Abstract
Biological processes occur in complex environments containing a myriad of potential interactors. Unfortunately, limitations on the sensitivity of biophysical techniques normally restrict structural investigations to purified systems, at concentrations that are orders of magnitude above endogenous levels. Dynamic nuclear polarization (DNP) can dramatically enhance the sensitivity of NMR spectroscopy and enable structural studies in biologically complex environments. Here we applied DNP NMR to investigate the structure of a protein containing both an environmentally sensitive folding pathway and an instrinsically disordered region, the yeast prion protein Sup35. We added an exogenously-prepared isotopically-labeled protein to deuterated lysates, rendering the biological environment “invisible” and enabling highly efficient polarization transfer for DNP. In this environment, structural changes occurred in a region known to influence biological activity but intrinsically disordered in purified samples. Thus, DNP makes structural studies of proteins at endogenous levels in biological contexts possible and such contexts can influence protein structure.
INTRODUCTION
Structural investigations of biomolecules are typically confined to in vitro systems under limited conditions. Although investigations yield invaluable insights, such experiments can never capture all aspects of complex biological environments. Proteins must fold into their active conformations in complex environments. This situation becomes perilous when considering proteins that must attain a particular conformation, but whose energetic folding landscapes are rather flat or have several local minima. In these cases, the environment can clearly influence the conformation by favoring one pathway over another. Such decisions can have striking biological consequences, as is the case for a variety of protein folding diseases(Dobson, 2001). The effect of environment becomes even more critical when considering the substantial fraction of the human proteome that encodes disordered proteins(Dunker et al., 2001). Intrinsically disordered proteins (IDPs) are important components of the cellular signaling machinery, allowing the same polypeptide to undertake different interactions with different consequences (Wright & Dyson, 2015). Yet structural characterization of these domains is notoriously difficult(Uversky, 2013).
Yeast prions present both of these structural challenges as they have both environmentally-sensitive protein folding landscapes as well as intrinsically disordered regions. Yeast prions have provided a paradigm shift in our understanding of heritable biological information. They allow specific biological traits to be encoded and inherited solely though self-templating protein conformations. When a protein switches to its prion conformation, its function changes. This altered function is passed from generation to generation by conformational self-templating and catalyzed division of the template to daughter cells. The most extensively studied yeast prion, [PSI+](Cox, 1965), is an amyloid conformer of the translation termination factor Sup35. In purified amyloid fibrils of the prion domain of Sup35, called NM, the N terminal domain (N) adopts a beta-sheet-rich amyloid conformation while the adjacent middle domain (M) is intrinsically disordered(Frederick et al., 2014; Krishnan and Lindquist, 2005; Luckgei et al., 2013; Toyama et al., 2007). However, this is unlikely to be the case in vivo: the M domain is known to interact with many other biomolecules, including protein remodeling factors that regulate prion inheritance. As a consequence, mutations in the M domain(Helsen and Glover, 2012; Liu et al., 2002), or changes in the levels of protein chaperones (eg. Hsp70) and protein-remodeling factors (eg. Hsp104)(Kiktev et al., 2012; Masison et al., 2009; Tuite et al., 2011) have profound effects on prion propagation. NM also physically associates with protein chaperones(Allen, 2004), and at least one chaperone binding site has been localized to the M domain of NM(Helsen and Glover, 2012). Finally, a host of genetic data suggest that protein-based inheritance is sensitive to the combination and stoichiometry of many other proteins, meaning that isolated study of prion structure can offer at best only partial insight into this paradigm shifting biology.
Interest in prions is highlighted by the fact that similar structural transitions figure in the pathologies of a wide variety of human diseases. Prion strains were first described for the mammalian prion protein, PrP (Chien et al., 2004; Prusiner et al., 1998) and polymorphic amyloid forms have been reported for a variety of amyloidogenic protein associated with neurodegenerative disease (Guo et al., 2013; Kodali et al., 2010; Nekooki-Machida et al., 2009; Petkova et al., 2005). Upon structural characterization, only a portion of the protein is sequestered into the amyloid core. The amyloid cores of these fibers are flanked by intrinsically disordered regions(Heise et al., 2005; Helmus et al., 2008; Wasmer et al., 2009). More recently, amyloid forms of such proteins were demonstrated to have prion-like self-templating dispersion properties in vivo (Jucker and Walker, 2013; Polymenidou and Cleveland, 2012; Watts et al., 2013).
Nuclear magnetic resonance (NMR) is a powerful spectroscopic method for studying molecular structure and dynamics. A key strength of this technique is that it can be used to study non-crystalline, amorphous samples. Indeed, there have been a handful of high-profile in-cell NMR studies (Barbieri et al., 2013; Freedberg and Selenko, 2014; Inomata et al., 2009; Reckel et al., 2012; Sakakibara et al., 2009; Selenko et al., 2006; Vaiphei et al., 2011). These studies suggest that while protein structure can be perturbed, it is largely unchanged by the cellular context. However, these studies employed solution state NMR to detect proteins at concentrations two or more orders of magnitude above endogenous levels inside cells, radically altering endogenous stoichiometries. Because solution state NMR is limited by molecular tumbling times (which depend upon molecular size and solvent viscosity), the minority of the protein that might interact with cellular components would likely be undetectable. Moreover, because this population would comprise a small fraction of the total biomolecule, it would be difficult, if not impossible, to detect the resulting signal loss. Solid state NMR is not limited by molecular correlation times in this way. Instead, solid-state NMR is limited by its low sensitivity. Dynamic nuclear polarization (DNP) has the potential to alleviate this limitation by dramatically increasing the sensitivity of NMR spectroscopy, through the transfer of the large spin polarization that is associated with unpaired electrons to nearby nuclei(Abragam, 1983; Slichter, 1990). Theoretically, DNP can reduce experimental times by more than five orders of magnitude; an experiment that would require decades without DNP can be collected in a day with DNP. However, just as for other structural biology techniques, DNP sensitivity enhancements are critically dependent on experimental conditions(Ni et al., 2013) and sample composition(Akbey et al., 2010; 2013; Takahashi et al., 2014) and the specificity of NMR is critically dependent upon the choice of isotopic labeling(Wang et al., 2013). There is growing interest in application of DNP to complex systems. Several groups have applied DNP to investigate membrane proteins that were overexpressed to high levels in bacteria and have directly examined both concentrated membrane fractions and whole cells (Jacso et al., 2012; Renault et al., 2012; Yamamoto et al., 2014). We report conditions that enable high polarization transfer efficiencies in biologically complex environments. These are large enough to allow the characterization of a single protein at endogenous concentrations in its native environment. Structural methods to investigate either intrinsically disordered proteins or environmentally sensitive protein folding are limited. Here we present a generalizable approach for investigation of both of these challenging structural puzzles that lie at the heart of both fundamental biological questions and human diseases. Moreover we demonstrate that including the biological context can influence protein structure.
