Abstract
Epigenetic regulation of gene expression has been suggested to play a critical role in the development of alcoholic hepatitis (AH). Although it has been shown that ethanol-induced damage in hepatocytes resulted from a change in methionine metabolism causes global gene expression changes in hepatocytes, the role of the epigenetic machinery in such processes has, however, been barely investigated. 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are major molecules of epigenetic DNA modification that play an important role in the control of gene expression. Using antibodies against 5mC and 5hmC, the DNA methylation in patients with AH was examined by immunohistochemistry and quantified by morphometric image analysis. The immunoreactivity intensity of 5mC in patients with AH was significantly higher than that seen in normal controls. While there was a trend of decreased 5-hmC in patients with AH, the difference between patients with AH and normal control was not significant. Our study suggests that aberrant DNA-methylation is associated pathogenesis of AH.
Keywords: alcoholic hepatitis, DNA methylation, 5-methylycytosine, 5-hydroxymethylcytosine
Introduction
DNA methylation is one of the major epigenetic mechanisms that regulate gene expression and thereby modify biological functions or cause disorders (Bernstein et al., 2007). This predominant epigenetic modification in mammals is catalyzed by DNA methyltransferases (DNMTs). DNA methylation involves transfer of a methyl group to cytosine bases at the C5 position of CG dinucleotides, referred to as CpG dinucleotides (Cheng and Blumenthal, 2008; Esteller, 2007). While 80% of CpG dinucleotides are in methylated state, unmethylated CpG residues in promoter regions of constitutively active genes are referred to as CpG islands (Esteller, 2007). Hypomethylation and hypermethylation of DNA in promoter regions are known to facilitate gene expression and to suppress gene expression, respectively, and are often associated with human diseases (Feinberg, 2007; Gopalakrishnan et al., 2008). Epigenetic regulation of gene expression has been suggested to play a critical role in the development of alcoholic hepatitis (AH). Epigenetic regulation may mediate the effects of environmental factors including diet and alcohol (Mandrekar, 2011). S-adenosylmethionine (SAMe) is the main methyl donor to connect DNA methylation.
In the previous studies, we identified DNA methylation-associated epigenetic regulation in liver biopsies from patients with AH and NASH and mice fed with diethyl 1-1, 4-dehydro-2,4,6-trimethyl-1,5-pyridine carboxylate (DDC), correlating with Mallory–Denk Body (MDB) formation (Liu et al., 2014a; Liu et al., 2014b; Oliva et al., 2009). MDBs are large eosinophilic hepatocellular cytoplasmic protein aggregates which are characteristic hallmarks of alcoholic steatohepatitis but they also occur in a variety of other liver diseases such as non-alcoholic steatohepatitis, Wilson's disease, chronic cholestasis, or hepatocellular carcinoma, etc. (Strnad et al., 2013; Zatloukal et al., 2007). Importantly, there were a downregulation of Ufmylation in AH, which is important for protein modification and an involvement in various diseases including cancer (Kim et al., 2013; Komatsu et al., 2004; Lemaire et al., 2011), in alcoholic and nonalcoholic steatohepatitis patients, and mice that were fed with DDC (Liu et al., 2014b). We also found a good correlation between the methylation of the promoter region with the transcriptional silencing of Ufmylation in liver biopsies from patients with AH and NASH and that DNMT1 and DNMT3B mRNA levels were significantly upregulated in these biopsies (Liu et al., 2014a).
To better understand the epigenetic regulation of DNA methylation and its possible involvement in the development of AH, we analyzed epigenetic modifications included 5mC and 5hmC in liver biopsies from patients with AH.
2. Materials and Methods
2.1. Biopsies
Human archived formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had AH (n = 3-5) were obtained from Harbor UCLA hospital archives. In all the cases MDBs were present in some hepatocytes of liver tissue sections from patients with AH but were not present in the normal control livers (Control; n=3). The biopsy sections were cut 4 μm thick.
