Abstract
In the last years, epigenetic processes have emerged as a promising area of complex diseases research. DNA methylation measured in Long Interspersed Nucleotide Element 1 (LINE-1) sequences has been considered a surrogate marker for global genome methylation. New findings have suggested the potential involvement of epigenetic mechanisms in Type 2 diabetes (T2DM) as a crucial interface between the effects of genetic predisposition and environmental influences. Our study evaluated whether global DNA methylation predicted increased risk from T2DM or other carbohydrate metabolism disorders in a cohort study. We used a prospective cohort intervention study and a control group. We collected phenotypic, anthropometric, biochemical, and nutritional information from all subjects. Global LINE-1 DNA methylation was quantified by pyrosequencing technology. Subjects that did not improve their carbohydrate metabolism status showed lower levels of global LINE-1 DNA methylation (63.9 ± 1.7 vs. 64.7 ± 2.4) and they practiced less intense physical activity (5.8% vs. 21.5%). Logistic regression analyses showed a significant association between LINE-1 DNA methylation and metabolic status after adjustment for sex, age, BMI, and physical activity. Our study showed that lower LINE-1 DNA methylation levels were associated with a higher risk metabolic status worsening, independent of other classic risk factors. This finding highlights the potential role for epigenetic biomarkers as predictors of T2DM risk or other related metabolic disorders.
Keywords: cohort study, diabetes, DNA methylation, LINE-1, peripheral blood cell
Abbreviations
- LINE-1
Long Interspersed Nucleotide Element 1
- T2DM
Type 2 diabetes mellitus
- CVD
cardiovascular diseases
- IFG
impaired fasting glucose
- IGT
impaired glucose tolerance
- BMI
body mass index
Introduction
Epigenetic modifications are heritable changes in gene function that are not caused by variation in DNA sequence.1 In mammals, methylation involves addition of methyl groups to cytosine to form 5-methyl-cytosine (5 mC).2 About one-third of DNA methylation occurs in repetitive elements, representing a large portion of the human genome. Long Interspersed Nucleotide Element 1 (LINE-1) is the most abundant family of non-long terminal repeat retrotransposons found in the genome.3 CpG sites in repetitive elements, like LINEs, are largely methylated in normal somatic tissue suppressing most of their transposition activity.4,5 DNA methylation measured in LINE-1 sequences has been considered a surrogate marker for global genome methylation.6
Although most epigenetic studies have been focused on cancer,7,8, previous studies have suggested that DNA methylation in these LINE-1 elements may play a role in other complex human diseases, such as cardiovascular disease or diabetes.9-12 However, there are still many unresolved issues about the role of epigenetic changes in the study of complex diseases.
LINE-1 methylation is also susceptible to be modified by environmental factors (e.g., diet, exercise, smoking, and air pollution exposure) or DNA sequence variation.13 It is well known that several of these environmental exposures are associated with risk of complex diseases, such as Type 2 diabetes mellitus (T2DM).
Type 2 diabetes mellitus is a metabolic disorder influenced by interactions between environmental and genetic factors. Environmental changes can affect the phenotype directly or through epigenetic mechanisms that provide an interface with the genome.14 New findings suggest the potential involvement of epigenetic mechanisms in T2DM as a crucial interface between the effects of genetic predisposition and environmental influences.15 However, until now, only few studies have documented impaired DNA methylation events in T2DM.16
Recent studies prompt that LINE-1 DNA methylation might be useful in predicting the risk of common complex diseases, such as T2DM and cardiovascular diseases (CVD).17
This study forms part of a wider research project about lifestyle changes to achieve an improved metabolic status. The present study was performed on a population-based cohort including subjects 40 y old and older, suffering carbohydrate metabolism disorders (impaired fasting glucose (IFG), impaired glucose tolerance (IGT), or Type 2 diabetes mellitus (T2DM)), selected from the general population of 2 semi-urban towns of the south of Spain.
Our study evaluated whether global DNA methylation (LINE-1) predicted increased risk from T2DM or other carbohydrate metabolism disorders in a prospective study.
Results
Characteristics of the study subjects
Baseline and follow-up characteristics of the participants are shown according to their glycemic status one year after the intervention. A variable was created with 2 categories: 1) subjects with a carbohydrate metabolism disorder (IFG, IGT, or T2DM) at baseline and who showed an improved status metabolic one year later, and 2) subjects who did not change or worsened their glycemic status after one year.