RESULTS
NM adopts an amyloid form in cell lysates at low concentrations
We first confirmed the NM protein adopted its active conformation at endogenous concentrations in a native environment. Previous studies have employed extensive serial dilution and propagation in purified in vitro conditions (Frederick et al., 2014). To ensure that the exogenously added protein was faithfully templated by the prion conformers from the cell lysate, we probed its structural state using semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)(Bagriantsev et al., 2006; Halfmann and Lindquist, 2008). NM did not form amyloid in lysates from cells that do not harbor the [PSI+] prion form of Sup35 (Figure 1B). In contrast, NM was templated into an amyloid form by both purified pre-formed fibers and lysates from cells that harbored the prion. We determined the concentration of templated, exogenously added NM was ~1 μM by immunoblot (Figure 1C), in good agreement with previously reported endogenous Sup35 concentrations of 2.5 to 5 μM (Ghaemmaghami et al., 2003). In this way, we prepared samples of isotopically-labeled NM amyloids at endogenous levels in a complex biological environment.
Figure 1. NM adopts an amyloid form in cell lysates at low concentrations.
(A) Prion status is maintained for yeast grown on deuterated media, indicating that the [PSI+] protein folding phenotypes were robust to growth in a deuterated environment. Phenotypically prion minus [psi−] (red) or prion plus [PSI+] (pink) yeasts were grown to mid-log phase in media made with either H2O or D2O and then spotted onto a ¼ YPD plate. Plates were incubated at 30 °C for 1 week.
(B) Amyloid formation of purified NM-His6 was visualized by SDD-AGE using an anti-His6 antibody in prion minus ([psi−]) cell lysates that do not contain endogenous prion templates, in the presence of 2% (w/w) purified amyloid seeds and in prion containing ([PSI+]) cellular lysates that contain endogenous prion templates. As with the endogenous prion, boiling (b) destroyed the templated amyloid aggregates.
(C) NM templated into the amyloid form in yeast cell lysates is not degraded and present at endogenous levels. Full length endogenous Sup35 runs at 100 kDa and is visualized with an antibody specific to the C-terminal domain. Cellular lysates both with and without exogenously added NM-His6 as well as a concentration gradient of purified NM-His6 were boiled in 2% SDS to denature any higher order aggregates and separated by SDS-PAGE before Western blotting with an antibody specific for the His6 epitope. NM-His6 runs at 55 kDa. Endogenous concentrations of Sup35 is between 2.5 and 5μM. The ECL signal for NM-His6 in lysates is less intense than that of purified NM-His6 at a concentration of 1.2 μM, indicating that the concentration of exogenously added NM in the NMR sample is below 1.2 μM.
Sensitivity and specificity of DNP MAS NMR
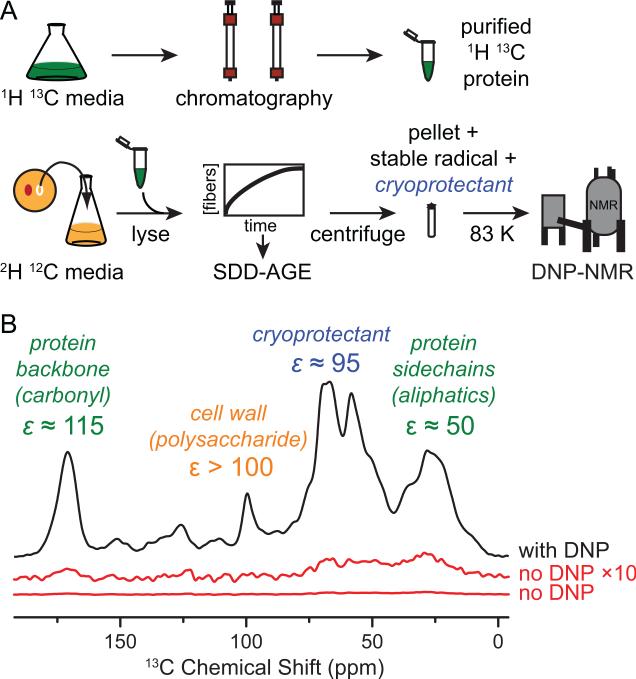
Having established that NM adopts an amyloid conformation in cellular lysates, we prepared recombinant, 1H, 13C-labeled NM and added it to cell lysates that had been grown in deuterated media with carbon isotopes in natural abundance. This created a spectroscopically-active prion protein in an NMR silent cellular background. We prepared the sample for DNP magic angle spinning (MAS) NMR by addition of cryoprotectant (glycerol) and a stable biradical TOTAPOL(Song et al., 2006). We collected 1D 13C{1H} cross polarization (CP) spectra of the cellular lysates both with and without microwave-driven polarization transfer from electrons to nuclei (DNP). Experiments using DNP resulted in significant signal enhancements relative to conventional NMR. DNP signal enhancements (ε) at 211 MHz were between 50- and 115- fold (Figure 2). The carbonyl carbon enhancements were similar to the maximal enhancements obtained for the reference system proline (ε = 130) for this instrumental configuration. This establishes that DNP MAS NMR is well-suited to study complex biological mixtures.
