Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma (NHL). Patients with DLBCL harboring MYC aberrations concurrent with BCL2 or/and BCL6 aberrations constitute a specific group with extremely poor outcome. In this study, we retrospectively investigated the incidence and prognosis of MYC, BCL2, and BCL6 aberrations with DLBCL patients in Chinese population. We applied fluorescence in situ hybridization and immunohistochemical analysis in 246 DLBCL patients. The results showed that patients with MYC or BCL2 copy number aberration (CNA) had significantly worse overall survival (OS) and progression-free survival (PFS) than negative cases (P < 0.0001). Patients with both MYC and BCL2 CNA had similar outcomes to those with classic double hit lymphoma or protein double expression lymphoma (MYC and BCL2/BCL6 coexpression). By multivariate analysis, MYC CNA, BCL2 CNA and double CNA were the independent worse prognostic factors. In conclusions, patients with MYC or BCL2 CNA constituted a unique group with extremely poor outcome and may require more aggressive treatment regimens.
Keywords: MYC, BCL2, copy number aberration, diffuse large B-cell lymphoma, pathology
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL), which is the most common group of non-Hodgkin lymphomas (NHL), accounts for 30%–40% of all lymphomas. The International Prognostic Index (IPI) is a clinical score that segregates DLBCL patients into four prognostic groups with distinct survival durations [1]. However, the differences in clinical features and treatment responses can also be affected by genetic and molecular features [2]. Genetic alterations can be of pivotal importance in establishing the correct diagnosis and predicting the course of disease [3].
MYC translocation, a biological hallmark of Burkitt lymphoma, can also be detected in DLBCL and B cell lymphoma unclassifiable with features intermediate between DLBCL and Burkitt lymphoma [4–6]. The t(14;18) translocation juxtaposes BCL2 to the immunoglobulin heavy chain gene (IGH) enhancer, resulting in BCL2 protein overexpression and inhibition of apoptosis [7]. This translocation is found in 80%–90% of follicular lymphoma and 20%–30% of de novo DLBCL cases [8]. It was reported that MYC concurrent with BCL2 or/and BCL6 translocations in DLBCL, called double-hit lymphoma or triple-hit lymphoma (DHL/THL), determines highly aggressive clinical behavior with extremely poor outcome and resistance to chemotherapy [9–11]. In addition, protein expression (such as MYC and BCL2 or BCL6) also had important prognostic value with or without gene aberrations [12–16]. However, little attention has been paid to copy number aberration (CNA) of genes associated with DHL. Therefore, we investigated the incidence and prognosis of MYC and BCL2 CNA in a population based study. Importantly, we compared the prognostic differences of double CNA with classic DHL and protein double expression (MYC and BCL2/BCL6 coexpression) lymphoma (DEL) and indicated the special value of double CNA which might be an important supplement to the DHL system.
RESULTS
The incidence of CNA, gene rearrangement and protein expression
Among the 246 DLBCL patients diagnosed in the First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital, fluorescence in situ hybridization (FISH) analysis was successfully performed in 240 cases and immunohistochemistry (IHC) analysis in 246 cases. Among 240 cases analyzed by FISH, MYC CNA was detected in 18 cases (7.5%), which was less frequent than MYC rearrangement {13.7% [33/240] cases had MYC translocation. Among these, 36.4% [12/33] cases accompanied IG [50% (6/12) each for IGH and IGL]}. BCL2 CNA was observed in 65 (27.1%) cases, which was more common than IGH/BCL2 rearrangement (12.5%, 30/240). In the 240 patients, 9 cases (3.8%) were identified as having CNA of both MYC and BCL2 (double CNA), which was a little more frequent than classic DHL (2.9%, 7/240) in this study.
At the protein level, the incidence of MYC, BCL2, and BCL6 expression was 36.6% (90/246), 57.3% (141/246), and 65.9% (162/246), respectively. Among these, 26.0% (64/246) showed coexpression of MYC and BCL2, and 22.8% (56/246) showed coexpression of MYC and BCL6.
The distribution of CNA, rearrangement and expression of MYC (Table 1a) and BCL2 (Table 1b) was showed in Table 1. Both MYC CNA (gain: r = 0.208, P = 0.002; amplification : r = 0.083, P = 0.340; gain plus amplification: r = 0.213, P = 0.001) and MYC rearrangement (r = 0.253, P < 0.001) were associated with MYC expression. A trend of association were observed between MYC CNA (gain plus amplification: r = 0.116, P = 0.082) and MYC rearrangement. BCL2 CNA (gain: r = 0.397, P < 0.0001; amplification: r = 0.154, P = 0.029; gain plus amplification: r = 0.358, P < 0.0001) but not BCL2 rearrangement (r = 0.124, P = 0.055) was associated with BCL2 expression. No association was found between BCL2 CNA and BCL2 rearrangement (r = 0.004, P = 0.956).
Table 1a. The distribution of MYC CNA, MYC rearrangement and MYC expression.
| Number of variables | MYC-R+ | MYC-R− | MYC+ | MYC− | |
|---|---|---|---|---|---|
| MYC CNA+ | gain | 4 | 9 | 10 | 3 |
| amplification | 1 | 4 | 3 | 2 | |
| MYC CNA− | 28 | 194 | 74 | 148 | |
| MYC CNA* | 3 | 3 | |||
| MYC-R+ | 22 | 11 | |||
| MYC-R− | 65 | 142 | |||
| MYC-R* | 3 | 3 | |||
Abbreviation: R: rearrangement; CNA: copy number aberration; MYC+: MYC expression; +: positive; *: no results.
Table 1b. The distribution of BCL2 CNA, BCL2 rearrangement and BCL2 expression.
| Number of variables | BCL2-R+ | BCL2-R− | BCL2+ | BCL2− | |
|---|---|---|---|---|---|
| BCL2 CNA+ | gain | 6 | 40 | 38 | 1 |
| amplification | 2 | 17 | 18 | 8 | |
| BCL2 CNA− | 22 | 153 | 81 | 94 | |
| BCL2 CNA* | 4 | 2 | |||
| BCL2-R+ | 22 | 8 | |||
| BCL2-R− | 115 | 95 | |||
| BCL2-R* | 4 | 2 | |||
Abbreviations: R: rearrangement; CNA: copy number aberration; BCL2+: BCL2 expression; +: positive; *: no results.
The association between gene CNA and clinical characteristics
Clinical characteristics analyzed for patients with gene CNA included age, sex, clinical stages, serum lactate dehydrogenase (LDH) level, performance status of Eastern Cooperative Oncology Group (ECOG PS), sites of extranodal involvement, IPI, B symptoms and cell of origin (COO). (Table 2). MYC CNA was associated with older age (P = 0.047) and higher IPI score (> 2) (P = 0.028). BCL2 CNA was associated with older age (P = 0.005), poorer ECOG PS (≥ 2) (P = 0.016) and non-GCB preference (P = 0.005). Double MYC and BCL2 CNA (MC+BC+) was associated with older age (P = 0.010).
