Abstract
Staphylococci are frequent human commensals and some species can cause disease. Staphylococcus aureus in particular is a dangerous human pathogen. In staphylococci, the ability to sense the bacterial cell density, or quorum, and to respond with genetic adaptations is due to one main system, which is called accessory gene regulator (Agr). The extracellular signal of Agr is a post-translationally modified peptide containing a thiolactone structure. Under conditions of high cell density, Agr is responsible for the increased expression of many toxins and degradative exoenzymes, and decreased expression of several colonization factors. This regulation is important for the timing of virulence factor expression during infection and the development of acute disease, while low activity of Agr is associated with chronic staphylococcal infections, such as those involving biofilm formation. Accordingly, drugs inhibiting Agr are being evaluated for their capacity to control acute forms of S. aureus infection.
Keywords: Staphylococcus aureus, Staphylococcus epidermidis, quorum-sensing, toxins, biofilm, Agr, LuxS
Introduction
Staphylococci, including Staphylococcus aureus and the coagulase-negative staphylococci, the most common of which is Staphylococcus epidermidis, have been implicated as causative agents in a variety of human infections, including infections involving the skin and soft tissue, the blood stream, the respiratory system, the skeletal system, as well as infections involving implanted medical devices. Due to their ability to aptly regulate a wide armamentarium of virulence factors, including toxins, degradative enzymes, antimicrobial resistance genes, and immune evasion mechanisms, staphylococci not only are able to successfully secure infectious niches in a wide range of organ systems, but are also successful pathogens in both acute and chronic human infections (Lowy, 1998; Otto, 2009).
Among the regulatory mechanisms that ensure timely adaptation of staphylococcal physiology to the environment, quorum-sensing stands out as one of the most intensely studied and probably most important mechanisms for the control of pathogenesis. Quorum-sensing is population density-dependent and environment-dependent gene regulation that occurs through cell-cell communication (Waters and Bassler, 2005). The role of two regulatory systems in staphylococci, the accessory gene regulator (Agr) system and the LuxS system, are discussed herein. Agr is considered the prototype quorum-sensing regulator system in Gram-positive bacteria (Kleerebezem et al., 1997), while the role of LuxS as a quorum-sensing system in staphylococci is less certain.
The Agr system
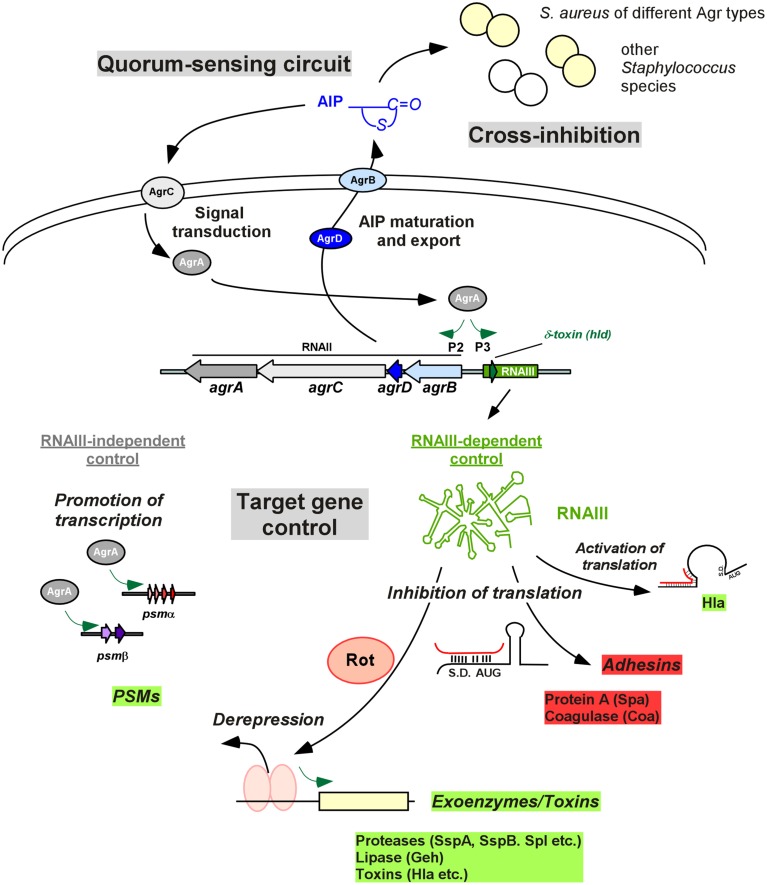
The agr locus is 3.5 kb in size and consists of two divergent transcriptional units, RNAII and RNAIII, whose transcription is driven by the P2 and P3 promoters, respectively (Peng et al., 1988; Kornblum et al., 1990; Figure 1). The RNAII locus contains four genes, agrB, agrD, agrC and agrA (Kornblum et al., 1990; Figure 1). The agrD transcript encodes a peptide precursor of the extracellular quorum signal of Agr, called autoinducing peptide (AIP) (Ji et al., 1995). The AIP in its mature form is 7–9 amino acids long and contains a characteristic thiolactone (or in Staphylococcus intermedius, lactone) ring between the centrally located cysteine and the C terminus (Ji et al., 1997, 2005; Otto et al., 1998; Novick and Geisinger, 2008). The agrB gene product is a transmembrane endopeptidase that is responsible for the introduction of the thiolactone modification, C-terminal cleavage, and export of the AIP (Saenz et al., 2000; Zhang et al., 2002; Zhang and Ji, 2004); in the extracellular milieu the AIP receives final trimming by the type I signal peptidase SspB (Kavanaugh et al., 2007). The agrC and agrA genes encode a two-component signal transduction system involving a histidine kinase sensor AgrC, a transmembrane protein that is phosphorylated upon the binding of AIP (Lina et al., 1998), and its associated response regulator AgrA (Novick et al., 1995; Queck et al., 2008). Upon being activated by AgrC-dependent phosphorylation, AgrA binds to the P2 promoter region for RNAII and the P3 promoter region for RNAIII (Koenig et al., 2004), as well as the promoters controlling expression of the PSMα and PSMβ peptides (Novick et al., 1995; Queck et al., 2008; Figure 1).