Figure 2. Dynamic Nuclear Polarization (DNP) enhances NMR signals in cellular lysates.
(A) Preparation of samples for DNP MAS NMR of proteins at endogenous levels in biological environments. The protein of interest is expressed on isotopically-enriched media and purified. The cellular background comes from cells grown on media containing D2O. The cells are lysed and the isotopically labeled protein is added exogenously to whole lysate. The mixture is pelleted, the pellet is resuspended in a matrix containing stable radical and cryoprotectant, and the mixture is frozen for analysis by DNP MAS NMR.
(B) One dimensional 13C{1H} spectra both with (black) and without DNP enhancement by microwaves (red). Dynamic nuclear polarization gave large signal enhancements (ε) for uniformly 1H 13C labeled NM in a deuterated matrix of cellular lysates containing a 60:30:10 (v/v) mixture of d8-glycerol:D2O:H2O and 10 mM TOTAPOL at 211 MHz / 140 GHz with ω/2π = 4.3 kHz and a sample temperature of 83 K. See also Figure S2.
DNP enhances the NMR signal of all 13C atoms in the sample. Interestingly, in samples with the uniformly 13C-labeled protein at endogenous concentrations, the 13C content from its natural abundance is an order of magnitude larger than that of added NM protein. However, because the natural abundance of 13C is 1.1%, only 0.01% of the 13C sites in the cell lysate were adjacent to another 13C site. Conversely, all the 13C sites in the exogenously added uniformly 13C labeled NM had adjacent 13C sites. To isolate 13C signals from NM and filter out background 13C signals from the cell lysates, we collected one-bond 13C-13C dipolar recoupled correlation spectra using proton driven spin diffusion (PDSD)(Szeverenyi et al., 1982). In this 2-dimensional experiment, on-diagonal peaks report on all 13C sites in the sample while off-diagonal peaks, or cross-peaks, report only on 13C sites that are directly bonded to another 13C site. To determine the contributions of cell lysates to the 13C-13C correlation spectra, we used signals from β1,3-glucan, a major cell wall component that is well-resolved from protein signals. As expected, the ratio of the cross-peak C1-C2 signal intensity relative to the diagonal C1 signal for β1,3-glucan was 2.5 ± 2% of that for yeast grown on uniformly 13C-enriched glucose. However, the ratio of the protein carbonyl carbon (C’) – carbon alpha (Cα) cross peak signal intensity relative to the diagonal C’ signal intensity for the protein backbone region was ten fold higher (21 ± 2%) for the natural abundance sample containing added 13C NM than the ratio for the β1,3-glucan region. The protein signal was an order of magnitude larger than the lysate background expected from natural abundance, establishing that the cross-peak signals in the 13C-13C correlation spectra report on the added NM and not on 13C in the cellular lysates. To completely eliminate any concerns about the contribution of natural abundance 13C from the cellular lysates, samples of prion-containing yeasts for structural investigations were grown with 13C-depleted (99.9% 12C) glucose as the carbon source, further reducing the 13C cross-peak intensity from the cellular lysates by two orders of magnitude. Thus, the combination of DNP with this isotopic labeling scheme provides the sensitivity and specificity to observe a protein at endogenous levels in a biologically complex native environment.
To investigate the structural influence of cellular lysates on NM amyloid assembly, we compared spectra of NM fibers at endogenous levels in cellular lysates to spectra of purified lysatetemplated NM fibers (Frederick et al., 2014). We conducted these experiments at higher magnetic fields (700MHz rather than 211 MHz) to achieve significant improvements in spectral resolution (Barnes et al., 2012; Michaelis et al., 2014). We collected a one-bond 13C-13C dipolar-assisted rotational resonance (DARR)(Takegoshi et al., 2001) correlation spectrum on 1 mg of cryoprotected, purified NM fibrils in six hours. For 10 μg of NM fibrils in unlabeled cellular lysates we collected a one-bond 13C-13C DARR spectrum for one week. As expected, no cross peaks for β1,3 glucan were present in spectra of cellular lysates grown in depleted 13C glucose. Inhomogeneous line broadening due to experimental temperatures required for DNP (83 K) potentially counteracts any gain in spectral resolution from higher magnetic fields. Thus, we compared spectra of purified NM fibers under DNP conditions to spectra of purified NM fibers at room temperature. In both samples, most of the resonances overlapped due to the number of sites and highly degenerate amino acid composition of this protein, a common feature of prion proteins(Frederick et al., 2014). Nonetheless, the line widths of isolated side chain sites in the DNP spectra at 83 K are similar to those of room temperature spectra (Figure S1). This establishes that DNP conditions did not compromise resolution gains at high magnetic fields, consistent with several other recent reports for cryogenic experiments on amyloid proteins(Debelouchina et al., 2010; Linden et al., 2011; Lopez Del Amo et al., 2013)
Native environments structure intrinsically disordered regions
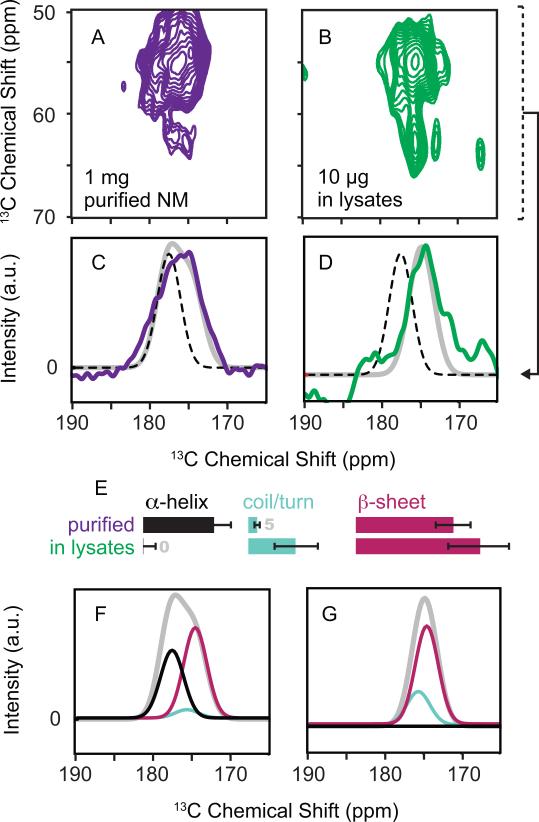
Thus poised, we sought to determine the structural influences of the biological context on NM. The NMR chemical shift is a sensitive indicator of the secondary structure of the protein backbone. To investigate effects of lysates on NM secondary structure, we compared the backbone chemical shifts in the presence and absence of cellular lysates. To isolate signals from backbone C’-Cα sites, we projected the Cα region of the one-bond 13C-13C DARR correlation spectra into one dimension (Figure 3A, 3B). We fit the carbonyl region of the projections to a sum of three Gaussians which described the chemical shift distributions for alpha helices, random coil and beta sheet conformations(Wang, 2002) (Figure 3). At 283 K, NM fibers experience motion over a broad range of time scales, (Frederick et al., 2014)). The rigid regions of NM fibers at 283 K had a chemical shift distribution consistent with a mix of turns and sheets (Figure S3). Cryoprotected NM fibers at 83 K had a chemical shift distribution that was dramatically shifted towards alpha helical values, consistent with sequence-based secondary structural predictions for the M domain(Chou and Fasman, 1974; Cuff et al., 1998; Kumar, 2013). This change is likely a result of secondary structural stabilization effects from the low experimental temperature and the cryoprotectant (Mehrnejad et al., 2011; Vagenende et al., 2009). In contrast, cryoprotected NM fibers that had been polymerized in cellular lysates had a chemical shift distribution that was dramatically shifted away from alpha helical values and towards beta sheet values (Figure 3). Thus, the cellular context had a profound effect on protein secondary structure.