Table 2. Clinical features with MYC, BCL2 and double CNA.
| Characteristics | MYC CNA+ | MYC CNA− | P value | BCL2 CNA+ | BCL2 CNA− | P value | MC+BC+ | MC−BC− | P value |
|---|---|---|---|---|---|---|---|---|---|
| No. of cases (%) | No. of cases (%) | No. of cases (%) | |||||||
| Age (years) | 18 | 222 | 65 | 175 | 9 | 166 | |||
| ≤ 60 | 7 (38.9) | 139 (62.6) | 0.047 | 30 (46.2) | 116 (66.3) | 0.005 | 2 (22.2) | 111 (66.9) | 0.010 |
| > 60 | 11 (61.1) | 83 (37.3) | 35 (53.8) | 59 (33.7) | 7 (77.8) | 55 (33.1) | |||
| Sex | 18 | 222 | 65 | 175 | 9 | 166 | |||
| Male | 14 (77.8) | 134 (60.4) | 0.144 | 46 (70.8) | 102 (58.3) | 0.077 | 6 (66.7) | 94 (56.6) | 0.734 |
| Female | 4 (22.2) | 88 (39.6) | 19 (29.2) | 73 (41.7) | 3 (33.3) | 72 (43.4) | |||
| Stage | 18 | 213 | 63 | 168 | 9 | 159 | |||
| III–IV | 12 (66.7) | 110 (51.6) | 0.220 | 30 (47.6) | 92 (54.8) | 0.333 | 4 (44.4) | 84 (52.8) | 0.738 |
| I–II | 6 (33.3) | 103 (48.4) | 33 (52.4) | 76 (45.2) | 5 (55.6) | 75 (47.2) | |||
| LDH | 18 | 213 | 63 | 168 | 9 | 159 | |||
| Elevated | 9 (50.0) | 90 (42.3) | 0.524 | 32 (50.8) | 67 (39.9) | 0.136 | 4 (44.4) | 62 (39.0) | 0.739 |
| Normal | 9 (50.0) | 123 (57.7) | 31 (49.2) | 101 (60.1) | 5 (55.6) | 97 (61.0) | |||
| ECOG PS | 18 | 222 | 65 | 175 | 9 | 166 | |||
| ≥ 2 | 4 (22.2) | 36 (16.2) | 0.512 | 17 (26.2) | 23 (13.1) | 0.016 | 2 (22.2) | 21 (12.7) | 0.336 |
| < 2 | 14 (77.8) | 186 (83.8) | 48 (73.8) | 152 (86.9) | 7 (77.8) | 145 (87.3) | |||
| Extranodal involvement | 18 | 213 | 63 | 170 | 9 | 159 | |||
| ≥ 2 | 4 (22.2) | 44 (20.7) | 0.772 | 15 (23.8) | 35 (25.6) | 0.595 | 2 (22.2) | 31 (19.5) | 1.000 |
| < 2 | 14 (77.8) | 169 (79.3) | 48 (76.2) | 135 (79.4) | 7 (77.8) | 128 (80.5) | |||
| IPI | 18 | 213 | 63 | 168 | 9 | 159 | |||
| 3–5 | 9 (50.0) | 53 (24.9) | 0.028 | 19 (30.2) | 43 (22.6) | 0.486 | 4 (44.4) | 38 (23.9) | 0.230 |
| 0–2 | 9 (50.0) | 160 (75.1) | 44 (69.8) | 125 (74.4) | 5 (55.6) | 121 (76.1) | |||
| B symptoms | 18 | 222 | 65 | 175 | 9 | 166 | |||
| Positive | 5 (27.8) | 79 (35.6) | 0.504 | 27 (41.5) | 57 (32.6) | 0.196 | 3 (33.3) | 55 (33.1) | 1.000 |
| Negative | 13 (72.2) | 143 (64.4) | 38 (58.5) | 118 (67.4) | 6 (66.7) | 111 (66.9) | |||
| COO (Hans) | 18 | 222 | 65 | 175 | 9 | 166 | |||
| GCB | 5 (27.8) | 93 (41.9) | 0.241 | 17 (26.2) | 81 (46.3) | 0.005 | 2 (22.2) | 78 (47.0) | 0.183 |
| Non-GCB | 13 (72.2) | 129 (58.0) | 48 (73.8) | 94 (53.7) | 7 (77.8) | 88 (53.0) | |||
Abbreviations: CNA: copy number aberration; MC+BC+: MYC CNA concurrent with BCL2 CNA; MC−BC−: negative for both MYC CNA and BCL2 CNA; COO: cell of origin; ECOG PS: performance status of Eastern Cooperative Oncology Group; GCB: germinal-center B-cell type; IPI: International Prognostic Index; LDH: lactate dehydrogenase.
Survival analysis
Patients characteristics
In the present era of rituximab, we carried out subset analysis of 141 patients who treated with R-CHOP-like therapies. The median follow-up time was 30 months (3–112 months). The clinical features of the patients were listed in Table 3.
Table 3. The clinical features of the 141 patients who treated with R-CHOP-like therapies.
| Characteristics | No. of cases (%) |
|---|---|
| Age (years) | |
| ≤ 60 | 82 (58.2) |
| Male | 92 (65.2) |
| Stage III–IV | 75 (53.2) |
| Elevated LDH | 58 (41.1) |
| ECOG PS ≥ 2 | 29 (20.6) |
| Extranodal sites ≥ 2 | 34 (24.1) |
| IPI score of 3–5 | 48 (34.0) |
| B symptoms | 52 (36.9) |
| COO (Hans) | |
| GCB | 63 (44.7) |
| Non-GCB | 78 (55.3) |
| Treatment | |
| R-CHOP | 72 (51.1) |
| R-DA-EPOCH | 22 (15.6) |
| R-CHOP-likeζ | 47 (33.3) |
| Prophylactic CNS treatment§ | 25 (17.7) |
| Radiation¶ | 12 (8.6) |
| Treatment response | |
| CR(u) | 112 (79.4) |
| PR | 14 (10.0) |
| SD/PD | 15 (10.6) |
Cases who received multiple regimens because of the following events: disease progression, cardiotoxicity of doxorubicin, accompanied hemophagocytic syndrome and extremely poor ECOG PS. The R-CHOP-like regimens including R-CDOP, R-CEOP and R-mini-CHOP.
Cases of with an increased risk of CNS events (paranasal sinus, testicular, bone marrow involvement) received 4–8 cycles of intrathecal methotrexate and/or cytarabine during the course of treatment.
Cases with localized lesion received radiotherapy alone or radioimmunotherapy.
Abbreviations: COO: cell of origin; CR(u): complete remission (unconfirmed); DLBCL: diffuse large B-cell lymphoma; ECOG PS: performance status of Eastern Cooperative Oncology Group; GCB: germinal-center B-cell type; IPI: International Prognostic Index; LDH: lactate dehydrogenase; PR: partial remission; SD/PD: stable disease/progression of disease.
Prognosis of MYC or BCL2 CNA
The incidences of MYC and BCL2 CNA were 7.1% (10/141) and 24.1% (34/141), respectively. The median percentages of cells with MYC and BCL2 CNA were 4% (2%–60%) and 5% (1%–80%), respectively. We then divided the MYC or BCL2 CNA patients into gain (3–4 copies) and amplification (≥ 5 copies), and no survival differences were observed between the two groups (Figure 1A–1D). The presence of MYC CNA was significantly associated with worse OS (median OS, 17.8 months vs not reached, P < 0.0001) (Figure 2A) and PFS (median PFS, 8.0 months vs not reached, P < 0.0001) (Figure 2B). The presence of BCL2 CNA was significantly associated with worse OS (median OS, 29.4 months vs not reached, P < 0.0001) (Figure 2C) and PFS (median PFS, 13.4 months vs not reached, P < 0.0001) (Figure 2D). In the current study, the best percentages of cells with CNA that predict outcome were 10% for both MYC and BCL2. By multivariate analysis, BCL2 CNA was an independent prognostic factor for both OS and PFS while MYC CNA was an independent prognostic factor for OS (Table 4).