At baseline, there were no differences between the 2 groups except for physical activity and levels of LINE-1 DNA methylation (Table 1). Subjects that did not improve their carbohydrate metabolism status showed lower levels of global DNA methylation (63.9 ± 1.7 vs. 64.7 ± 2.4) and practiced less intense physical activity (5.8% vs. 21.5%). After the intervention, significant differences were found in BMI, weight, and glucose levels [both fasting and post oral glucose tolerance test (OGTT; Table 1]. There was no difference in the dietary pattern between both groups.
Table 1.
Baseline and follow-up characteristics of the study subjects
| Baseline |
After one year |
|||||
|---|---|---|---|---|---|---|
| Improvement | Same or worsening | P value | Improvement | Same or worsening | P value | |
| Age (years) | 52.18 ± 8.2 | 53.8 ± 7.6 | NS | |||
| Sex (male/female)(%) | 46.2 vs 53.8 | 42.2 vs 57.8 | NS | |||
| Group (control/intervention) | 51.8/48.2 | 48/52 | NS | |||
| Weight (kg) | 79.1 ± 13.6a | 81.7 ± 15.2b | NS | 76.6±12.8a | 80.6 ± 14.7b | 0.03 |
| BMI (Kg/m2) | 30.2 ± 4.7a | 31.3 ± 5.2b | NS | 29.2±4.3a | 30.9 ± 5.2b | 0.01 |
| WHR | 0.94 ± 0.08 | 0.93 ± 0.08 | NS | 0.94±0.08 | 0.94 ± 0.07 | NS |
| Fasting glucose (mmol/l) | 5.92 ± 0.74c | 6.0 ± 1.0d | NS | 5.27±0.6c | 6.08 ± 0.93d | <0.001 |
| Alcohol consumption (%) Every day | 27.4 | 31.1 | NS | |||
| Smoking (%) Yes | 29.2 | 23.8 | NS | |||
| Physical activity (%) | 0.001 | |||||
| Slight | 33.8 | 47.1 | ||||
| Moderate | 44.6 | 47.1 | ||||
| Intense | 21.5 | 5.8 | ||||
| Mediterranean Diet Pattern (%) | 53.1 | 47.4 | NS | |||
| Global methylation (%5mC) | 64.7 ± 2.4 | 63.9 ± 1.7 | 0.003 | |||
aP value before vs. after intervention < 0.001.
bP value before vs. after intervention < 0.001.
cP value before vs. after intervention = 0.001.
dP value before vs. after intervention = NS.
Regarding to subjects’ characteristics and LINE-1 methylation, we only identified a statistical association for gender: levels of LINE-1 DNA methylation were higher in males than females (65.9 ± 1.6 vs. 64.8 ± 2.0; P = 0.01).
Association between global DNA methylation and carbohydrate metabolism disorders
To evaluate the relationship between LINE-1 methylation levels and the risk of metabolic status worsening, a regression logistic analysis was performed. We found that age, sex, and BMI at baseline were not significantly associated with the risk of metabolic status worsening (Table 2). The group to which they belonged (control or intervention) was included in all models as a potential confounding variable. Regression logistic analyses were done including known predictors of T2DM, such as physical activity, smoking, alcohol drinking, fasting glucose, and family history of diabetes mellitus. Only physical activity and LINE-1 DNA methylation levels showed a statistically significant association (OR = 4,7; p = 0.005 and OR = 0,817; p = 0.021, respectively; Table 2). Subjects with low baseline LINE-1 methylation levels exhibited a higher risk of T2DM or other related metabolic disorders (IFG, IGT, or both) during follow-up (Table 2).
Table 2.
Association of LINE-1 methylation with carbohydrate metabolism disorders after unadjusted and adjusted logistic regression models
| OR | IC 95% | P value | |
|---|---|---|---|
| Model 1 | |||
| Age (years) | 1.01 | 0.98–1.05 | NS |
| Sex (male vs. female) | 0.85 | 0.48–1.52 | NS |
| BMI (Kg/m2) | 1.01 | 0.96–1.07 | NS |
| Group (control vs. intervention) | 0.85 | 0.46–1.58 | NS |
| Model 2a | |||
| Physical activity | |||
| Slight vs intense | 4.74 | 1.61–13.90 | 0.005 |
| Moderate vs intense | 3.14 | 1.11–8.88 | 0.031 |
| Model 3b | |||
| LINE-1 methylation levels (%5mC) | 0.81 | 0.68–0.97 | 0.021 |
Dependent variable: metabolic status after one year (0: improvement, 1: same or worsening).
aAdjusted for age, gender, baseline BMI, and group.
bAdjusted for age, gender, group, BMI, and physical activity at baseline.
Finally, to test if there was a trend according to LINE-1 methylation levels, a logistic regression analysis was done using only DNA methylation as an independent variable. Our study showed that participants with lower LINE-1 methylation (below the 25th percentile) had an increased risk of worsening their carbohydrate metabolism versus subjects with values above the 75th percentile (Table 3).