Figure 3. The secondary structure of NM fibers in cellular lysates differs from the secondary structure of in vitro templated NM.
(A and B) Carbonyl carbon region of 13C-13C correlation spectra at 700 MHz using DNP MAS NMR of (A) cryoprotected purified NM fibers acquired in 6 hours and (B) cryoprotected NM fibers assembled in the presence of cellular lysates acquired in 1 week.
(C and D) Examination of the carbonyl carbon (C’) region of the spectra in projections of the Cα region (50-70 ppm indicated by dotted bracket) reveals the secondary structural composition of the protein backbone. The projection eliminates signals from non-backbone sites, such as the carbonyl moieties in the amino acid side chains like Asn and Gln. Dotted black lines indicate the expected chemical shift values for alpha helical conformations of the protein backbone and highlight a large shift away from alpha helical character for NM in lysates (D). The gray line represents the best-fitted solution to three Gaussian distributions describing the expected chemical shifts for the three possible secondary structural motifs: alpha helices (177.8 ± 1.5 ppm), random coils and turns (175.6 ± 1.5 ppm) and beta sheets (175.4 ± 1.55 ppm) (Wang 2002). Fits to a sum of these three Gaussian distributions gave standard estimates of error of 0.84 (C) and 0.93 (D). Residuals are plotted in Figure S3.
(E) Relative secondary structure contributions (in percent) as determined by intensity of each Gaussian distribution for the protein backbone of purified NM fibers (top) and NM fibers assembled in lysates (bottom). The error bars represent the standard error for the fitted intensity of each of the Gaussian distribution.
(F and G) The fitted intensities for alpha helices (black), random coils and turns (light blue) and beta sheets (magenta) are plotted with the fits (gray) from C and D.
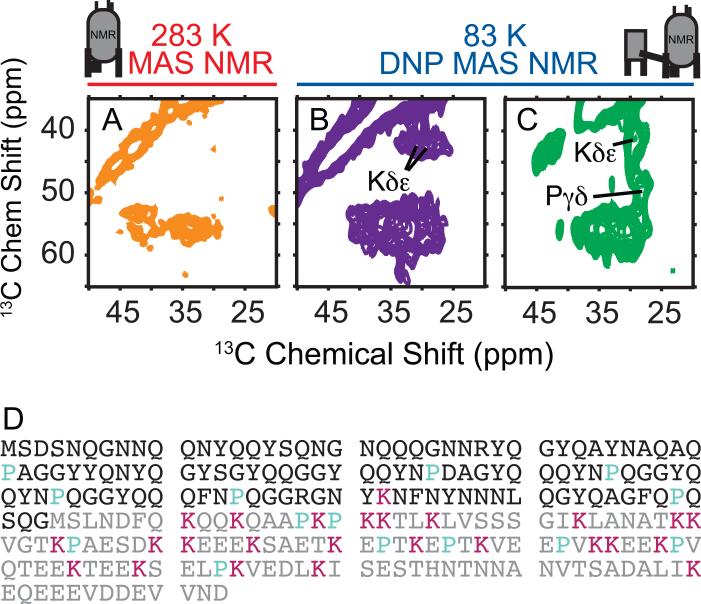
The NMR chemical shift is sensitive indicator of chemical identity and structural conformation (Wang, 2002). To determine which amino acid types undergo changes in their secondary structure in cellular contexts, we therefore compared the aliphatic region of the 13C-13C correlation spectra because this region reports on the chemically diverse amino acid side chains. The amyloid core of NM is largely composed of N, Q and Y residues. In purified room temperature samples, the 13C-13C correlation spectra was consistent with an amyloid core containing N, Q and Y residues in rigid beta sheet and turn conformations. Changes in the secondary structure at the alpha carbon for N, Q and Y from a beta sheet or random coil conformation to an alpha helical conformation result in an average change in chemical shift of ~4 ppm (Wang, 2002). The average chemical shift values for this region of the spectra at 83 K for both purified NM and NM in cell lysates were the same as those for the room temperature sample, consistent with the amyloid character of NM being maintained at low temperatures and unperturbed by a biological context (Figure 4 and Figure S4). Thus, the secondary structural changes (Figure 3) were not derived from a structural rearrangement of the amyloid core.
Figure 4. Complex biological environments restructure intrinsically disordered protein regions.
