C, and extension for 1 min at 68°C. The PCR-amplified DNA fragment was digested with HindIII enzyme (New England Biolabs).
The edited allele, fitm-2(av41), was sequence confirmed from three independent lines. PCR was performed as described in the previous paragraph, followed by clean up using a MinElute PCR Clean-Up kit (Qiagen) and sequenced using the fitm-2 sequencing primer (FITM-2 seq; Table S2).
Worm BODIPY staining
fitm-2 heterozygous and homozygous worms were harvested as L4 larvae and adults. Animals were incubated in M9 buffer containing BODIPY 493/503 at 1 μg/ml concentration for 15–20 min at RT. Subsequent to three M9 washes, worms were imaged live.
Determination of terminal phenotypes of homozygous fitm-2(av41) worms
Nongreen L4 animals were picked from fitm-2(av41)/hT2 plates and placed on separate plates for brood size determination. Mothers were transferred daily until they no longer laid embryos. Hatched and unhatched embryos were counted; those that hatched were followed daily to determine their terminal phenotype.
Online supplemental material
Fig. S1 shows formation of nascent LDs upon induction of Dga1p, localization of yeast FIT proteins, and a time course of nascent LD formation. Fig. S2 shows that LD wrapping in FIT mutants is not related to autophagy. Fig. S3 shows that the rates of neutral lipid mobilization in FIT mutants and wild-type cells are similar. Table S1 shows the strains and plasmids used in this study. Table S2 shows oligonucleotides used. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505067/DC1.
Supplementary Material
Acknowledgments
We thank R. Schneiter, J. Hinshaw, A. Toulmay, and P. Carvalho for critically reading the text. Wethank D.J. Dickinson, B. Goldstein, and S. Shrestha for providing materials and S. Mitani and A. Fabritius for generating C. elegans mutants.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. V. Choudhary is supported by fellowship from Swiss National Science Foundation (grants PA00P3_145358 and P300P3_158454).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FIT
- fat storage-inducing transmembrane
- gRNA
- guide RNA
- HPF
- high-pressure freezing
- LD
- lipid droplet
- SE
- sterol ester
- TAG
- triacylglycerol
- TEM
- transmission electron microscope
References
- Adeyo O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., and Goodman J.M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055. 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A.K., and Garg A.. 2004. Seipin: a mysterious protein. Trends Mol. Med. 10:440–444. 10.1016/j.molmed.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Andersson L., Boström P., Ericson J., Rutberg M., Magnusson B., Marchesan D., Ruiz M., Asp L., Huang P., Frohman M.A., et al. . 2006. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J. Cell Sci. 119:2246–2257. 10.1242/jcs.02941 [DOI] [PubMed] [Google Scholar]
- Arribere J.A., Bell R.T., Fu B.X., Artiles K.L., Hartman P.S., and Fire A.Z.. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., and Daum G.. 2003. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278:23317–23323. 10.1074/jbc.M302577200 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., and Daum G.. 2005. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280:37301–37309. 10.1074/jbc.M507261200 [DOI] [PubMed] [Google Scholar]
- Bernales S., McDonald K.L., and Walter P.. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4:e423 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D., Januszewski T., Chen Y., Hill J., Markin V.S., Zhao Y., Gilpin C., Chapman K.D., Anderson R.G., and Goodman J.M.. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173:719–731. 10.1083/jcb.200511125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branicky R., Desjardins D., Liu J.L., and Hekimi S.. 2010. Lipid transport and signaling in Caenorhabditis elegans. Dev. Dyn. 239:1365–1377. [DOI] [PubMed] [Google Scholar]
- Brasaemle D.L., and Wolins N.E.. 2012. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 287:2273–2279. 10.1074/jbc.R111.309088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G.M., and Han G.S.. 2009. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284:2593–2597. 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V., Jacquier N., and Schneiter R.. 2011. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: implications for the biogenesis of lipid droplets. Commun. Integr. Biol. 4:781–784. 10.4161/cib.17830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T., Athenstaedt K., and Daum G.. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta. 1771:299–309. 10.1016/j.bbalip.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Wang H., Fu X., Bielby C., and Yang H.. 2009. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem. J. 424:61–67. [DOI] [PubMed] [Google Scholar]