2.2. Immunohistochemical staining
FFPE tissue slides were triple stained for 5-mC (mouce anti-5’-methylcytosine (Millipore, Temecula CA); Rabbit anti-5’-hydroxymethylcytosine (Active Motif, Carlsbad, CA).
The slides were examined with a Nikon 400 fluorescent microscope. The fluorescence intensity of staining of the protein of interest was measured quantitatively using 40× objective and a standard exposure time of 800 ms using a Nikon 400 fluorescent microscope with three filters (FITC-green, Texas Red, and Tri-Color), and the Nikon morphometric system. The results were displayed as a graph attached to the immunofluorescent photography using a screen snip.
2.5. Statistical analysis
Statistical significance was determined using the t-test. p values less than 0.05 were considered statistically significant. All data were presented as the mean ± S.E.M.
3. Results
3.1. Immunoreactivity of 5mC was elevated in patients with AH
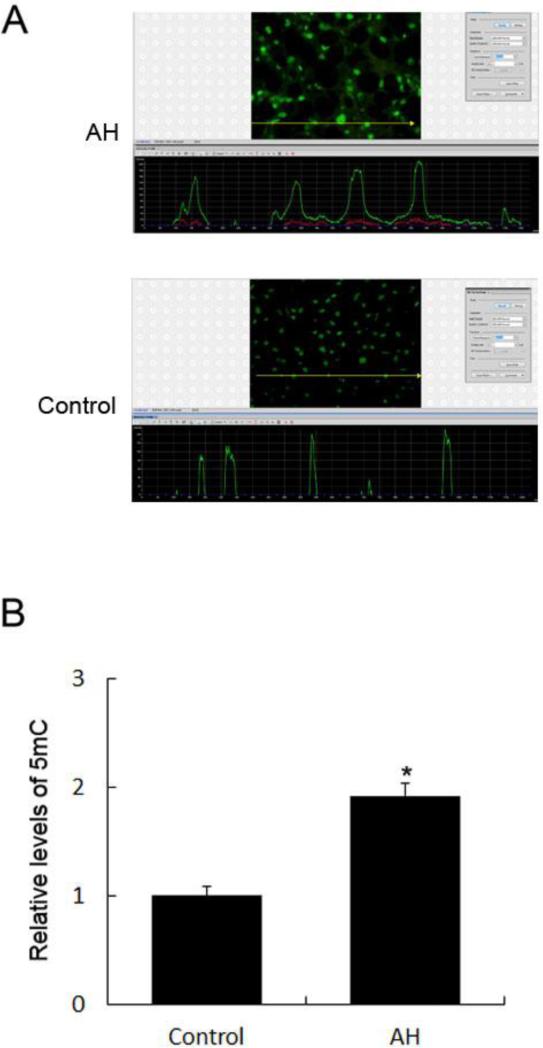
Using antibodies against 5-methylcytosine (5mC), we analyzed whether the level of 5mC changes in human AH. To that end, liver biopsies stemming from AH with or without liver fibrosis were included. In the normal control livers, no alcoholic, viral hepatitis, or diabetes was involved. The DNA methylation in patients with AH was examined by immunohistochemistry and quantified by morphometric image analysis. The immunoreactivity intensity of 5mC in patients with AH was 192±11% of that seen in normal controls (p<0.01, n=4) (Fig. 1).
Fig. 1. Increased immunoreactivity of 5mC in the livers of patients with AH.
A, The liver sections from patients with AH and controls were stained with an antibody against 5mC (top picture). The liver sections from patients with AH stained with greater intensity for 5mC compared to the controls. The bottom picture shows the fluorescence intensity measurement of the immunostaining of 5mC. A reprehensive screen snip was obtained from the morphometric screen (at 271× magnification). The fluorescence intensity was traced along the yellow line in the top panel and shown as a green tracer in the bottom picture. B, Quantification of fluorescence intensity of the immunostaining of 5mC. Data represent mean values ± S.E.M. *p<0.01.