(A, B and C) Side chains of NM fibers in cellular lysates have a different chemical environment than in vitro templated NM. Aliphatic region of (A) purified NM fibers at 283 K in protonated assembly buffer (B) purified NM fibers at 83 K in 60% glycerol and (C) NM fibers at 83 K in 60% glycerol templated into the amyloid form in the presence of cellular lysates. See also Figure S1 and Figure S4.
(D) Amino acid sequence of NM with positions of lysines (magenta) and proline (cyan) highlighted. The N domain is black and the M domain is gray.
We next compared the aliphatic region of the 13C-13C correlation spectra to determine if amino acid types found in the M domain of NM were affected the biological contexts. In purified room temperature spectra of NM, the 13C-13C correlation spectra was consistent with previous findings that established that the M domain is highly dynamic with random coil character(Frederick et al., 2014; Luckgei et al., 2013); side chain resonances for the amino acid types found only in the M domain were absent. At 83 K, cross-peaks for methyl-bearing amino acids such as threonine, valine, isoleucine and leucine found in the M domain were absent from both spectra due to temperature-dependent dynamically-mediated relaxation of methyl-bearing amino acid side chains at this temperature(Bajaj et al., 2009; Beshah et al., 1987). However, at 83 K, lysine Cδ-Cε and proline Cγ-Cε cross peaks were present in the DNP MAS NMR spectra of both purified NM and of NM in cell lysates. Unlike the amyloid core residues, these amino acid types had very different chemical environments with differences in chemical shift of 5 ppm or greater depending on whether or not the fibers were templated in cellular lysates (Figure 4). Proline residues are present throughout the sequence of NM, while lysine residues are found only in the M domain (Figure 4D) localizing the regions experiencing large structural changes to the M domain. There are 25 lysine residues in the M domain of NM that contribute to the signal, all of which have different chemical environments and therefore different chemical shifts. The dramatic change in the shape of the lysine Cδ-Cε cross-peak indicates that a large proportion of the lysine side chains have a dramatically altered chemical environment in cellular lysates, indicating the majority of the M domain is involved. This establishes that the M domain, which contains chaperone-binding sites critical for faithful prion inheritance, makes many interactions with such components in vivo.
Multiple lines of evidence reveal that chaperone proteins directly interact with NM fibers. For example, the Hsp70 chaperone proteins Ssa1p and Ssa2p interact with NM aggregates (Allen, 2004), are among the top one hundred most highly expressed proteins (Ghaemmaghami et al., 2003) and the major components of amyloid aggregates isolated from yeast (Bagriantsev et al., 2008). In prion containing cells, NM forms membrane-free cellular structures with specific cellular localizations (Tyedmers et al., 2010). Within these structures, NM amyloid fibers are deposited in highly ordered arrays of regularly spaced fibrils. These arrays consist of bundles of fibers organized by inter-fibrils structures that are thought to be an Hsp70 because cells lacking Hsp70 can no longer form ordered arrays.(Saibil et al., 2012) This organization may be important for the faithful inheritance of the prion by daughter cells or for mitigating the toxicity that is otherwise associated with protein aggregation. The direct observation of NM structure in its biological context indicates that these organizing protein-protein interactions are mediated through the M domain of the protein via the adoption of a beta sheet secondary structure by the majority of this otherwise intrinsically disordered region. This work suggests that disordered regions that are often observed in purified fibril samples may be intimately involved with cellular components to create a self-organization mechanism that coordinates fiber deposition.
DISCUSSION
Application of high-field DNP MAS NMR methodology to a challenging biological system allowed us to pursue a scientific question that was previously impossible due to limits in instrumental sensitivity. Without DNP, these experiments would not be possible. With DNP MAS NMR we detected prion fibrils that had been assembled in a complex cellular environment containing all of the potential organizing protein components, such as chaperones, at their endogenous levels and stoichiometries. We established such fibers are structurally distinct from purified fibers in a region that is intrinsically disordered and highly dynamic in purified systems. The cellular environment structures an intrinsically disordered region. Sup35 is not unique; over a third of encoded proteins are predicted to be intrinsically disordered (Dunker et al., 2001). Indeed, intrinsically disordered protein regions have important roles in many biological processes, yet their structural characterization is notoriously difficult. Using DNP NMR, we can directly observe a protein of interest in its biological context. We found that the intrinsically disordered domain makes many direct interactions with cellular components. For NM, this suggests the M domain may be responsible for mediating interactions with the inter-fiber structures involved in prion fibril bundle organization visualized using in-vivo cryo-tomography (Saibil et al., 2012).
Our results demonstrate not only that structural studies of proteins in their native contexts are possible, but also that the native context can and does have a dramatic influence on protein structure. We anticipate that our methodology will enable structural investigations of heterotypic quaternary interactions between a protein of interest and cellular constituents. The methods described in this work can be extended to further investigations of protein conformation in biologically relevant environments. For example, protein structures can be determined in cellular contexts that have been modified, either genetically by deletion or overexpression of a protein or by the addition of small molecule agonists. Moreover, because the protein of interest is prepared exogenously, the full suite of specific isotopic enrichment schemes can be employed(Jaipuria et al., 2012) or segmentally isotopically labeled proteins can be used to obtain atomic level structural insights for otherwise crowded spectra(Volkmann and Iwaï, 2010). These approaches will be particularly useful for structural investigations of protein folding and mis-folding in native and perturbed environments. There are a large number of protein folding diseases and work across many fields of study is continually uncovering genetic, physical and chemical modulators of their pathobiology. Our approach will allow direct observation of the structural consequence of such modulators. Thus, this work provides the framework to answer structural questions about the toxic and non-toxic conformations of disease-associated proteins in a way that is directly informed by genetic backgrounds and biological phenotypes. This will allow us to investigate how genetic backgrounds modify the energetic landscape of protein folding and will enable tight coupling of genotypes, phenotypes and environments with specific structural arrangements.