Figure 1. Overall survival and progression-free survival of cases grouped according to gain and amplification with MYC 1A–1B and BCL2 1C–1D in the R-CHOP-like group.
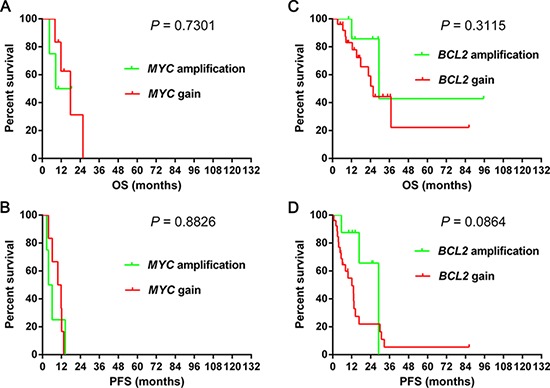
Abbreviations: OS: overall survival; PFS: progression-free survival.
Figure 2. Overall survival and progression-free survival of cases grouped according to MYC CNA 2A–2B, BCL2 CNA 2C–2D and double CNA 2E–2F.
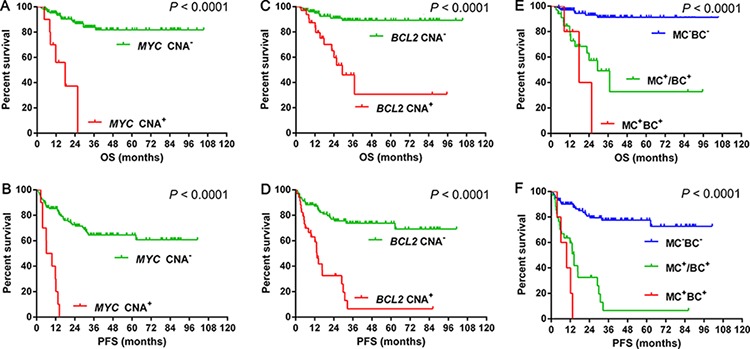
Abbreviations: CNA: copy number aberration; OS: Overall survival; PFS: progression-free survival; MC: MYC CNA; BC: BCL2 CNA; MC+BC+: MYC CNA concurrent with BCL2 CNA; MC+/BC+: MYC CNA or BCL2 CNA; MC−BC−: negative for both MYC CNA and BCL2 CNA.
Table 4a. Univariate and multivariate analysis with OS.
| Variates | Univariate analysis (OS) | Multivariate analysis (OS) | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| ENI | 2.484 | 1.496–4.126 | 0.0004 | 2.988 | 1.396–6.399 | 0.0048 |
| LDH | 2.746 | 1.661–4.541 | < 0.0001 | 1.502 | 0.777–2.903 | 0.2262 |
| Stage | 1.558 | 0.950–2.553 | 0.0787 | 0.674 | 0.325–1.399 | 0.2900 |
| IPI | 2.801 | 1.714–4.576 | < 0.0001 | 0.809 | 0.343–1.908 | 0.6279 |
| ECOG PS ≥ 2 | 2.552 | 1.486–4.383 | 0.0007 | 1.603 | 0.833–3.085 | 0.1580 |
| MYC+ | 5.515 | 3.339–9.108 | < 0.0001 | 3.127 | 1.649–5.929 | 0.0005 |
| BCL2+ | 2.086 | 1.243–3.500 | 0.0054 | 0.934 | 0.465–1.875 | 0.8471 |
| BCL6+ | 0.665 | 0.415–1.066 | 0.0905 | 0.792 | 0.434–1.443 | 0.4458 |
| Non-GCB | 0.640 | 0.387–1.059 | 0.0825 | 0.710 | 0.335–1.502 | 0.3705 |
| MYC-R+ | 4.715 | 2.873–7.739 | < 0.0001 | 7.527 | 2.358–24.031 | 0.0007 |
| DHL | 1.724 | 1.116–2.563 | 0.0070 | 1.595 | 1.005–2.534 | 0.0478 |
| MYC CNA+ | 5.344 | 2.790–10.234 | < 0.0001 | 3.058 | 1.227–7.620 | 0.0164 |
| BCL2 CNA+ | 3.808 | 2.373–6.111 | < 0.0001 | 2.619 | 1.417–4.840 | 0.0002 |
| MC+BC+ | 3.504 | 2.460–4.991 | < 0.0001 | 2.414 | 1.510–3.860 | 0.0021 |
Abbreviations: OS: overall survival; HR: hazard; ENI: extranodal involvement; LDH: serum lactate dehydrogenase; IPI: International Prognostic Index; ECOG PS: performance status of Eastern Cooperative Oncology Group; +: positive; GCB: germinal-center B-cell; R: rearrangement; DHL: double hit lymphoma; CNA: copy number aberration; MC+BC+: double CNA of MYC and BCL2.
Table 4b. Univariate and multivariate analysis with PFS.
| Variates | Univariate analysis (PFS) | Multivariate analysis (PFS) | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| ENI | 2.064 | 1.387–3.071 | 0.0004 | 2.193 | 1.263–3.807 | 0.0053 |
| LDH | 1.945 | 1.363–2.777 | 0.0002 | 1.199 | 0.765–1.879 | 0.4286 |
| Stage | 1.590 | 1.109–1.280 | 0.0116 | 1.161 | 0.727–1.852 | 0.5320 |
| IPI | 2.010 | 1.374–2.939 | 0.0003 | 0.632 | 0.339–1.177 | 0.1482 |
| ECOG PS≥2 | 1.842 | 1.195–2.841 | 0.0057 | 1.415 | 0.869–2.304 | 0.1630 |
| B symptoms | 1.781 | 1.255–2.526 | 0.0012 | 1.230 | 0.788–1.922 | 0.3618 |
| MYC+ | 3.411 | 2.401–4.845 | <0.0001 | 2.287 | 1.505–3.474 | 0.0001 |
| BCL2+ | 1.789 | 1.237–2.587 | 0.0020 | 1.027 | 0.666–1.583 | 0.9048 |
| Non-GCB | 0.692 | 0.481–0.997 | 0.0479 | 0643 | 0.405–1.020 | 0.0607 |
| MYC-R+ | 3.205 | 2.143–4.793 | <0.0001 | 2.528 | 1.508–4.236 | 0.0004 |
| DHL | 1.610 | 1.180–2.196 | 0.0027 | 1.466 | 0.999–2.151 | 0.0504 |
| MYC CNA+ | 3.521 | 2.058–6.022 | <0.0001 | 1.610 | 0.873–2.969 | 0.1271 |
| BCL2 CNA+ | 2.827 | 1.972–4.052 | <0.0001 | 2.066 | 1.349–3.165 | 0.0008 |
| MC+BC+ | 2.613 | 1.981–3.446 | <0.0001 | 2.067 | 1.341–3.186 | 0.0010 |
Abbreviations: PFS: progression-free survival; HR: hazard; ENI: extranodal involvement; LDH: serum lactate dehydrogenase; IPI: International Prognostic Index; ECOG PS: performance status of Eastern Cooperative Oncology Group; +: positive; GCB: germinal-center B-cell; R: rearrangement; DHL: double hit lymphoma; CNA: copy number aberration; MC+BC+: double CNA of MYC and BCL2.