3.2. Immunoreactivity of 5hmC did not change significantly in patients with AH
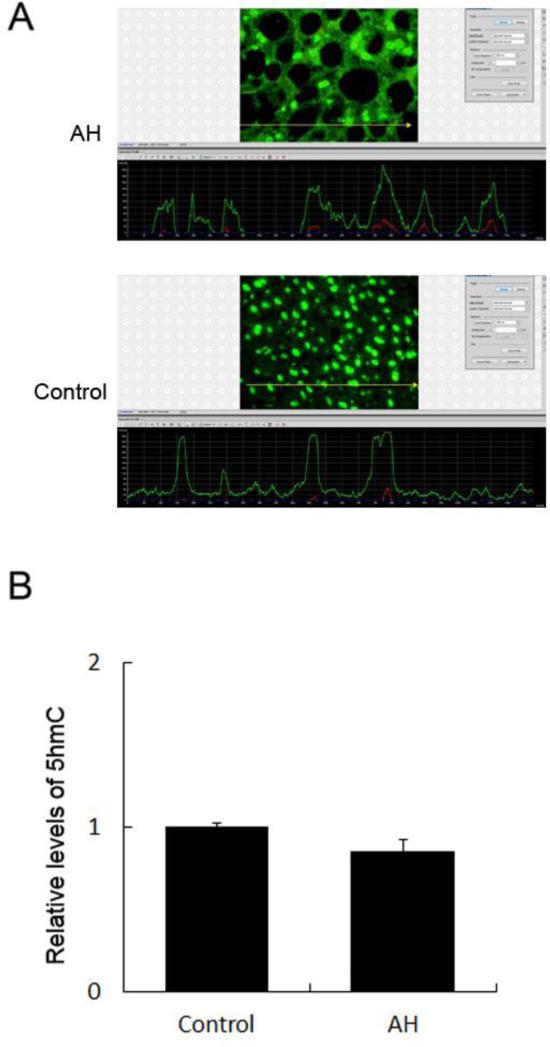
We next examined the level of 5hmC in patients with AH, and normal controls. While there was trend of decreased 5hmC in patients with AH, the difference between patients with AH and normal control was not significant (Fig. 2B).
Fig. 2. Immunoreactivity of 5hmC in the livers of patients with AH and controls.
A, The liver sections from patients with AH and controls were stained with an antibody against 5hmC (top picture). The liver sections from patients with AH stained with greater intensity for 5hmC compared to the controls. The bottom picture shows the fluorescence intensity measurement of the immunostaining of 5hmC. A reprehensive screen snip was obtained from the morphometric screen (at 271× magnification). The fluorescence intensity was traced along the yellow line in the top panel and shown as a green tracer in the bottom picture. B, Quantification of fluorescence intensity of the immunostaining of 5hmC. Data represent mean values ± S.E.M.
4. Discussion
In the present study, we investigated DNA methylation in the livers of AH patients by examining the levels of 5mC and 5hmC. We showed that the immunoreactivity intensity of 5mC in patients with AH was significantly higher than that seen in normal controls. Our data is consistent with a previous report showing significant increase of genomic DNA methylation in patients with alcoholism (Bonsch et al., 2004). The increased 5mC can be explained by the fact that the mRNA levels of DNMT1 and DNMT3B, enzymes that catalyze methylation of mammalian genomic DNA, were significantly upregulated in AH and NASH biopsies (Liu et al., 2014a). The functional consequence of increased 5mC is hypermethylation of DNA in promoter regions of certain genes leading to suppress gene expression. One pathway can be affected by increased 5mC in liver diseases is Ufmylation. We reported recently downregulation of Ufmylation in human alcoholic and nonalcoholic steatohepatitis, and mice fed with DDC (Liu et al., 2014b). In addition, the DNA methylation levels of Ufm1, Ufc1 and UfSP1, the components of Ufmylation pathway, in the promoter CpG region were significantly increased both in AH and NASH patients compared to normal subjects (Liu et al., 2014a). We also showed there was no significant decrease of 5hmC in patients with AH. Our data may indicate differential regulation of 5mC and 5hmC in AH.