Figure 1.
Agr control in Staphylococcus. The quorum-sensing circuit is shown at the top left. The AIP signal is produced from the AgrD precursor by AgrB maturation and export. At a certain threshold concentration, AIP activates the AgrC-AgrA two-component system and phosphorylated AgrA activates transcription from the P2 promoter, resulting in auto-feedback regulation. An important feature of Agr is group specificity, resulting in cross-inhibition, i.e., inhibition of Agr activity, in strains belonging to other Agr specificity groups and other species (top right). Target gene control is shown at the bottom. Most Agr targets are regulated via RNAIII, whose transcription is increased by AgrA, via the P3 promoter. RNAIII also contains the gene for delta-toxin (hld). RNAIII controls target genes by base pairing with 5′UTRs, in most cases inhibiting translation. This is used for direct inhibition of Agr-inhibited target genes such as protein A, while inhibition of translation of the repressor Rot leads to de-repression of transcription of many of the classical Agr target toxins, such as alpha-toxin. AgrA also increases transcription of the psmα and psmβ operons, encoding PSM peptides, in an RNAIII-independent mode of Agr target gene regulation. S.D., Shine/Dalgarno sequence; AUG, start codon of controlled gene.
Although agr is generally conserved within the staphylococci, variations in the sequences of agrB, agrC, agrD lead to the production of AIPs with varied signaling specificities, allowing for activation of self and cross-inhibition of Agr groups of non-self (Ji et al., 1997; Otto et al., 2001; Olson et al., 2014), a phenomenon that might be reflective of evolutionary selective pressures (Novick and Geisinger, 2008). Besides the AIP, Agr can also be activated by a variety of other regulators, such as SarA (Heinrichs et al., 1996) or SrrAB (Yarwood et al., 2001), and environmental factors such as glucose concentration or pH (Regassa et al., 1992).
RNAIII-dependent gene regulation
RNAIII is the intracellular effector molecule of the Agr system responsible for the control of Agr targets (Novick et al., 1993). It is also a messenger RNA, containing the hld gene for delta-toxin (or delta-hemolysin) (Janzon et al., 1989). The primary mechanism by which RNAIII controls target gene expression is by antisense base pairing with 5′ untranslated regions (5′ UTRs), forming RNA duplexes. Except for in the case of alpha-toxin, in which RNAIII may also act as a post-transcriptional activator (Morfeldt et al., 1995), RNAIII usually blocks translation (Boisset et al., 2007). By this direct mechanism, RNAIII inhibits the production of a series of predominantly surface proteins, such as protein A and others. Furthermore, RNAIII blocks translation of the repressor of toxin (Rot) protein, which belongs to the staphylococcal accessory regulator (Sar) transcriptional regulator family (McNamara et al., 2000; Saïd-Salim et al., 2003; Boisset et al., 2007). Rot binds to the promoter region of many exoproteins and toxins, blocking their transcription. These mechanisms allow for prompt density-dependent up-regulation of enterotoxins, alpha-toxin, leukocidins, degradative exoenzymes and down-regulation of surface proteins. Moreover, genome-wide analyses of gene expression indicate that in addition to the control of virulence determinants, Agr control also comprises a series of metabolic targets (Dunman et al., 2001; Yao et al., 2006; Cheung et al., 2011). These general physiological adaptations may assist to adapt bacterial physiology to the changed requirements during infection.