Prognosis of combining MYC CNA (MC) and BCL2 CNA (BC)
The incidence of double CNA was 3.5%(5/141) in the R-CHOP-like group. The presence of double CNA was associated with worse OS (median OS, MC+BC+ vs. MC+/BC+: 17.8 vs 29.4 months, P = 0.307; MC+BC+ vs. MC−BC−: 17.8 months vs. not reached, P < 0.0001; MC+/BC+ vs. MC−BC−: 29.4 months vs. not reached, P < 0.0001) (Figure 2E) and PFS (median PFS, MC+BC+ vs. MC+/BC+: 9.7 vs. 14.4 months, P = 0.096; MC+BC+ vs. MC−BC−: 9.7 months vs. not reached, P < 0.0001; MC+/BC+ vs. MC−BC−: 14.4 months vs. not reached, P < 0.0001) (Figure 2F). By multivariate analysis, MCBC was an independent prognostic factor for both OS and PFS (Table 4).
Survival differences with MYC CNA, MYC rearrangement and MYC expression
The incidences of MYC rearrangement and MYC expression were 13.5% (19/141) and 29.8% (42/141), respectively. Patients with MYC CNA had similar OS (median OS: MYC CNA vs. MYC rearrangement: 17.8 vs. 29.4 months; P = 0.177; MYC CNA vs. MYC expression: 17.8 vs. 24.1 months, P = 0.180; MYC rearrangement vs. MYC expression: 29.4 vs. 24.1 months, P = 0.910) (Figure 3A) to cases with MYC rearrangement or MYC expression. Patients with MYC CNA tended to have a worse PFS (median PFS: MYC CNA vs. MYC rearrangement: 8.0 vs. 14.4 months; P = 0.019; MYC CNA vs. MYC expression: 8.0 vs. 13.4 months, P = 0.023; MYC rearrangement vs. MYC expression: 14.4 vs. 13.4 months; P = 0.972) (Figure 3B) than cases with MYC rearrangement or MYC expression.
Figure 3. Overall survival and progression-free survival in cases grouped according to MYC CNA, MYC gene rearrangement and MYC expression 3A–3B.
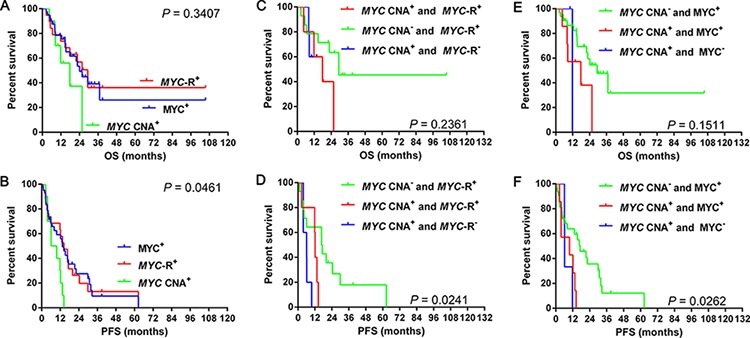
Overall survival and progression-free survival in cases grouped according to MYC CNA or rearrangement alone and MYC CNA along with rearrangement 3C–3D and MYC CNA or protein expression alone and MYC CNA along with protein expression 3E–3F.
Abbreviations: CNA: copy number aberration; R: rearrangement; OS: overall survival; PFS: progression-free survival.
Besides, we analyzed the survival differences among cases of MYC CNA, MYC rearrangement alone and MYC CNA along with rearrangement. Similar OS (median OS: MYC CNA alone vs. MYC rearrangement alone vs. MYC CNA along with rearrangement: not reached vs. 29.4 vs. 17.8 months, P = 0.236) were recognized among the three groups (Figure 3C, Table 5a). However, patients with MYC rearrangement alone tended to have longer PFS than cases with MYC CNA alone (median PFS: 17.0 vs. 6.1 months, P = 0.054) (Figure 3D, Table 5a) and MYC CNA along with rearrangement (median PFS: 17.0 vs. 12.2 months, P = 0.062) (Figure 3D, Table 5a). Patients with MYC CNA alone had worse PFS (median PFS: 6.1 vs. 12.2 months, P = 0.034) than cases with MYC CNA along with rearrangement (Figure 3D, Table 5a). We then analyzed the survival differences among cases of MYC CNA, MYC protein expression alone and MYC CNA along with protein expression. Patients with MYC CNA along with MYC expression tended to have shorter OS (median OS: MYC CNA along with MYC expression vs. MYC expression alone: 17.8 vs. 29.4 months, P = 0.060; MYC CNA along with MYC expression vs. MYC CNA alone: 17.8 vs. 11.8 months, P = 0.819) than cases with MYC expression alone but not MYC CNA alone (Figure 3E–3F, Table 5b). Patients with MYC CNA along with MYC expression had significantly shorter PFS (median PFS: MYC CNA along with MYC expression vs. MYC expression alone: 9.7 vs. 17.2 months, P = 0.018; MYC CNA along with MYC expression vs. MYC CNA alone: 9.7 vs. 6.2 months, P = 0.490) than cases with MYC expression alone but not MYC CNA alone (Figure 3E–3F, Table 5b). Similar OS (median OS: 11.8 vs. 29.4 months, P = 0.364) and PFS (median PFS: 6.2 vs. 17.2 months, P = 0.108) were observed between patients with MYC CNA alone and MYC expression alone (Figure 3E–3F, Table 5b).
Table 5a. The survival differences among MYC CNA+ alone, MYC rearrangement alone and MYC CNA+ along with MYC rearrangement.
| Survival | MYC CNA+ and MYC-R+ | MYC-R+ alone | MYC CNA+ alone | |||
|---|---|---|---|---|---|---|
| χ2 | P value | χ2 | P value | χ2 | P value | |
| Overall survival | ||||||
| MYC CNA+ and MYC-R+ | 2.819 | 0.093 | 0.066 | 0.797 | ||
| MYC-R+ alone | 2.819 | 0.093 | 0.339 | 0.560 | ||
| MYC CNA+ alone | 0.066 | 0.797 | 0.339 | 0.560 | ||
| Progression-free survival | ||||||
| MYC CNA+ and MYC-R+ | 3.481 | 0.062 | 4.499 | 0.034 | ||
| MYC-R+ alone | 3.481 | 0.062 | 3.400 | 0.054 | ||
| MYC CNA+ alone | 4.499 | 0.034 | 3.400 | 0.054 | ||
Abbreviations: CNA: copy number aberration; R: rearrangement; +: positive.
Table 5b. The survival differences among MYC CNA+alone, MYC expression alone and MYC CNA+ along with MYC expression.
| Survival | MYC CNA+ and MYC+ | MYC+ alone | MYC CNA+ alone | |||
|---|---|---|---|---|---|---|
| χ2 | P value | χ2 | P value | χ2 | P value | |
| Overall survival | ||||||
| MYC CNA+ and MYC+ | 3.547 | 0.060 | 0.053 | 0.819 | ||
| MYC+ alone | 3.547 | 0.060 | 0.825 | 0.364 | ||
| MYC CNA+ alone | 0.053 | 0.819 | 0.825 | 0.364 | ||
| Progression-free survival | ||||||
| MYC CNA+ and MYC+ | 5.608 | 0.018 | 0.477 | 0.490 | ||
| MYC+ alone | 5.608 | 0.018 | 2.585 | 0.108 | ||
| MYC CNA+ alone | 0.477 | 0.490 | 2.585 | 0.108 | ||
Abbreviations: CNA: copy number aberration; MYC+: MYC expression; +: positive.