EXPERIMENTAL PROCEDURES
Sample preparation
Both untagged NM and C-terminally His6 tagged NM were expressed and purified as described elsewhere(Serio et al., 1999). Uniformly-labeled 13C NM samples were prepared by growing BL21(DES)-Rosetta Escherichia coli in the presence of M9 media with 2 g L−1 D-glucose 1H,13C6 (Cambridge Isotope Labs, Cambridge, MA). Purified, lysate-templated NM seeds for the purified fiber sample were prepared as described elsewhere(Frederick et al., 2014), using cell lysates from a strong [PSI+] yeast strain. One milligram of purified denatured 13C-labeled NM was diluted 120-fold out of 6 M GdHCL into 4 mL of lysis buffer (see below) containing 0.02 mg preformed fibers. The reaction was allowed to polymerize for 24 hours at 4 °C and fibers were collected by ultracentrifugation at 430,000 × g for 1 hour. Bradford analysis revealed that removal of the supernatant decreased the total protein content of the sample by one third. The pellet was resuspended in 60:30:10 (v/v/v) mixture of 13C-depleted d8-glycerol (99.9% 12C):D2O:H2O(Lange et al., 2012) containing 10 mM of the stable biradical TOTAPOL(Corzilius et al., 2014; Song et al., 2006).
Cell lysate samples for DNP
Phenotypically strong [PSI+] yeast were grown in a 20 mL culture volume at 30 °C to mid-log phase in YPD media made with protonated carbon sources and 100% D2O. Because we use protonated carbon sources, the final deuteration level for the lysates is estimated to be 70%(Leiting et al., 1998). Cells maintained their [PSI+] status in deuterated media (Figure 1A). Cells were collected by centrifugation (5 min, 4000 × g) and washed once with water and once with D2O. Pellets were suspended in 200 μL of lysis buffer (50 mM Tris-HCl pH 7.4, 200 mM NaCl, 2 mM TCEP, 5% d8-13C depleted glycerol, 1 mM EDTA, 5 ug/mL of aprotinin and leupeptin and 100 μg/mL Roche protease inhibitor cocktail. Lysis buffer was 80% (v/v) D2O.) Cells were lysed by bead beating with 500 μm acid washed glass beads for 8 minutes at 4 °C. After bead beating, the bottom of the eppendorf tube was punctured with a 22G needle and the entire lysate mixture was transferred to a new tube. Purified denatured 13C-labeled NM was diluted 150-fold out of 6 M GdHCl to a final concentration of 5 μM and the mixture was allowed to polymerize, quiescent, at 4 °C for 24 hours. Unassembled NM was removed by centrifugation at 20000 x g for 1 hour at 4 °C and removal of the supernatant. The ~30 μL pellet was resuspended in 30 μL of 100% d8-13C depleted glycerol containing 20 mM TOTAPOL and transferred to a 4 mm sapphire rotor. The final radical concentration was 10 mM(Corzilius et al., 2014) and the glycerol concentration was 60% (Lange et al., 2012). The cell lysate sample for high field DNP was made analogously, except that yeast cells were grown in SD-CSM media made with D2O and 2% (w/v) protonated 13C-depleted glucose (99.9% 12C, Cambridge Isotope Labs) as the carbon source. Uniform 13C labeled samples were grown using U-13C glucose (99% Cambridge Isotope Labs) as the carbon source. The final sample volume was 20 μL and the sapphire rotor had a 3.2 mm diameter.
Immunohistochemistry
Cell lysate samples were made as described above, except NM-His6 was substituted for NM. SDD-AGE was performed as described(Halfmann and Lindquist, 2008), and NM was visualized using an anti-His6 antibody. Cell lysates were fractionated by SDS-PAGE, transferred to nitrocellulose and probed with both anti-His6 and anti-Sup35 antibodies. For SDD-AGE analysis we prepared cellular lysates as described above and added 5 μM purified denatured NM-His6 to reactions containing cellular lysates from prion minus ([psi−]) cultures, purified NM fibers prepared in isolation (2% seeding w/w), and cellular lysates from prion plus ([PSI+]) cultures. For Western blot analysis, lysate samples were denatured by incubation at 95 °C for 10 minutes in the presence of 2% SDS before fractionation to denature amyloid aggregates. Secondary antibodies were coupled to horseradish peroxidase. Blots were visualized by a standard ECL analysis.
Spectroscopy
DNP MAS NMR experiments were performed on custom-designed home-built instruments, consisting of a 212 MHz (1H, 5 T) and a 697 MHz (1H, 16.4 T) NMR spectrometer (courtesy of Dr. David Ruben, Francis Bitter Magnet Laboratory, MIT) equipped with custom-built 140 and 460 GHz gyrotrons (Joye et al., 2006) (i.e., high power microwave devices generating up to 12 W), respectively. DNP MAS NMR spectra were recorded on home-built 4 mm (211 MHz) quadruple resonance (1H, 13C, 15N & e−) or 3.2 mm (700 MHz) triple resonance (1H, 13C & e−) cryogenic probes equipped with Kel-F stators (Revolution NMR, Fort Collins, CO). Microwaves were guided to the sample via circular overmoded waveguide in which the inner surface has been corrugated to reduce mode conversion and ohmic losses. Sample temperatures were maintained below 85 K, with spinning frequencies of ωr/2π = 4.3 – 10 kHz.