RNAIII-independent gene regulation
While RNAIII was long believed to represent the only mechanism by which Agr controls target genes, it was found more recently that the response regulator AgrA not only binds to the P2 and P3 promoters, but also directly up-regulates transcription of the phenol-soluble modulin psmα and psmβ operons by binding to the respective promoter sequences (Queck et al., 2008). The phenol-soluble modulins (PSMs) are a family of staphylococcal peptide toxins that also include the delta-toxin (Cheung et al., 2014a). As the delta-toxin gene is embedded within RNAIII, it has been speculated that evolution of RNAIII around the delta-toxin gene connected an evolutionarily more ancient regulatory network consisting of AgrA and possibly quorum-sensing control of psm genes to the control of further virulence determinants via RNAIII and Rot (Queck et al., 2008).
The impact of Agr on acute infection and toxicity
In S. aureus, the up-regulation of virulence factors by Agr is necessary for disease progression in several animal models of acute infection, including infective endocarditis (Cheung et al., 1994), skin and soft tissue infections (Wright et al., 2005; Cheung et al., 2011), pneumonia (Heyer et al., 2002; Bubeck Wardenburg et al., 2007a; Montgomery et al., 2010), and septic arthritis and osteomyelitis (Abdelnour et al., 1993; Gillaspy et al., 1995). Conversely, down-regulation by Agr of PSMs and microbial surface components has been implicated in enhanced biofilm formation and bacterial colonization of indwelling medical devices (see below) (Vuong et al., 2000, 2004; Wang et al., 2011; Periasamy et al., 2012). Moreover, Agr dysfunction is correlated with persistent S. aureus bacteremia (Fowler et al., 2004). In general, the cell density/Agr-dependent up-regulation of toxins and degradative exoenzymes on the one hand and the down-regulation of surface components on the other is supposed to reflect the differential temporal requirements for specific virulence determinants during the course of a bacterial infection: in the beginning of an infection, low cell density and consequential low expression of Agr results in increased production of surface components required for the initial colonization of tissues. Once this is established, bacteria grow to higher cell densities, requiring additional food sources and increased protection from host defenses, which is accomplished by Agr-dependent up-regulation of degradative exoenzymes and toxins. Notably, this timing also ascertains that production of toxins, many of which are pro-inflammatory, is delayed until the growing bacterial colonies can deal with attacks by host defenses, which likely explains the findings particularly related to the role of Agr and Agr-controlled factors during S. aureus bacteremia (Fowler et al., 2004; Cheung et al., 2014b). Finally, Agr-controlled expression of virulence determinants is energy-consuming and there are several recent reports indicating that this needs to be balanced with the expression of antibiotic resistance in antibiotic-resistant strains (Joo et al., 2010; Rudkin et al., 2014). Specifically, subinhibitory concentrations of antibiotics have been shown to increase Agr expression (Joo et al., 2010; Paulander et al., 2013; Rudkin et al., 2014), imposing a fitness cost, which has been speculated to drive the observed formation of Agr-dysfunctional mutants in strains isolated from hospital infections (Traber et al., 2008; Paulander et al., 2013).
Agr-controlled virulence determinants
One of the most prominent Agr-regulated toxins is alpha-toxin (alpha-hemolysin, Hla), which is encoded by the hla locus. It is a 319 amino acid beta-barrel pore-forming toxin that binds to the disintegrin and metalloprotease 10 (ADAM10) receptor on the host cell membrane (Wilke and Bubeck Wardenburg, 2010; Berube and Bubeck Wardenburg, 2013). Hla has been implicated in multiple staphylococcal infections in humans. Isogenic hla mutants consistently demonstrated reduced disease severity when compared to the wild-type strain when examined in animal models involving pneumonia (Bubeck Wardenburg et al., 2007a,b), skin and soft tissue infection (Kennedy et al., 2010; Kobayashi et al., 2011), and endovascular infection (Powers et al., 2012).
The bi-component leukocidins constitute another family of generally Agr-controlled pore-forming toxins, which includes Panton-Valentine Leukocidin (PVL), γ-hemolysin, LukDE, and LukGH (LukAB) (Queck et al., 2008; Cheung et al., 2011; Alonzo and Torres, 2014). While PVL is now believed to play a more limited role in severe staphylococcal infections than previously thought (Otto, 2011), LukED has emerged as an important factor controlling S. aureus disease development (Gravet et al., 1998; Alonzo et al., 2012). LukED causes lysis of phagocytic cells by targeting the CCR5 chemokine receptor on lymphocytes, macrophages and dendritic cells (Alonzo et al., 2013). Furthermore, LukGH (LukAB) appears to play a key role in S. aureus pathogenesis, as it is the only S. aureus toxin in addition to the PSMα peptides that contributes to lysis after phagocytosis (DuMont et al., 2013).