DNA hypomethylation in promoter regions can facilitate gene expression and has been linked to ALD. The effects of alcohol on hepatic methionine metabolism and the role of methionine adenosyltransferase (MAT) and SAMe, the predominant methyl donor, in alcohol-associated liver cancer has been well documented (French, 2013; Varela-Rey et al., 2013). Lee et al., reported that patients with alcoholic hepatitis show reduced expression of the MAT1A gene that encodes the MAT isoenzymes, which led to lower hepatic SAMe levels (Lee et al., 2004). Patients with alcoholic cirrhosis exhibit decreased hepatic MAT activity and SAMe formation (Lu and Mato, 2008). The association of ALD and DNA hypomethylation was further supported by findings that SAMe prevented MDB induction by DDC refeeding (Li et al., 2008) and that betaine, a methyl donor, significantly prevents the downregulation of Ufmylation (Li et al., 2011). The alcohol-induce decline of SAMe may lead to DNA hypomethylation in the promoter regions of specific genes needs to be further identified.
It has been demonstrated that inflammation plays a critical role in the pathogenesis of ALD (Gao et al., 2008; Gao et al., 2011; Peng et al., 2014; Wang et al., 2012). Our previous studies demonstrated there was an upregulation of the MyD88-dependent TLR4/NFκB pathway in hepatocytes where MDBs formed (Liu et al., 2014c). There was an increase in the activity of the complement system in the liver of patients with alcoholic hepatitis (Shen et al., 2014).
As an appropriate gene expression pattern in immune cells is required for the immune response, epigenetic regulatory mechanisms plays an important role in immunity (Feinberg, 2007). SAMe has been shown to decrease TNF-alpha expression in LPS-stimulated Kupffer cells at the transcriptional level by downregulation of NF-kappaB promoter activity (Veal et al., 2004). Interestingly, SAMe did not lead to DNA methylation at the most common CpG target sites in the TNF-alpha promoter but was mainly mediated by MTA and SAMe at the level of histone methylation (Ara et al., 2008; Veal et al., 2004). Aberrant DNA-methylation occurred in the liver of (48)Ti-exposed mice where radiation-induced chronic oxidative stress and inflammation (Jangiam et al., 2015). 5mC increased in a dose-dependent pattern at 1 week and 1 month but only at the highest dosage (0.5 Gy) at 6 month. In contrast, 5hmC decreased in a dose-dependent patter at all time-points., and persistent aberrant DNA-methylation occurred in the liver of (48)Ti-exposed mice The cross-talk between alcohol-induced inflammation and alternation in DNA methylation remains to be elucidated in future. In summary, our study supports the view that aberrant DNA-methylation is associated with pathogenesis of AH.
Acknowledgments
This work was supported by grants from NIH (NIAAA U01AA021898) and P50-1999 Morphology Core.