13C{1H} cross polarization(Pines, 1973) spectra were acquired with a contact time of 1.5 ms. Recycle delays were chosen as TB (polarization buildup time constant) × 1.26 (Figure S2), yielding optimum sensitivity per unit of time. These were 4.6 s and 8 s for 211 MHz and 700 MHz, respectively. A series of 13C-13C DARR spectra were recorded using either a mixing period of 6 or 15 ms, 64-512 co-added transients and, between 60 and 100 t2 increments. All data were acquired using high-power TPPM 1H decoupling (ƔB1> 83 kHz). Enhancements at 211 MHz are reported in Figure 2 and those at 700 MHz were estimated at −8 to −10. DNP enhancements at both fields were about 80% of the maximal enhancements recorded on a standard sample of proline. Experimental data were processed using RNMR (1D) or NMRpipe(Delaglio et al., 1995) (2D) and analyzed using Sparky(Goddard and Kneller, 2006). 13C NMR data were referenced to adamantane(Morcombe and Zilm, 2003) (40.49 ppm at room temperature) and KBr was used to set the magic angle.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Lindquist and Griffin groups for valuable discussions and comments during the course of this research. S.L. is an investigator of the Howard Hughes Medical Institute. K.K.F. was supported by the Life Science Research Foundation as an HHMI fellow. V.K.M. is grateful to the Natural Sciences and Engineering Research Council of Canada and the Government of Canada for a Banting postdoctoral fellowship. B.C. was supported by the Deutsche Forschungsgemeinschaft (research fellowship CO 802/1-1). This work was supported by US National Institutes of Health Grants GM-025874 to S.L. and, EB-001960, EB-002804 and EB-002026 to R.G.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, K.K.F.; Methodology, K.K.F.; Investigation, K.K.F., A.J., V.K.M., B.C. and T-C.O. Writing – Original Draft, K.K.F.; Writing – Review and Editing, K.K.F., V.K.M. and S.L.; Funding, S.L. and R.G.G.; Supervision, K.K.F, S.L., and R.G.G.
No competing financial interests have been declared.
REFERENCES
- Abragam A. The principles of nuclear magnetism. Clarendon Press; 1983. [Google Scholar]
- Akbey Ü, Franks WT, Linden A, Lange S, Griffin RG, van Rossum B-J, Oschkinat H. Dynamic nuclear polarization of deuterated proteins. Angew. Chem. Int. Ed. 2010;49:7803–7806. doi: 10.1002/anie.201002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbey Ü, Franks WT, Linden A, Orwick-Rydmark M, Lange S, Oschkinat H. Topics in Current Chemistry. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. Dynamic Nuclear Polarization Enhanced NMR in the Solid-State. pp. 181–228. [DOI] [PubMed] [Google Scholar]
- Allen KD. Hsp70 Chaperones as Modulators of Prion Life Cycle: Novel Effects of Ssa and Ssb on the Saccharomyces cerevisiae Prion [PSI+]. Genetics. 2004;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Meth Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Bajaj VS, van der Wel PCA, Griffin RG. Observation of a Low-Temperature, Dynamically Driven Structural Transition in a Polypeptide by Solid-State NMR Spectroscopy. J Am Chem Soc. 2009;131:118–128. doi: 10.1021/ja8045926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L, Bertini I, Luchinat E, Secci E, Zhao Y, Banci L, Aricescu AR. atomic- resolution monitoring of protein maturation in live human cells by nMr. Nat Chem Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AB, Markhasin E, Daviso E, Michaelis VK, Nanni EA, Jawla SK, Mena EL, DeRocher R, Thakkar A, Woskov PP, et al. J Magn Reson. 2012;224:1–7. doi: 10.1016/j.jmr.2012.08.002. Journal of Magnetic Resonance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshah K, Olejniczak ET, Griffin RG. Deuterium NMR study of methyl group dynamics in L-alanine. J. Chem. Phys. 1987;86:4730. [Google Scholar]
- Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Corzilius B, Andreas LB, Smith AA, Ni QZ, Griffin RG. Paramagnet induced signal quenching in MAS-DNP experiments in frozen homogeneous solutions. J Magn Reson. 2014;240:113–123. doi: 10.1016/j.jmr.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS. Ψ, A cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Debelouchina GT, Bayro MJ, van der Wel PCA, Caporini MA, Barnes AB, Rosay M, Maas WE, Griffin RG. Dynamic nuclear polarization-enhanced solid-state NMR spectroscopy of GNNQQNY nanocrystals and amyloid fibrils. Phys Chem Chem Phys. 2010;12:5911–5919. doi: 10.1039/c003661g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dobson C. The structural basis of protein folding and its links with human disease. Phil Trans Royal Society London, B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol. 2014;21:295–305. doi: 10.1016/j.chembiol.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg DI, Selenko P. Live cell NMR. Annu Rev Biophys. 2014;43:171–192. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky. University of California; 2006. [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW, Glover JR. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem. 2012;287:542–556. doi: 10.1074/jbc.M111.302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Takeuchi T, Futaki S, Ito Y, Hiroaki H, et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- Jacso T, Franks WT, Rose H, Fink U, Broecker J, Keller S, Oschkinat H, Reif B. Characterization of membrane proteins in isolated native cellular membranes by dynamic nuclear polarization solid-state NMR spectroscopy without purification and reconstitution. Angew. Chem. Int. Ed. 2012;51:432–435. doi: 10.1002/anie.201104987. [DOI] [PubMed] [Google Scholar]
- Jaipuria G, Krishnarjuna B, Mondal S, Dubey A, Atreya HS. Amino acid selective labeling and unlabeling for protein resonance assignments. Adv. Exp. Med. Biol. 2012;992:95–118. doi: 10.1007/978-94-007-4954-2_6. [DOI] [PubMed] [Google Scholar]
- Joye CD, Griffin RG, Hornstein MK, Kan-Nian Hu, Kreischer KE, Rosay M, Shapiro MA, Sirigiri JR, Temkin RJ, Woskov PP. Operational characteristics of a 14-W 140-GHz gyrotron for dynamic nuclear polarization. IEEE Trans. Plasma Sci. 2006;34:518–523. doi: 10.1109/TPS.2006.875776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Patterson JC, Muller S, Bariar B, Pan T, Chernoff YO. Regulation of Chaperone Effects on a Yeast Prion by Cochaperone Sgt2. Mol Cell Biol. 2012;32:4960–4970. doi: 10.1128/MCB.00875-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TA. CFSSP: Chou and Fasman Secondary Structure Prediction Server. Wide Spectrum. 2013;1:15–19. [Google Scholar]