PSMs are a family of peptide toxins (Cheung et al., 2014a) that stand out among Agr-controlled virulence factors as the only ones that are under direct control by AgrA (Queck et al., 2008), emphasizing a likely central role in staphylococcal physiology that is underlined by their role in the non-infectious lifestyle of staphylococcal commensals (Cheung et al., 2014a). Among the PSMs, those encoded in the psmα locus of S. aureus, most notably PSMα3, are strongly pro-inflammatory and lytic to a variety of cell types, including neutrophils, macrophages, osteoblasts, and erythrocytes (Wang et al., 2007; Rasigade et al., 2013). As a consequence, PSMα peptides have a strong impact on acute types of S. aureus infection, such as skin and soft tissue infections, sepsis, and osteomyelitis (Wang et al., 2007; Cassat et al., 2013).
Many secreted enzymes are under regulation by Agr. Several proteases in particular are very strongly Agr-controlled and in addition to representing virulence factors in their own right (by contributing to the degradation of tissue and host defense proteins such as antimicrobial peptides), they can shape the composition of the pathogen secretome and thus its pathogenic properties (Cheung et al., 2011; Kolar et al., 2013).
Among the generally Agr-down-regulated surface proteins, protein A (Spa) stands out, due to its multifactorial impact on pathogenesis in addition to the fact that it appears to be consistently Agr-controlled (Cheung et al., 2011). In contrast, the general dogma of negative control by Agr of other surface proteins does not hold true in all strains (Cheung et al., 2011). Protein A has been implicated in infection models involving multiple organ systems, including pneumonia (Heyer et al., 2002; Bubeck Wardenburg et al., 2007a; Montgomery et al., 2010), septic arthritis (Palmqvist et al., 2002), and bloodstream infection (Date et al., 2014). It has a role in triggering pro-inflammatory mediators in the lung parenchyma through activation of the TNF-α receptor (TNFR1) (Gómez et al., 2004), and also has the capacity to inactivate the host humoral immune response upon release from the staphylococcal envelope (Becker et al., 2014) by binding the Fcγ domain of IgG to block Fc receptor-mediated opsonophagocytosis (Peterson et al., 1977), and also by binding to the Fab domain of VH3-clan IgM and triggering apoptosis (Goodyear et al., 2006; Pauli et al., 2014).
Agr regulation of biofilms and biofilm-associated infection
Biofilm formation is thought to occur through several stages: (1) attachment of microorganisms to abiotic or host matrix protein-coated surfaces and aggregation into multicellular structures, (2) proliferation/maturation, and (3) detachment/dispersal (Otto, 2008). Initially speculated to lead to biofilm dispersal, based on the observation that agr mutants showed extended biofilm formation in both S. aureus and S. epidermidis (Vuong et al., 2000, 2003, 2004), the detailed mechanisms by which Agr impacts biofilm development have recently become clear.
Agr controls proteases, which in vitro impact biofilm extension by degrading protein components of the biofilm matrix (Lauderdale et al., 2009). However, the in vivo relevance of proteases in staphylococcal biofilm formation is debatable (Otto, 2013). The only biofilm-structuring and dispersal mechanism whose relevance has been confirmed in vivo, and in experiments using biological fluids, is based on PSM surfactant function (Wang et al., 2011; Periasamy et al., 2012; Dastgheyb et al., 2015). In a process that is largely independent of the mode of biofilm formation, i.e., the composition of the biofilm matrix, PSMs structure biofilms by forming channels and lead to the dispersal of cells from the biofilm. This process leads to the dissemination of biofilm-associated infection in vivo and is responsible for the extensive biofilm formation that is observed under conditions of low Agr activity, such as in synovial fluid during joint infection (Otto, 2013; Dastgheyb et al., 2015). Thus, the impact of Agr on biofilm-associated infection is divergent: Agr is necessary for biofilm structuring and the dissemination of biofilm infection, but dysfunction of Agr leads to enhanced biofilm formation, which may be advantageous for the bacteria under those conditions. Accordingly, strains with a dysfunctional Agr system are often isolated from infections on indwelling devices (Vuong et al., 2004; Traber et al., 2008) and colonizing strains (Shopsin et al., 2008). These strains have lost the capacity to disseminate in the patient's body as well as to other individuals (Shopsin et al., 2010).
LuxS
Described in Vibrio spp., the regulatory effect of luxS was discovered in the context of bioluminescence regulation, and since has been recognized as a widely utilized quorum-sensing system among bacteria, prompting speculation of its role in interspecies communication (Bassler et al., 1997; Bassler, 2002). The LuxS system employs an autoinducer called AI-2, which is a furanosyl borate diester molecule (Chen et al., 2002). Several phenotypes, such as capsule synthesis, biofilm formation, antibiotic susceptibility, and virulence, have been linked to AI-2 regulation in S. aureus (Zhao et al., 2010; Yu et al., 2012; Xue et al., 2013). Similarly, in S. epidermidis, the luxS gene was found to impact the expression of a series of genes, including biofilm exopolysaccharide biosynthesis (Li et al., 2008). Furthermore, a luxS mutant demonstrated increased virulence in a catheter-related murine infection model (Xu et al., 2006). This finding was later corroborated under both in vitro and in vivo experimental conditions in S. aureus, where luxS exerted control of biofilm growth through the icaR locus (Yu et al., 2012). However, the role of LuxS as a quorum-sensing regulatory system in staphylococci remains under debate. In fact, it has been argued that the impact of a luxS deletion on the observed phenotypes in staphylococci may be due only to its primary role in metabolism, where it is an integral part of the S-adenosyl methionine cycle (Winzer et al., 2002; Doherty et al., 2006).