- Goh V.J., and Silver D.L.. 2013. The lipid droplet as a potential therapeutic target in NAFLD. Semin. Liver Dis. 33:312–320. 10.1055/s-0033-1358521 [DOI] [PubMed] [Google Scholar]
- Goh V.J., Tan J.S., Tan B.C., Seow C., Ong W.Y., Lim Y.C., Sun L., Ghosh S., and Silver D.L.. 2015. Postnatal deletion of Fat storage-inducing Transmembrane Protein 2 (FIT2/FITM2) causes lethal enteropathy. J. Biol. Chem. 10.1074/jbc.M115.676700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D.A., and Silver D.L.. 2014. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Crit. Rev. Biochem. Mol. Biol. 49:304–326. 10.3109/10409238.2014.931337 [DOI] [PubMed] [Google Scholar]
- Gross D.A., Zhan C., and Silver D.L.. 2011. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA. 108:19581–19586. 10.1073/pnas.1110817108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Walther T.C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J.S., Vale R.D., Walter P., and Farese R.V.. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 453:657–661. 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman I.Z., Liu P., Zehmer J.K., Luby-Phelps K., Jo Y., Anderson R.G., and DeBose-Boyd R.A.. 2010. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285:19288–19298. 10.1074/jbc.M110.134213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S.A., Kohlwein S.D., and Carman G.M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190:317–349. 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K., Nikawa J., Kodaki T., Ishizu H., and Yamashita S.. 1994. Cloning and sequence of the SCS3 gene which is required for inositol prototrophy in Saccharomyces cerevisiae. J. Biochem. 116:1317–1321. [DOI] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Jacquier N., Mishra S., Choudhary V., and Schneiter R.. 2013. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J. Cell Sci. 126:5198–5209. 10.1242/jcs.131896 [DOI] [PubMed] [Google Scholar]
- Kadereit B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., and Silver D.L.. 2008. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA. 105:94–99. 10.1073/pnas.0708579105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Barbosa A.D., Sembongi H., Mari M., Han G.S., Reggiori F., Carman G.M., and Siniossoglou S.. 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell. 24:2124–2133. 10.1091/mbc.E13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia H., Duelund L., Pakkanen K.I., and Ipsen J.H.. 2010. Triglyceride blisters in lipid bilayers: implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS ONE. 5:e12811 10.1371/journal.pone.0012811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köffel R., Tiwari R., Falquet L., and Schneiter R.. 2005. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell. Biol. 25:1655–1668. 10.1128/MCB.25.5.1655-1668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S.D. 2010. Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285:15663–15667. 10.1074/jbc.R110.118356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S., Chao J.T., Tavassoli S., Wong A.K., Choudhary V., Young B.P., Loewen C.J., and Prinz W.A.. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12:e1001969 10.1371/journal.pbio.1001969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V. Jr., Ory D.S., and Schaffer J.E.. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100:3077–3082. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T. Jr., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. BSCL Working Group . 2001. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28:365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Martin S., and Parton R.G.. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378. 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- Miranda D.A., Kim J.H., Nguyen L.N., Cheng W., Tan B.C., Goh V.J., Tan J.S., Yaligar J., Kn B.P., Velan S.S., et al. . 2014. Fat storage-inducing transmembrane protein 2 is required for normal fat storage in adipose tissue. J. Biol. Chem. 289:9560–9572. 10.1074/jbc.M114.547687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K.. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- Moir R.D., Gross D.A., Silver D.L., and Willis I.M.. 2012. SCS3 and YFT2 link transcription of phospholipid biosynthetic genes to ER stress and the UPR. PLoS Genet. 8:e1002890 10.1371/journal.pgen.1002890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325–438. 10.1016/S0163-7827(01)00013-3 [DOI] [PubMed] [Google Scholar]