Survival differences with BCL2 CNA, BCL2 rearrangement and BCL2 expression
The incidences of BCL2 rearrangement and BCL2 expression were 14.2% (20/141) and 51.1% (72/141), respectively. Patients with BCL2 CNA showed decreased OS (median OS: BCL2 CNA vs. BCL2 rearrangement: 29.4 months vs. not reached; P = 0.204; BCL2 CNA vs. BCL2 expression: 29.4 months vs. not reached, P = 0.019; BCL2 rearrangement vs. BCL2 expression: both not reached, P = 0.764) (Figure 4A) and PFS (median PFS: BCL2 CNA vs. BCL2 rearrangement: 13.4 vs. 21.4 months; P = 0.048; BCL2 CNA vs. BCL2 expression: 13.4 vs. 32.6 months, P = 0.003; BCL2 rearrangement vs. BCL2 expression: 21.4 vs. 32.6 months; P = 0.958) (Figure 4B) than cases with BCL2 rearrangement or BCL2 expression.
Figure 4. Overall survival and progression-free survival in cases grouped according to BCL2 CNA, BCL2 gene rearrangement and BCL2 expression 4A–4B.
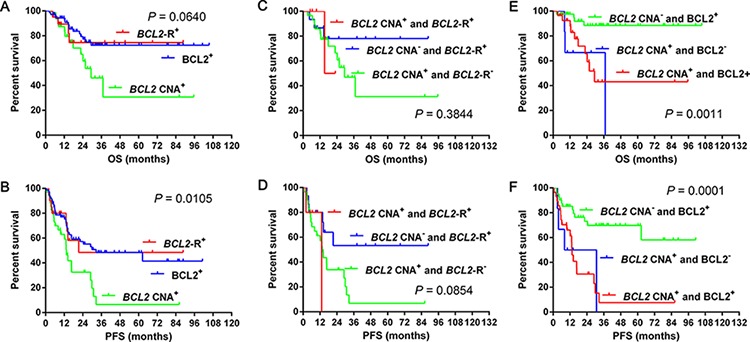
Overall survival and progression-free survival in cases grouped according to BCL2 CNA or rearrangement alone and BCL2 CNA along with rearrangement 4C–4D and BCL2 CNA or protein expression alone and BCL2 CNA along with protein expression 4E–4F.
Abbreviations: CNA: copy number aberration; R: rearrangement; OS: overall survival; PFS: progression-free survival.
Meanwhile, we analyzed the survival differences among cases of BCL2 CNA, BCL2 rearrangement alone and BCL2 CNA along with rearrangement. Similar OS was observed among above three groups (median OS: BCL2 CNA alone vs. BCL2 rearrangement alone vs. BCL2 CNA along with rearrangement: 29.4 months vs. not reached vs. 19.1 months, P = 0.384) (Figure 4C, Table 6a). Patients with BCL2 CNA alone had worse PFS than cases with BCL2 rearrangement alone (median PFS: BCL2 CNA alone vs. BCL2 rearrangement alone: 13.4 months vs. not reached, P = 0.032) while patients with BCL2 CNA along with rearrangement showed similar PFS to cases with BCL2 CNA (median PFS: 13.2 vs. 13.4 months, P = 0.753) or BCL2 rearrangement alone (median PFS: 13.2 months vs. not reached, P = 0.265) (Figure 4D, Table 6a). We then analyzed the survival differences among cases of BCL2 CNA, protein expression alone and BCL2 CNA along with protein expression. Patients with BCL2 CNA along with BCL2 expression and BCL2 CNA alone showed worse OS (median OS: BCL2 CNA along with BCL2 expression vs. BCL2 CNA alone vs. BCL2 expression alone: 29.4 vs 37.0 months vs. not reached, P = 0.002 for both) and PFS (median PFS: BCL2 CNA along with BCL2 expression vs. BCL2 expression alone: 13.4 months vs. not reached, P < 0.001; BCL2 CNA alone vs. BCL2 expression alone: 19.5 months vs. not reached, P = 0.011) than cases with BCL2 expression alone (Figure 4E–4F, Table 6b). Patients with BCL2 CNA along with BCL2 expression had similar OS (median OS: 29.4 vs. 37.0 months, P = 0.587) and PFS (median PFS: 13.4 vs. 19.5 months, P = 0.899) to cases with BCL2 CNA alone (Figure 4E–4F, Table 6b).
Table 6a. The survival differences among BCL2 CNA+ alone, BCL2 rearrangement alone and BCL2 CNA+ along with BCL2 rearrangement.
| Survival | BCL2 CNA+ and BCL2-R+ | BCL2-R+ alone | BCL2 CNA+ alone | |||
|---|---|---|---|---|---|---|
| χ2 | P value | χ2 | P value | χ2 | P value | |
| Overall survival | ||||||
| BCL2 CNA+ and BCL2-R+ | 0.082 | 0.775 | 1.888 | 0.169 | ||
| BCL2-R+ alone | 0.082 | 0.775 | 0.015 | 0.901 | ||
| BCL2 CNA+ alone | 0.015 | 0.901 | 1.888 | 0.169 | ||
| Progression-free survival | ||||||
| BCL2 CNA+ and BCL2-R+ | 1.245 | 0.265 | 0.099 | 0.753 | ||
| BCL2-R+ alone | 1.245 | 0.265 | 4.596 | 0.032 | ||
| BCL2 CNA+ alone | 0.099 | 0.753 | 4.596 | 0.032 | ||
Abbreviations: CNAs: copy number aberration; R: rearrangement; +: positive.
Table 6b. The survival differences among BCL2 CNA+ alone, BCL2 expression alone and BCL2 CNA+ along wvith BCL2 expression.
| Survival | BCL2 CNA+ and BCL2+ | BCL2+ alone | BCL2 CNA+ alone | |||
|---|---|---|---|---|---|---|
| χ2 | P value | χ2 | P value | χ2 | P value | |
| Overall survival | ||||||
| BCL2 CNA+ and BCL2+ | 9.847 | 0.002 | 0.295 | 0.587 | ||
| BCL2+ alone | 9.847 | 0.002 | 9.959 | 0.002 | ||
| BCL2 CNA +alone | 0.295 | 0.587 | 9.959 | 0.002 | ||
| Progression-free survival | ||||||
| BCL2 CNA+ and BCL2+ | 15.868 | < 0.001 | 0.016 | 0.899 | ||
| BCL2+ alone | 15.868 | < 0.001 | 6.505 | 0.011 | ||
| BCL2 CNA+ alone | 0.016 | 0.899 | 6.505 | 0.011 | ||
Abbreviations: CNA: copy number aberration; BCL2+: BCL2 expression; +: positive.