Therapeutic approaches targeting staphylococcal quorum-sensing
There has been continued interest in targeting Agr for the development of anti-staphylococcal drugs (Khan et al., 2015). One such effort exploits the cross-inhibiting activity of Agr AIPs and includes altering the AIPs through amino acid substitution, relocation of the amino acid side chains, removal of the exocyclic tail, and AIP polymerization, in order to enhance the stability of the AIPs and make them more resistant to proteolysis (Mayville et al., 1999; Affas et al., 2001; Lyon et al., 2002; Melamed Yerushalmi et al., 2013; Tal-Gan et al., 2013; Khan et al., 2015). Furthermore, monoclonal antibodies against a specific AIP were effective in suppressing Agr expression in an Agr strain of a different Agr specificity group and protected mice against a lethal S. aureus challenge of that strain (Park et al., 2007). Finally, there have been promising recent approaches targeting the AgrA response regulator by a small molecule, savirin (Sully et al., 2014). Notably, in that study the virulence-suppressing activity did not result in any development of resistance, which is an often claimed but infrequently tested supposed advantage of anti-virulence drugs.
However, a noticeable problem with targeting Agr remains the dichotomy of Agr control. Due to the phenotype of biofilm enhancement in agr mutants, which has been confirmed using a cross-inhibiting AIP (Vuong et al., 2004), the impact of Agr-inhibiting drugs such as those derived from cross-inhibiting AIPs, on chronic and biofilm-associated forms of staphylococcal infection would be counterproductive. Due to the predominant involvement of coagulase-negative staphylococci in biofilm infections (Otto, 2004), Agr blockers would certainly not be used for infections by those strains. Nevertheless, Agr inhibitors may form valuable drugs for the inhibition of acute toxicity in many S. aureus infections, possibly to be given together with conventional antibiotics (Khan et al., 2015).
Conclusion
Quorum-sensing regulation in staphylococci is mainly due to the Agr locus, which regulates a wide variety of virulence determinants in addition to metabolic genes. Accordingly, there is a strong impact of Agr on many types of staphylococcal disease. In acute types of disease, Agr generally enhances pathogenesis by increasing expression of aggressive virulence determinants such as toxins and degradative exoenzymes. In contrast, Agr has a more complicated role during chronic infections, as mutants in Agr show increased biofilm formation, but decreased potential to disseminate, in addition to being correlated with enhanced success during persistent bacteremia. Compared to Agr, the role of LuxS in staphylococcal quorum-sensing is less well understood.
Clearly, there is renewed recent interest in targeting Agr for drug development, which appears a valid option for acute disease types. However, going forward, the often divergent roles of Agr in various types of staphylococcal infection need to be understood better to establish a solid scientific basis underscoring the applicability of quorum-sensing blockers for the treatment of staphylococcal disease.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health (grant ZIA AI000904-14).
References
- Abdelnour A., Arvidson S., Bremell T., Ryden C., Tarkowski A. (1993). The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61, 3879–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affas M. D. P. Z., Reynolds C., Holden M. T., Wood S. J., Saint S., Cockayne A., et al. (2001). Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41, 503–512. 10.1046/j.1365-2958.2001.02539.x [DOI] [PubMed] [Google Scholar]
- Alonzo F., III, Benson M. A., Chen J., Novick R. P., Shopsin B., Torres V. J. (2012). Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 83, 423–435. 10.1111/j.1365-2958.2011.07942.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F., III, Kozhaya L., Rawlings S. A., Reyes-Robles T., DuMont A. L., Myszka D. G., et al. (2013). CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55. 10.1038/nature11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F., III, Torres V. J. (2014). The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230. 10.1128/MMBR.00055-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L. (2002). Small talk. Cell-to-cell communication in bacteria. Cell 109, 421–424. 10.1016/S0092-8674(02)00749-3 [DOI] [PubMed] [Google Scholar]
- Bassler B. L., Greenberg E. P., Stevens A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S., Frankel M. B., Schneewind O., Missiakas D. (2014). Release of protein A from the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, 1574–1579. 10.1073/pnas.1317181111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube B. J., Bubeck Wardenburg J. (2013). Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins (Basel) 5, 1140–1166. 10.3390/toxins5061140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., et al. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366. 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck Wardenburg J., Bae T., Otto M., Deleo F. R., Schneewind O. (2007b). Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13, 1405–1406. 10.1038/nm1207-1405 [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J., Patel R. J., Schneewind O. (2007a). Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect. Immun. 75, 1040–1044. 