Abbreviations
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- AH
alcoholic hepatitis
- ALD
alcoholic liver disease
- DNMTs
DNA methyltransferases
- FFPE
formalin-fixed, paraffin-embedded
- HCC
hepatocellular carcinoma
- MDBs
Mallory–Denk bodies
- NASH
non-alcoholic steatohepatitis
- SAMe
S-adenosylmethionine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ara AI, Xia M, Ramani K, Mato JM, Lu SC. S-adenosylmethionine inhibits lipopolysaccharide-induced gene expression via modulation of histone methylation. Hepatology. 2008;47:1655–1666. doi: 10.1002/hep.22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. Journal of neural transmission. 2004;111:1611–1616. doi: 10.1007/s00702-004-0232-x. [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature reviews. Genetics. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- French SW. Epigenetic events in liver cancer resulting from alcoholic liver disease. Alcohol research : current reviews. 2013;35:57–67. [PMC free article] [PubMed] [Google Scholar]
- Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- Gao B, Seki E, Brenner DA, Friedman S, Cohen JI, Nagy L, Szabo G, Zakhari S. Innate immunity in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2011;300:G516–525. doi: 10.1152/ajpgi.00537.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutation research. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. International journal of radiation biology. 2015;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- Kim CH, Nam HS, Lee EH, Han SH, Cho HJ, Chung HJ, Lee NS, Choi SJ, Kim H, Ryu JS, Kwon J, Kim H. Overexpression of a novel regulator of p120 catenin, NLBP, promotes lung adenocarcinoma proliferation. Cell cycle. 2013;12:2443–2453. doi: 10.4161/cc.25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. The EMBO journal. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcoholism, clinical and experimental research. 2004;28:173–181. doi: 10.1097/01.ALC.0000108654.77178.03. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E, Cnop M, Schuit F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PloS one. 2011;6:e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bardag-Gorce F, Dedes J, French BA, Amidi F, Oliva J, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li XM, Caudill M, Malysheva O, Bardag-Gorce F, Oliva J, French BA, Gorce E, Morgan K, Kathirvel E, Morgan T, French SW. Betaine feeding prevents the blood alcohol cycle in rats fed alcohol continuously for 1 month using the rat intragastric tube feeding model. Experimental and molecular pathology. 2011;91:540–547. doi: 10.1016/j.yexmp.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Gong M, French BA, Li J, Tillman B, French SW. Mallory-Denk Body (MDB) formation modulates Ufmylation expression epigenetically in alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH). Experimental and molecular pathology. 2014a;97:477–483. doi: 10.1016/j.yexmp.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Tillman B, French BA, French SW. Ufmylation and FATylation pathways are downregulated in human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC, where Mallory-Denk bodies (MDBs) form. Experimental and molecular pathology. 2014b;97:81–88. doi: 10.1016/j.yexmp.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Tillman B, Morgan TR, French BA, French SW. TLR3/4 signaling is mediated via the NFkappaB-CXCR4/7 pathway in human alcoholic hepatitis and non-alcoholic steatohepatitis which formed Mallory-Denk bodies. Experimental and molecular pathology. 2014c;97:234–240. doi: 10.1016/j.yexmp.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Mato JM. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. Journal of gastroenterology and hepatology. 2008;23(Suppl 1):S73–77. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World journal of gastroenterology : WJG. 2011;17:2456–2464. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Dedes J, Li J, French SW, Bardag-Gorce F. Epigenetics of proteasome inhibition in the liver of rats fed ethanol chronically. World J Gastroenterol. 2009;15:705–712. doi: 10.3748/wjg.15.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, French BA, Tillman B, Morgan TR, French SW. The inflammasome in alcoholic hepatitis: Its relationship with Mallory-Denk body formation. Experimental and molecular pathology. 2014;97:305–313. doi: 10.1016/j.yexmp.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, French BA, Liu H, Tillman BC, French SW. Increased activity of the complement system in the liver of patients with alcoholic hepatitis. Experimental and molecular pathology. 2014;97:338–344. doi: 10.1016/j.yexmp.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Nuraldeen R, Guldiken N, Hartmann D, Mahajan V, Denk H, Haybaeck J. Broad spectrum of hepatocyte inclusions in humans, animals, and experimental models. Compr Physiol. 2013;3:1393–1436. doi: 10.1002/cphy.c120032. [DOI] [PubMed] [Google Scholar]
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol research : current reviews. 2013;35:25–35. [PMC free article] [PubMed] [Google Scholar]
- Veal N, Hsieh CL, Xiong S, Mato JM, Lu S, Tsukamoto H. Inhibition of lipopolysaccharide-stimulated TNF-alpha promoter activity by S-adenosylmethionine and 5′-methylthioadenosine. American journal of physiology. Gastrointestinal and liver physiology. 2004;287:G352–362. doi: 10.1152/ajpgi.00316.2003. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Gao B, Zakhari S, Nagy LE. Inflammation in alcoholic liver disease. Annu Rev Nutr. 2012;32:343–368. doi: 10.1146/annurev-nutr-072610-145138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]