- Murphy D.J., and Vance J.. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24:109–115. 10.1016/S0968-0004(98)01349-8 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., and Fujimoto T.. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta. 1791:399–407. 10.1016/j.bbalip.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Suzuki M., and Fujimoto T.. 2014. Open questions in lipid droplet biology. Chem. Biol. 21:86–96. 10.1016/j.chembiol.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Richter C.M., and Kopito R.R.. 2013. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. USA. 110:1345–1350. 10.1073/pnas.1213738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H.E., Lee C.Y., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., and Seydoux G.. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks L.W., Bottema C.D., Rodriguez R.J., and Lewis T.A.. 1985. Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 111:333–346. 10.1016/S0076-6879(85)11020-7 [DOI] [PubMed] [Google Scholar]
- Ploegh H.L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 448:435–438. 10.1038/nature06004 [DOI] [PubMed] [Google Scholar]
- Pol A., Gross S.P., and Parton R.G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., and Rapoport T.A.. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. 10.1083/jcb.150.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robenek H., Buers I., Hofnagel O., Robenek M.J., Troyer D., and Severs N.J.. 2009. Compartmentalization of proteins in lipid droplet biogenesis. Biochim. Biophys. Acta. 1791:408–418. 10.1016/j.bbalip.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Sandager L., Gustavsson M.H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., and Stymne S.. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277:6478–6482. 10.1074/jbc.M109109200 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S. 2013. Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim. Biophys. Acta. 1831:575–581. 10.1016/j.bbalip.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Skinner J.R., Shew T.M., Schwartz D.M., Tzekov A., Lepus C.M., Abumrad N.A., and Wolins N.E.. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284:30941–30948. 10.1074/jbc.M109.013995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer D., Humbel B.M., and Chiquet M.. 2008. Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem. Cell Biol. 130:877–889. 10.1007/s00418-008-0500-1 [DOI] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger R.H., and Scherer P.E.. 2010. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol. Metab. 21:345–352. 10.1016/j.tem.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaden D.L., Gohil V.M., Gu Z., and Greenberg M.L.. 2005. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338:162–164. 10.1016/j.ab.2004.11.020 [DOI] [PubMed] [Google Scholar]
- van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., and Kohlwein S.D.. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301. 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wältermann M., Hinz A., Robenek H., Troyer D., Reichelt R., Malkus U., Galla H.J., Kalscheuer R., Stöveken T., von Landenberg P., and Steinbüchel A.. 2005. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 55:750–763. 10.1111/j.1365-2958.2004.04441.x [DOI] [PubMed] [Google Scholar]
- Walther P., and Ziegler A.. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 208:3–10. 10.1046/j.1365-2818.2002.01064.x [DOI] [PubMed] [Google Scholar]
- Walther T.C., and Farese R.V. Jr. 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791:459–466. 10.1016/j.bbalip.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Haas J.T., Walther T.C., and Farese R.V. Jr. 2014. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29:39–45. 10.1016/j.ceb.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Brasaemle D.L., and Bickel P.E.. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484–5491. 10.1016/j.febslet.2006.08.040 [DOI] [PubMed] [Google Scholar]
- Wright R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Yang L., Ding Y., Chen Y., Zhang S., Huo C., Wang Y., Yu J., Zhang P., Na H., Zhang H., et al. . 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53:1245–1253. 10.1194/jlr.R024117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hashmi S., Cheema F., Al-Nasser N., Bakheet R., Parhar R.S., Al-Mohanna F., Gaugler R., Hussain M.M., and Hashmi S.. 2013. Regulation of lipoprotein assembly, secretion and fatty acid β-oxidation by Krüppel-like transcription factor, klf-3. J. Mol. Biol. 425:2641–2655. 10.1016/j.jmb.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Yu H., Yin X., Sabour P.M., Chen W., and Gong J.. 2014. Lactobacillus zeae protects Caenorhabditis elegans from enterotoxigenic Escherichia coli-caused death by inhibiting enterotoxin gene expression of the pathogen. PLoS One. 9:e89004 10.1371/journal.pone.0089004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.