Survival differences with double CNA, classic DHL and DEL
Double CNA had similar OS (median OS: double CNA vs. classic DHL: 17.8 vs. 14.7 months, P = 0.850; double CNA vs. DEL: 17.8 vs. 24.1 months, P = 0.425; classic DHL vs. DEL: 14.7 vs. 24.1 months, P = 0.571) (Figure 5A) and PFS (median PFS: double CNA vs. classic DHL: 9.7 vs. 6.0 months, P = 0.338; double CNA vs. DEL: 9.7 vs. 13.4 months, P = 0.127; classic DHL vs. DEL: 6.0 vs. 13.4 months, P = 0.086) (Figure 5B) to classic DHL or DEL (MYC and BCL2). Double CNA also had similar OS (median OS: double CNA vs. classic-DHL: 17.8 vs. 14.7 months, P = 0.850; double CNA vs. DEL: 17.8 vs. 29.4 months, P = 0.366; classic DHL vs. DEL: 14.7 vs. 29.4 months, P = 0.583) (Figure 5C) and PFS (median PFS: double CNA vs. classic DHL: 9.7 vs. 6.0 months, P = 0.338; double CNA vs. DEL: 9.7 vs. 15.3 months, P = 0.071; classic DHL vs. DEL: 6.0 vs. 15.3 months, P = 0.098) (Figure 5D) to classic DHL or DEL (MYC and BCL6).
Figure 5. The differences of overall survival 5A–5B and progression-free survival 5C–5D in cases grouped according to double CNA, classic DHL and DEL.
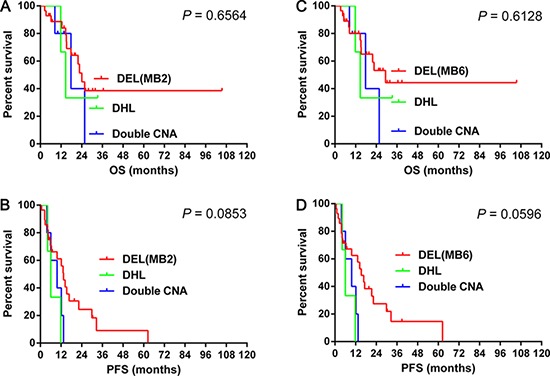
Abbreviations: CNA: copy number aberration; OS: overall survival; PFS: progression-free survival; DHL: double hit lymphoma; DEL: double expression lymphoma; MB2: double expression of MYC and BCL2 ; MB6: double expression of MYC and BCL6.
DISCUSSION
In this study, we performed a systematic investigation of the incidences and prognostic significances of MYC and BCL2 aberrations of DLBCL in a Chinese population. As far as we know, this is the first report to compare the prognosis among CNA, rearrangement and protein expression of MYC or BCL2. The results showed MYC or BCL2 CNA had more significant prognostic value than gene rearrangement or protein expression. It is noteworthy that we also compared the different prognostic values of double CNA with classic DHL and DEL. Double CNA had remarkable prognostic significance and might be an indispensable component of classic DHL.
The addition of rituximab to classic CHOP chemotherapy has significantly improved the outcome of DLBCL patients. However, more than one-third of the patients experience relapse and eventually die within 1–2 years [15]. It is necessary to identify such poor-risk patients who may benefit from alternative treatment strategies. Although gene expression profiling studies uncover biological heterogeneity with prognostic significance in DLBCL, the incorporation of this information into treatment algorithms awaits further investigation. This situation has motivated us to assess the clinical and prognostic significances of protein expression and rearrangement, such as MYC, BCL2, and BCL6, in a series of representative patients with DLBCL [17].
Recent studies have highlighted the importance of assessing MYC rearrangement in aggressive B-cell lymphomas, mainly DLBCL, as well as the detection of protein expression [3, 14, 18–20]. However, little knowledge has been investigated for CNA of genes associated with DHL, which might also be important for the prognosis of DLBCL. In the current study, we evaluated the clinical features, genetic characteristics, and prognostic significance of 246 cases of DLBCL with MYC or BCL2 CNA, rearrangement and protein expression.
It is noteworthy that we observed a series of patients with additional copies of MYC (7.5%) or BCL2 (27.1%), which were further confirmed by centromere 8 and BCL2 break apart probes. The attribute of MYC or BCL2 CNA, although not systematically investigated, has been reported in recent and previous studies [8, 16, 21, 22]. However, most studies did not analyze the prognostic significance of CNA [16, 21] or use a centromere reference and additional probe to distinguish polysomies or single gene break apart [8, 20]. Yoon et al. [22] previously observed increased copy number of MYC and BCL2 in 7.1% and 11.7% of DLBCL patients, respectively, more frequently in the non-GCB subtype. Our study showed that BCL2 but not MYC CNA was associated with the non-GCB subtype. However, the incidence of BCL2 CNA detected in our study was higher than theirs, probably due to different ethnic background or geographic variation. Stasik et al. [23] recently described a colorimetric in situ hybridization (CISH) method for detecting extra copies of the MYC gene in DLBCL and frequent occurrence of excess copies of discrete MYC signals (38%) in the context of diploidy or polyploidy of chromosome 8, which was correlated with increased mRNA signals and poor outcome. Although CISH might be more accurate to study MYC gene, it is not widely used. The most common and classic method to analyze MYC gene aberration is still conventional FISH. Additionally, conventional FISH is much easier to perform routinely. We confirmed these results and found that MYC CNA was associated with poor outcome [24, 25]. Similarly, another report had also indicated adverse effects on survival of MYC or BCL2 CNA [22]. However, they focused on MYC or BCL2 CNA alone, and not in combination (double CNA), which predicted worse OS and PFS, similar to classic DHL.
As is reported that MYC rearrangement was associated with decreased OS and PFS, [19, 26–28] while BCL2 rearrangement, in keeping with most reports [12, 29–31], was not predictive of both OS or PFS in patients treated with R-CHOP. Four recent studies evaluated the effect of MYC expression in DLBCL patients treated with R-CHOP [14, 15, 19, 27]. However, only one study, which was in line with ours, showed that MYC expression predicted poor survival [19]. The prognosis of BCL2 expression was more confusing in previous reports [16, 21]. Based on these results, we compared the differences among prognostic values of CNA, gene rearrangement and protein expression. Unexpectedly, patients with BCL2 CNA showed much shorter OS and PFS than those with BCL2 rearrangement or BCL2 expression. However, patients with MYC CNA showed similar OS and PFS to those with MYC rearrangement or MYC expression. These results indicated that CNA of MYC or BCL2 had significant prognostic value and should not be neglected.
Similar to previous researches [15, 19, 20, 27], we confirmed that patients with MYC and BCL2 coexpression (DEL) had extremely poor OS and PFS. Besides, we found that patients with MYC and BCL6 double expression predicted inferior outcome as well. Accordingly, we then compared the survival differences among double CNA, classic DHL and DEL. Surprisingly, no significant difference of OS or PFS was recognized among them, which indicated that double CNA had similar prognostic value to classic DHL and DEL and should not be overlooked in future studies.
In summary, patients with MYC or BCL2 CNA constituted another group of patients with extremely poor outcome. In multivariate analysis, MYC CNA (except for PFS), BCL2 CNA and double CNA were independent prognostic factors. Though the limited cases analyzed in our study, as far as we know, it is the first study comparing the different prognostic values of MYC and BCL2 at three distinct levels, which demonstrated the important prognosis of CNA. Thus, we suggest that patients with DLBCL harboring MYC or BCL2 CNA constituted a unique group with extremely poor outcome and may require more aggressive treatment regimens.