10.1128/IAI.01313-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat J. E., Hammer N. D., Campbell J. P., Benson M. A., Perrien D. S., Mrak L. N., et al. (2013). A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13, 759–772. 10.1016/j.chom.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. 10.1038/415545a [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Eberhardt K. J., Chung E., Yeaman M. R., Sullam P. M., Ramos, et al. (1994). Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94, 1815–1822. 10.1172/JCI117530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Joo H. S., Chatterjee S. S., Otto M. (2014a). Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 38, 698–719. 10.1111/1574-6976.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Kretschmer D., Duong A. C., Yeh A. J., Ho T. V., Chen Y., et al. (2014b). Production of an attenuated phenol-soluble modulin variant unique to the MRSA clonal complex 30 increases severity of bloodstream infection. PLoS Pathog. 10:e1004298. 10.1371/journal.ppat.1004298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Wang R., Khan B. A., Sturdevant D. E., Otto M. (2011). Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935. 10.1128/IAI.00046-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastgheyb S. S., Villaruz A. E., Le K. Y., Tan V. Y., Duong A. C., Chatterjee S. S., et al. (2015). Role of phenol-soluble modulins in formation of staphylococcus aureus biofilms in synovial fluid. Infect. Immun. 83, 2966–2975. 10.1128/IAI.00394-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date S. V., Modrusan Z., Lawrence M., Morisaki J. H., Toy K., Shah I. M., et al. (2014). Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J. Infect. Dis. 209, 1542–1550. 10.1093/infdis/jit668 [DOI] [PubMed] [Google Scholar]
- Doherty N., Holden M. T., Qazi S. N., Williams P., Winzer K. (2006). Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 188, 2885–2897. 10.1128/JB.188.8.2885-2897.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuMont A. L., Yoong P., Surewaard B. G., Benson M. A., Nijland R., van Strijp J. A., et al. (2013). Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect. Immun. 81, 1830–1841. 10.1128/IAI.00095-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., et al. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353. 10.1128/JB.183.24.7341-7353.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V. G., Jr., Sakoulas G., McIntyre L. M., Meka V. G., Arbeit R. D., Cabell C. H., et al. (2004). Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190, 1140–1149. 10.1086/423145 [DOI] [PubMed] [Google Scholar]
- Gillaspy A. F., Hickmon S. G., Skinner R. A., Thomas J. R., Nelson C. L., Smeltzer M. S. (1995). Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63, 3373–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez M. I., Lee A., Reddy B., Muir A., Soong G., Pitt A., et al. (2004). Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10, 842–848. 10.1038/nm1079 [DOI] [PubMed] [Google Scholar]
- Goodyear C. S., Sugiyama F., Silverman G. J. (2006). Temporal and dose-dependent relationships between in vivo B cell receptor-targeted proliferation and deletion-induced by a microbial B cell toxin. J. Immunol. 176, 2262–2271. 10.4049/jimmunol.176.4.2262 [DOI] [PubMed] [Google Scholar]
- Gravet A., Colin D. A., Keller D., Girardot R., Monteil H., Prévost G. (1998). Characterization of a novel structural member, LukE-LukD, of the bi-component staphylococcal leucotoxins family. FEBS Lett. 436, 202–208. 10.1016/S0014-5793(98)01130-2 [DOI] [PubMed] [Google Scholar]
- Heinrichs J. H., Bayer M. G., Cheung A. L. (1996). Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178, 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer G., Saba S., Adamo R., Rush W., Soong G., Cheung A., et al. (2002). Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infect. Immun. 70, 127–133. 10.1128/IAI.70.1.127-133.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzon L., Löfdahl S., Arvidson S. (1989). Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219, 480–485. 10.1007/BF00259623 [DOI] [PubMed] [Google Scholar]
- Ji G., Beavis R. C., Novick R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U.S.A. 92, 12055–12059. 10.1073/pnas.92.26.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Beavis R., Novick R. P. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030. 10.1126/science.276.5321.2027 [DOI] [PubMed] [Google Scholar]
- Ji G., Pei W., Zhang L., Qiu R., Lin J., Benito Y., et al. (2005). Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J. Bacteriol. 187, 3139–3150. 10.1128/JB.187.9.3139-3150.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H. S., Chan J. L., Cheung G. Y., Otto M. (2010). Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54, 4942–4944. 10.1128/AAC.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh J. S., Thoendel M., Horswill A. R. (2007). A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65, 780–798. 10.1111/j.1365-2958.2007.05830.x [DOI] [PubMed] [Google Scholar]
- Kennedy A. D., Bubeck Wardenburg J., Gardner D. J., Long D., Whitney A. R., Braughton K. R., et al. (2010). Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infect. Dis. 202, 1050–1058. 10.1086/656043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B. A., Yeh A. J., Cheung G. Y., Otto M. (2015). Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 24, 689–704. 10.1517/13543784.2015.1019062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Quadri L. E., Kuipers O. P., de Vos W. M. (1997). Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24, 895–904. 10.1046/j.1365-2958.1997.4251782.x [DOI] [PubMed] [Google Scholar]