- Lange S, Linden AH, Akbey Ü, Franks WT, Loening NM, van Rossum B-J, Oschkinat H. Journal of Magnetic Resonance. 2012;216:209–212. doi: 10.1016/j.jmr.2012.01.002. Journal of Magnetic Resonance. [DOI] [PubMed] [Google Scholar]
- Leiting B, Marsilio F, O'Connell JF. Predictable Deuteration of Recombinant Proteins Expressed in Escherichia coli. Anal Biochem. 1998 doi: 10.1006/abio.1998.2904. [DOI] [PubMed] [Google Scholar]
- Linden AH, Franks WT, Akbey Ü, Lange S, van Rossum B-J, Oschkinat H. Cryogenic temperature effects and resolution upon slow cooling of protein preparations in solid state NMR. J Biomol NMR. 2011;51:283–292. doi: 10.1007/s10858-011-9535-z. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci USA 99 Suppl. 2002;4:16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Del Amo J-M, Schneider D, Loquet A, Lange A, Reif B. Cryogenic solid state NMR studies of fibrils of the Alzheimer's disease amyloid-β peptide: perspectives for DNP. J Biomol NMR. 2013 doi: 10.1007/s10858-013-9755-5. [DOI] [PubMed] [Google Scholar]
- Luckgei N, Schütz AK, Bousset L, Habenstein B, Sourigues Y, Gardiennet C, Meier BH, Melki R, Böckmann A. The Conformation of the Prion Domain of Sup35 p in Isolation and in the Full-Length Protein. Angew. Chem. Int. Ed. 2013;52:12741–12744. doi: 10.1002/anie.201304699. [DOI] [PubMed] [Google Scholar]
- Masison DC, Kirkland PA, Sharma D. Influence of Hsp70s and their regulators on yeast prion propagation. Prion. 2009;3:65–73. doi: 10.4161/pri.3.2.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrnejad F, Ghahremanpour MM, Khadem-Maaref M, Doustdar F. Effects of osmolytes on the helical conformation of model peptide: molecular dynamics simulation. J. Chem. Phys. 2011;134:035104. doi: 10.1063/1.3530072. [DOI] [PubMed] [Google Scholar]
- Michaelis VK, Ong T-C, Kiesewetter MK, Frantz DK, Walish JJ, Ravera E, Luchinat C, Swager TM, Griffin RG. Topical Developments in High-Field Dynamic Nuclear Polarization. Isr. J. Chem. 2014;54:207–221. doi: 10.1002/ijch.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J Magn Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Ni QZ, Daviso E, Can TV, Markhasin E, Jawla SK, Swager TM, Temkin RJ, Herzfeld J, Griffin RG. High Frequency Dynamic Nuclear Polarization. Acc Chem Res. 2013;46:1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines A. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569. [Google Scholar]
- Reckel S, Lopez JJ, Löhr F, Glaubitz C, Dötsch V. In-cell solid-state NMR as a tool to study proteins in large complexes. Chembiochem. 2012;13:534–537. doi: 10.1002/cbic.201100721. [DOI] [PubMed] [Google Scholar]
- Renault M, Pawsey S, Bos MP, Koers EJ, Nand D, Tommassen van Boxtel R, Rosay M, Tommassen J, Maas WE, Baldus M. Solid-State NMR Spectroscopy on Cellular Preparations Enhanced by Dynamic Nuclear Polarization. Angew. Chem. Int. Ed. 2012;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- Saibil HR, Seybert A, Habermann A, Winkler J, Eltsov M, Perkovic M, Castaño-Diez D, Scheffer MP, Haselmann U, Chlanda P, et al. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1211976109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Wälchli M, et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;457:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc Natl Acad Sci USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL. Yeast prion [psi +] and its determinant, Sup35p. Meth Enzymol. 1999;309:649–673. doi: 10.1016/s0076-6879(99)09043-6. [DOI] [PubMed] [Google Scholar]
- Slichter CP. Principles of Magnetic Resonance. Springer Science & Business Media; 1990. [Google Scholar]
- Song C, Hu K-N, Joo C-G, Swager TM, Griffin RG. TOTAPOL: a biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media. J Am Chem Soc. 2006;128:11385–11390. doi: 10.1021/ja061284b. [DOI] [PubMed] [Google Scholar]
- Szeverenyi NM, Sullivan MJ, Maciel GE. Observation of spin exchange by two-dimensional fourier transform 13C cross polarization-magic-angle spinning. Journal of Magnetic Resonance (1969) 1982;47:462–475. [Google Scholar]
- Takahashi H, Fernández-de-Alba C, Lee D, Maurel V, Gambarelli S, Bardet M, Hediger S, Barra A-L, De Paëpe G. Optimization of an absolute sensitivity in a glassy matrix during DNP-enhanced multidimensional solid-state NMR experiments. J Magn Reson. 2014;239:91–99. doi: 10.1016/j.jmr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344:631–637. [Google Scholar]
- Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Marchante R, Kushnirov V. Fungal prions: structure, function and propagation. Top Curr Chem. 2011;305:257–298. doi: 10.1007/128_2011_172. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, Mccaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagenende V, Yap MGS, Trout BL. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- Vaiphei ST, Tang Y, Montelione GT, Inouye M. The use of the condensed single protein production system for isotope-labeled outer membrane proteins, OmpA and OmpX in E. coli. Mol. Biotechnol. 2011;47:205–210. doi: 10.1007/s12033-010-9330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann G, Iwaï H. Protein trans-splicing and its use in structural biology: opportunities and limitations. Mol Biosyst. 2010;6:2110–2121. doi: 10.1039/c0mb00034e. [DOI] [PubMed] [Google Scholar]
- Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, Hong M. Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proc Natl Acad Sci USA. 2013;110:16444–16449. doi: 10.1073/pnas.1316290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Schütz A, Loquet A, Buhtz C, Greenwald J, Riek R, Böckmann A, Meier BH. The molecular organization of the fungal prion HET-s in its amyloid form. Journal of Molecular Biology. 2009;394:119–127. doi: 10.1016/j.jmb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Caporini MA, Im S-C, Waskell L, Ramamoorthy A. Biochimica et Biophysica Acta. BBA - Biomembranes. 2014:1–8. doi: 10.1016/j.bbamem.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.