MATERIALS AND METHODS
Patients
All patients enrolled informed consent in accordance with requirements of the Declaration of Helsinki, and the research project was approved by the University and Institutional Review Boards. We retrospectively enrolled 246 adult patients with de novo DLBCL that had been diagnosed between February 2006 and January 2014 in the First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital. All cases were diagnosed according to World Health Organization (WHO) classification criteria. Cases were excluded if patients had a history of low grade B-cell lymphoma, primary cutaneous DLBCL, primary DLBCL of the central nervous system, primary mediastinal B-cell lymphoma or AIDS/HIV infection.
IHC
IHC (Figure 6) was performed on 4 μm sections with formalin-fixed paraffin-embedded (FFPE) specimens. Antibodies applied in the study including CD10 (clone 56C6; Dako, cut-off: 30%), MYC (clone Y69; Abcam, cut-off: 40%), BCL2 (clone 124; Dako, cut-off: 50%), BCL6 (clone LN22; Dako, cut-off: 30%), and MUM1 (clone MUM1p; Dako, cut-off: 30%). The COO was classified according to Hans algorithm [32].
Figure 6. The results of immunohistochemistry.
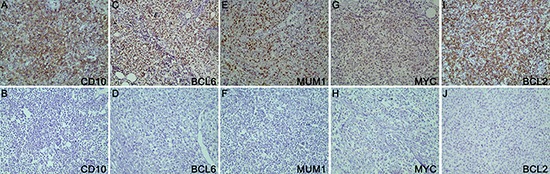
The proteins applied in this study were CD10 6A–6B, BCL6 6C–6D, MUM1 6E–6F, MYC 6G–6H and BCL2 6I–6J.
FISH
FISH was carried out according to manufacturer's instructions on FFPE tissue sections with the following probes: MYC dual-color, break apart translocation probe (Vysis LSI) and IGH/BCL2 dual-color, and dual fusion translocation probe (Vysis LSI). For cases with MYC translocation, the IGH/MYC/CEP 8 tri-color, dual fusion translocation probe (Vysis LSI), BCL6 dual-color break apart rearrangement probe (Vysis LSI), IG-kappa (IGK) DNA FISH probe, Split Signal (code Y5416; Dako) and IG-lambda (IGL) DNA FISH probe, Split Signal (code Y5412; Dako) were applied to further analyze on MYC concurrent gene rearrangements and partner genes. Cases with three or more BCL2 signals and normal IGH signals (BCL2/IGH probe) and without BCL2 gene break apart (BCL2 gene break apart probe) were considered as BCL2 CNA [33]. Cases with three or more MYC signals (IGH/MYC/CEP 8 tri-color, dual fusion translocation probe) along with two aqua signals of centromere 8 per nuclei were considered as MYC CNA [22]. Three or four copies of the gene studied was considered a gain, whereas more than four copies was considered as amplification (Figure 7) [34]. For probe signal scoring, a minimum of 200 interphase nuclei was examined. The cut-off levels for the probes were established by evaluating the split signal distribution in samples of reactive lymphoid tissues, calculating the mean number of split signals plus three times the standard deviation. The cut-off levels for positive values (mean of normal control ± 3 SD) were 14%, 5% and 7% for MYC break apart probe, IGH/BCL2 dual-fusion probe and BCL6 break apart probe, respectively.
Figure 7. CNA of MYC and BCL2.
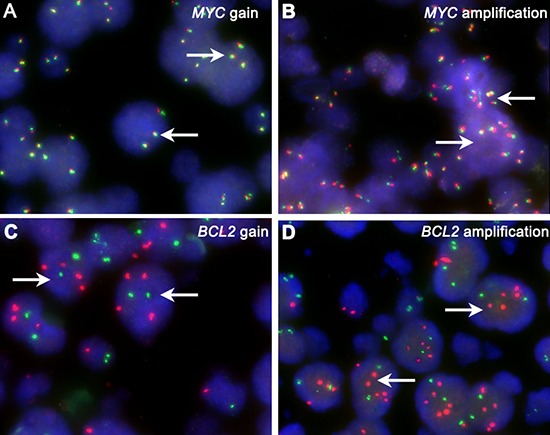
White arrows show MYC and BCL2 gain 7A, 7C and amplification 7B, 7D.
Abbreviation: CNA: copy number aberration.
Statistical analyses
OS and PFS were defined according to Cheson 2014 [35]. Statistical analyses were performed with use of SPSS software, version 20.0. The Chi-squared and Fisher exact tests were used to determine differences in the frequencies between groups. The Spearman test was used to analyze correlations between different variables. Survival curves were plotted by using Kaplan-Meier method and were compared by using log-rank test. For all tests, a probability value of less than 0.05 (2-sided) was considered statistically significant.
Acknowledgments
This study was supported by National Natural Science Foundation of China (30971296, 81170485, 81170488, 81370657, 81470328), Key Projects of Health Department of Jiangsu Province (K201108), Jiangsu Province's Medical Elite Program (RC2011169), National Public Health Grand Research Foundation (201202017), Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institute (JX10231801), Program for Development of Innovative Research Teams in the First Affiliated Hospital of Nanjing Medical University, Project of National Key Clinical Specialty. National Science & Technology Pillar Program (2014BAI09B12), and Project funded by Jiangsu Provincial Special Program of Medical Science (BL2014086).
REFERENCES
- 1.A predictive model for aggressive non-Hodgkin's lymphoma The International Non-Hodgkin's Lymphoma Prognostic Factors Project. The New England journal of medicine. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 2.Le Gouill S, Talmant P, Touzeau C, Moreau A, Garand R, Juge-Morineau N, Gaillard F, Gastinne T, Milpied N, Moreau P, Harousseau JL, Avet-Loiseau H. The clinical presentation and prognosis of diffuse large B-cell lymphoma with t(14, 18) and 8q24/c-MYC rearrangement. Haematologica. 2007;92:1335–1342. doi: 10.3324/haematol.11305. [DOI] [PubMed] [Google Scholar]
- 3.Aukema SM, Kreuz M, Kohler CW, Rosolowski M, Hasenclever D, Hummel M, Kuppers R, Lenze D, Ott G, Pott C, Richter J, Rosenwald A, Szczepanowski M, et al. Biological characterization of adult MYC-translocation-positive mature B-cell lymphomas other than molecular Burkitt lymphoma. Haematologica. 2014;99:726–735. doi: 10.3324/haematol.2013.091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, Hansmann ML, Haralambieva E, Harder L, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. The New England journal of medicine. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 5.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, Novak AJ, Dogan A, Ansell SM, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steven H, Swerdlow EC, Harris Nancy Lee, Jaffe Elaine S., Pileri Stefano A., Stein Harrld, Thiele Jürgen, Vardiman James W. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues. 4. Lyon: IARC; 2008. [Google Scholar]
- 7.Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Lin P, Fayad LE, Lennon PA, Miranda RN, Yin CC, Lin E, Medeiros LJ. B-cell lymphomas with MYC/8q24 rearrangements and IGH@BCL2/t(14, 18)(q32;q21): an aggressive disease with heterogeneous histology, germinal center B-cell immunophenotype and poor outcome. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:145–156. doi: 10.1038/modpathol.2011.147. [DOI] [PubMed] [Google Scholar]
- 9.Johnson NA, Savage KJ, Ludkovski O, Ben-Neriah S, Woods R, Steidl C, Dyer MJ, Siebert R, Kuruvilla J, Klasa R, Connors JM, Gascoyne RD, Horsman DE. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai RK, Sathanoori M, Van Oss SB, Swerdlow SH. Double-hit B-cell lymphomas with BCL6 and MYC translocations are aggressive, frequently extranodal lymphomas distinct from BCL2 double-hit B-cell lymphomas. The American journal of surgical pathology. 2013;37:323–332. doi: 10.1097/PAS.0b013e31826cebad. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Lin P, Young KH, Kanagal-Shamanna R, Yin CC, Medeiros LJ. MYC/BCL2 double-hit high-grade B-cell lymphoma. Advances in anatomic pathology. 2013;20:315–326. doi: 10.1097/PAP.0b013e3182a289f2. [DOI] [PubMed] [Google Scholar]