- Kobayashi S. D., Malachowa N., Whitney A. R., Braughton K. R., Gardner D. J., Long D., et al. (2011). Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J. Infect. Dis. 204, 937–941. 10.1093/infdis/jir441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig R. L., Ray J. L., Maleki S. J., Smeltzer M. S., Hurlburt B. K. (2004). Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186, 7549–7555. 10.1128/JB.186.22.7549-7555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar S. L., Ibarra J. A., Rivera F. E., Mootz J. M., Davenport J. E., Stevens S. M., et al. (2013). Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2, 18–34. 10.1002/mbo3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J., Kreiswirth B., Projan S. J., Ross H., Novick R. P. (1990). Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, in Molecular Biology of the Staphylococci, ed Novick R. P. (New York, NY: VCH Publishers; ), 373–401. [Google Scholar]
- Lauderdale K. J., Boles B. R., Cheung A. L., Horswill A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. 10.1128/IAI.01036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Villaruz A. E., Vadyvaloo V., Sturdevant D. E., Otto M. (2008). AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 8:4. 10.1186/1471-2180-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G., Jarraud S., Ji G., Greenland T., Pedraza A., Etienne J., et al. (1998). Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28, 655–662. 10.1046/j.1365-2958.1998.00830.x [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Lyon G. J., Wright J. S., Muir T. W., Novick R. P. (2002). Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41, 10095–10104. 10.1021/bi026049u [DOI] [PubMed] [Google Scholar]
- Mayville P., Ji G., Beavis R., Yang H., Goger M., Novick R. P., et al. (1999). Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U.S.A. 96, 1218–1223. 10.1073/pnas.96.4.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P. J., Milligan-Monroe K. C., Khalili S., Proctor R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182, 3197–3203. 10.1128/JB.182.11.3197-3203.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed Yerushalmi S., Buck M. E., Lynn D. M., Lemcoff N. G., Meijler M, M. (2013). Multivalent alteration of quorum sensing in Staphylococcus aureus. Chem. Commun. (Camb). 49, 5177–5179. 10.1039/c3cc41645c [DOI] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Daum R. S. (2010). Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE 5:e15177. 10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E., Taylor D., von Gabain A., Arvidson S. (1995). Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14, 4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Geisinger E. (2008). Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kornblum J., Ross H. F., Ji G., Kreiswirth B., et al. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248, 446–458. 10.1007/BF02191645 [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. E., Todd D. A., Schaeffer C. R., Paharik A. E., Van Dyke M. J., Büttner H., et al. (2014). Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196, 3482–3493. 10.1128/JB.01882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2004). Virulence factors of the coagulase-negative staphylococci. Front. Biosci. 9, 841–863. 10.2741/1295 [DOI] [PubMed] [Google Scholar]
- Otto M. (2008). Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228. 10.1007/978-3-540-75418-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2011). A MRSA-terious enemy among us: end of the PVL controversy? Nat. Med. 17, 169–170. 10.1038/nm0211-169 [DOI] [PubMed] [Google Scholar]
- Otto M. (2013). Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188. 10.1146/annurev-med-042711-140023 [DOI] [PubMed] [Google Scholar]
- Otto M., Echner H., Voelter W., Götz F. (2001). Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 1957–1960. 10.1128/IAI.69.3.1957-1960.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M., Sussmuth R., Jung G., Götz F. (1998). Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 424, 89–94. 10.1016/S0014-5793(98)00145-8 [DOI] [PubMed] [Google Scholar]
- Palmqvist N., Foster T., Tarkowski A., Josefsson E. (2002). Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33, 239–249. 10.1006/mpat.2002.0533 [DOI] [PubMed] [Google Scholar]
- Park J., Jagasia R., Kaufmann G. F., Mathison J. C., Ruiz D. I., Moss J. A., et al. (2007). Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 14, 1119–1127. 10.1016/j.chembiol.2007.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulander W., Nissen Varming A., Baek K. T., Haaber J., Frees D., Ingmer H. (2013). Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. MBio 3:e00459-12. 10.1128/mBio.00459-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli N. T., Kim H. K., Falugi F., Huang M., Dulac J., Henry Dunand C., et al. (2014). Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J. Exp. Med. 211, 2331–2339. 10.1084/jem.20141404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. (1988). Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S., Joo H. S., Duong A. C., Bach T. H., Tan V. Y., Chatterjee S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286. 10.1073/pnas.1115006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson P. K., Verhoef J., Sabath L. D., Quie P. G. (1977). Effect of protein A on staphylococcal opsonization. Infect. Immun. 