- 12.van Imhoff GW, Boerma EJ, van der Holt B, Schuuring E, Verdonck LF, Kluin-Nelemans HC, Kluin PM. Prognostic impact of germinal center-associated proteins and chromosomal breakpoints in poor-risk diffuse large B-cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4135–4142. doi: 10.1200/JCO.2006.05.5897. [DOI] [PubMed] [Google Scholar]
- 13.Akay OM, Aras BD, Isiksoy S, Toprak C, Mutlu FS, Artan S, Oner U, Gulbas Z. BCL2, BCL6, IGH, TP53, and MYC protein expression and gene rearrangements as prognostic markers in diffuse large B-cell lymphoma: a study of 44 Turkish patients. Cancer genetics. 2014;207:87–93. doi: 10.1016/j.cancergen.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, Liu WM, Visco C, Li Y, Miranda RN, Montes-Moreno S, Dybkaer K, Chiu A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green TM, Young KH, Visco C, Xu-Monette ZY, Orazi A, Go RS, Nielsen O, Gadeberg OV, Mourits-Andersen T, Frederiksen M, Pedersen LM, Moller MB. Immunohistochemical Double-Hit Score Is a Strong Predictor of Outcome in Patients With Diffuse Large B-Cell Lymphoma Treated With Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone. Journal of Clinical Oncology. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 16.Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, Lynch J, Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Campo E, Ott G, et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 17.Thieblemont C, Briere J. MYC, BCL2, BCL6 in DLBCL: impact for clinics in the future? Blood. 2013;121:2165–2166. doi: 10.1182/blood-2013-01-480392. [DOI] [PubMed] [Google Scholar]
- 18.Lin P, Medeiros LJ. The impact of MYC rearrangements and “double hit” abnormalities in diffuse large B-cell lymphoma. Current hematologic malignancy reports. 2013;8:243–252. doi: 10.1007/s11899-013-0169-y. [DOI] [PubMed] [Google Scholar]
- 19.Horn H, Ziepert M, Becher C, Barth TF, Bernd HW, Feller AC, Klapper W, Hummel M, Stein H, Hansmann ML, Schmelter C, Moller P, Cogliatti S, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 20.Valera A, Lopez-Guillermo A, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Espinosa I, Novelli S, Briones J, Mate JL, Salamero O, Sancho JM, Arenillas L, et al. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98:1554–1562. doi: 10.3324/haematol.2013.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iqbal J, Meyer PN, Smith LM, Johnson NA, Vose JM, Greiner TC, Connors JM, Staudt LM, Rimsza L, Jaffe E, Rosenwald A, Ott G, Delabie J, et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:7785–7795. doi: 10.1158/1078-0432.CCR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon SO, Jeon YK, Paik JH, Kim WY, Kim YA, Kim JE, Kim CW. MYC translocation and an increased copy number predict poor prognosis in adult diffuse large B-cell lymphoma (DLBCL), especially in germinal centre-like B cell (GCB) type. Histopathology. 2008;53:205–217. doi: 10.1111/j.1365-2559.2008.03076.x. [DOI] [PubMed] [Google Scholar]
- 23.Stasik CJ, Nitta H, Zhang W, Mosher CH, Cook JR, Tubbs RR, Unger JM, Brooks TA, Persky DO, Wilkinson ST, Grogan TM, Rimsza LM. Increased MYC gene copy number correlates with increased mRNA levels in diffuse large B-cell lymphoma. Haematologica. 2010;95:597–603. doi: 10.3324/haematol.2009.012864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valentino C, Kendrick S, Johnson N, Gascoyne R, Chan WC, Weisenburger D, Braziel R, Cook JR, Tubbs R, Campo E, Rosenwald A, Ott G, Delabie J, et al. Colorimetric in situ hybridization identifies MYC gene signal clusters correlating with increased copy number, mRNA, and protein in diffuse large B-cell lymphoma. American journal of clinical pathology. 2013;139:242–254. doi: 10.1309/AJCP2Z0TAGMUYJEB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Testoni M, Kwee I, Greiner TC, Montes-Moreno S, Vose J, Chan WC, Chiappella A, Baldini L, Ferreri AJ, Gaidano G, Mian M, Zucca E, Bertoni F. Gains of MYC locus and outcome in patients with diffuse large B-cell lymphoma treated with R-CHOP. British journal of haematology. 2011;155:274–277. doi: 10.1111/j.1365-2141.2011.08675.x. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen MO, Gang AO, Poulsen TS, Knudsen H, Lauritzen AF, Nielsen SL, Klausen TW, Norgaard P. MYC translocation partner gene determines survival of patients with large B-cell lymphoma with MYC- or double-hit MYC/BCL2 translocations. European journal of haematology. 2014;92:42–48. doi: 10.1111/ejh.12212. [DOI] [PubMed] [Google Scholar]
- 27.Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S, Scott DW, Tan KL, Steidl C, Sehn LH, Chan WC, Iqbal J, Meyer PN, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savage KJ, Johnson NA, Ben-Neriah S, Connors JM, Sehn LH, Farinha P, Horsman DE, Gascoyne RD. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 29.Copie-Bergman C, Gaulard P, Leroy K, Briere J, Baia M, Jais JP, Salles GA, Berger F, Haioun C, Tilly H, Emile JF, Banham AH, Mounier N, et al. Immuno-fluorescence in situ hybridization index predicts survival in patients with diffuse large B-cell lymphoma treated with R-CHOP: a GELA study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5573–5579. doi: 10.1200/JCO.2009.22.7058. [DOI] [PubMed] [Google Scholar]
- 30.Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, Dave S, Xiao L, Cao K, Zhu Q, Sherman S, Hans CP, Weisenburger DD, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. The American journal of pathology. 2004;165:159–166. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kramer MH, Hermans J, Wijburg E, Philippo K, Geelen E, van Krieken JH, de Jong D, Maartense E, Schuuring E, Kluin PM. Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma. Blood. 1998;92:3152–3162. [PubMed] [Google Scholar]
- 32.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 33.Barrans S, Crouch S, Smith A, Turner K, Owen R, Patmore R, Roman E, Jack A. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3360–3365. doi: 10.1200/JCO.2009.26.3947. [DOI] [PubMed] [Google Scholar]
- 34.Mossafa H, Damotte D, Jenabian A, Delarue R, Vincenneau A, Amouroux I, Jeandel R, Khoury E, Martelli JM, Samson T, Tapia S, Flandrin G, Troussard X. Non-Hodgkin's lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leukemia & lymphoma. 2006;47:1885–1893. doi: 10.1080/10428190600687547. [DOI] [PubMed] [Google Scholar]
- 35.Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Lister TA, Alliance AL, Lymphoma G, Eastern Cooperative Oncology G, European Mantle Cell Lymphoma C, Italian Lymphoma F, European Organisation for R, Treatment of Cancer/Dutch Hemato-Oncology G et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:3059–3068. doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]