15, 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. E., Kim H. K., Wang Y., Bubeck Wardenburg J. (2012). ADAM10 mediates vascular injury induced by Staphylococcus aureus alpha-hemolysin. J. Infect. Dis. 206, 352–356. 10.1093/infdis/jis192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H., Khan B. A., Sturdevant D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigade J. P., Trouillet-Assant S., Ferry T., Diep B. A., Sapin A., Lhoste Y., et al. (2013). PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS ONE 8:e63176. 10.1371/journal.pone.0063176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regassa L. B., Novick R. P., Betley M. J. (1992). Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60, 3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin J. K., Laabei M., Edwards A. M., Joo H. S., Otto M., Lennon K. L., et al. (2014). Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 1100–1107. 10.1128/AAC.01618-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz H. L., Augsburger V., Vuong C., Jack R. W., Götz F., Otto M. (2000). Inducible expression and cellular location of AgrB, a protein involved in the maturation of the staphylococcal quorum-sensing pheromone. Arch. Microbiol. 174, 452–455. 10.1007/s002030000223 [DOI] [PubMed] [Google Scholar]
- Saïd-Salim B., Dunman P. M., McAleese F. M., Macapagal D., Murphy E., McNamara P. J., et al. (2003). Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185, 610–619. 10.1128/JB.185.2.610-619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B., Drlica-Wagner A., Mathema B., Adhikari R. P., Kreiswirth B. N., Novick R. P. (2008). Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 198, 1171–1174. 10.1086/592051 [DOI] [PubMed] [Google Scholar]
- Shopsin B., Eaton C., Wasserman G. A., Mathema B., Adhikari R. P., Agolory S., et al. (2010). Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 1593–1599. 10.1086/656915 [DOI] [PubMed] [Google Scholar]
- Sully E. K., Malachowa N., Elmore B. O., Alexander S. M., Femling J. K., Gray B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. 10.1371/journal.ppat.1004174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y., Stacy D. M., Foegen M. K., Koenig D. W., Blackwell H. E. (2013). Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J. Am. Chem. Soc. 135, 7869–7882. 10.1021/ja3112115 [DOI] [PubMed] [Google Scholar]
- Traber K. E., Lee E., Benson S., Corrigan R., Cantera M., Shopsin B., et al. (2008). agr function in clinical Staphylococcus aureus isolates. Microbiology 154(Pt 8), 2265–2274. 10.1099/mic.0.2007/011874-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C., Gerke C., Somerville G. A., Fischer E. R., Otto M. (2003). Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718. 10.1086/377239 [DOI] [PubMed] [Google Scholar]
- Vuong C., Kocianova S., Yao Y., Carmody A. B., Otto M. (2004). Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505. 10.1086/424487 [DOI] [PubMed] [Google Scholar]
- Vuong C., Saenz H. L., Götz F., Otto M. (2000). Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693. 10.1086/317606 [DOI] [PubMed] [Google Scholar]
- Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- Wang R., Khan B. A., Cheung G. Y., Bach T. H., Jameson-Lee M., Kong K. F., et al. (2011). Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248. 10.1172/JCI42520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. M., Bassler B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- Wilke G. A., Bubeck Wardenburg J. (2010). Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. U.S.A. 107, 13473–13478. 10.1073/pnas.1001815107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K., Hardie K. R., Burgess N., Doherty N., Kirke D., Holden M. T., et al. (2002). LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148(Pt 4), 909–922. 10.1099/00221287-148-4-909 [DOI] [PubMed] [Google Scholar]
- Wright J. S., III, Jin R., Novick R. P. (2005). Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U.S.A. 102, 1691–1696. 10.1073/pnas.0407661102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Li H., Vuong C., Vadyvaloo V., Wang J., Yao Y., et al. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496. 10.1128/IAI.74.1.488-496.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Zhao L., Sun B. (2013). LuxS/AI-2 system is involved in antibiotic susceptibility and autolysis in Staphylococcus aureus NCTC 8325. Int. J. Antimicrob. Agents 41, 85–89. 10.1016/j.ijantimicag.2012.08.016 [DOI] [PubMed] [Google Scholar]
- Yao Y., Vuong C., Kocianova S., Villaruz A. E., Lai Y., Sturdevant D. E., et al. (2006). Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defense. J. Infect. Dis. 193, 841–848. 10.1086/500246 [DOI] [PubMed] [Google Scholar]
- Yarwood J. M., McCormick J. K., Schlievert P. M. (2001). Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183, 1113–1123. 10.1128/JB.183.4.1113-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Zhao L., Xue T., Sun B. (2012). Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 12:288. 10.1186/1471-2180-12-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gray L., Novick R. P., Ji G. (2002). Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277, 34736–34742. 10.1074/jbc.M205367200 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ji G. (2004). Identification of a staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J. Bacteriol. 186, 6706–6713. 10.1128/JB.186.20.6706-6713.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Xue T., Shang F., Sun H., Sun B. (2010). Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 78, 3506–3515. 10.1128/IAI.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]