Table 3.
Association of LINE-1 methylation with carbohydrate metabolism disorders after logistic regression analysis using only DNA methylation as an independent variable
| OR | IC 95% | P value | |
|---|---|---|---|
| LINE-1 methylation levels at baseline | 0.06 | ||
| Reference category >65.2 (above the 75th percentile) (%5mC) | 1 | ||
| 63.05–65.21 (%5mC) | 1.76 | 0.92–3.36 | 0.084 |
| <63.05 (below the 25th percentile) (%5mC) | 2.54 | 1.11–5.80 | 0.026 |
Dependent variable: metabolic status after one year (0: improvement;1: same or worsening).
Predictive value for global DNA methylation as risk marker
Finally, our study assessed the predictive value for LINE-1 DNA methylation in T2DM and related metabolic disorders. The areas under the curve (AUCs) for the different risk models were reported. The model based on classic risk factors (age, sex, and BMI) showed an AUC of 0.582 (0.491-0.673) and was not statistically significant (Fig. 1). However, the addition of physical activity and LINE-1 methylation measures improved the predictive capacity from 0.582 to 0.650. Although the improvement was small, it was statistically significant (p = 0.001). Figure 1 shows the AUCs for the 3 models.
Figure 1.
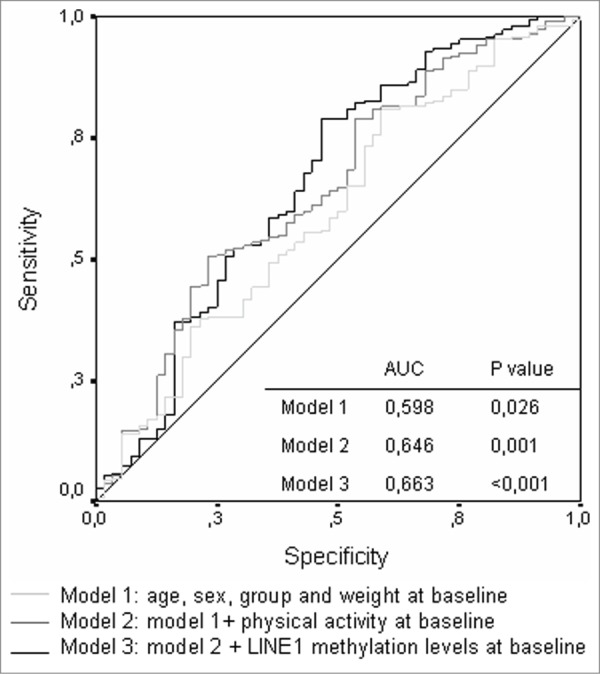
ROC curve for the different risk prediction models.
Discussion
Our study shows that lower LINE-1 DNA methylation is associated with a higher risk of worsening the carbohydrate metabolism in a prospective study. Our data show how global DNA methylation (measured in LINE-1 elements) could be considered as a risk factor for T2DM and related metabolic disorders, independently of other established risk factors. Furthermore, the predictive capacity for this new biomarker was measured.
LINE-1 DNA methylation correlates with global genomic DNA methylation. Hypomethylation in these elements increases their activity as retrotransposable sequences, which may induce genomic alterations and affect gene expression by a number of mechanisms.18,19 Previous studies have suggested that DNA methylation in these types of sequences may play a role in complex diseases other than cancer, such as cardiovascular diseases and diabetes.9,10,20
It is well known that DNA methylation is a dynamic process and it is affected by environmental factors, both from development in uterus (intrauterine development) and in adult life. Several of these environmental exposures are associated with the risk of complex diseases. Therefore, it is reasonable to postulate that global DNA methylation may provide a link between such environmental factors and T2DM.14 There is an increasing interest in the role of these epigenetic modifications as possible biomarkers and for their potential to explain inter-individual variability in phenotype.21
Global DNA methylation and T2DM are affected by common factors, such as age, sex, and diet, among others. Our study evaluated whether LINE-1 DNA methylation could be a biomarker of risk of T2DM or related metabolic disorders independently of classical risk factors.
Growing evidence supports that global DNA methylation is associated with age, although some controversies still remain.22 Several studies have shown a decrease of global DNA methylation (mainly measured in LINE-1 elements) with increasing age.23 However, other authors have not found any association.24 In our study, no relationship was found between both variables, but age was included as a possible confounder in all statistical analysis. Another risk factor related to global DNA methylation is gender. It was observed, as in other studies that women had lower DNA methylation levels than men.24 A few studies have investigated the association between physical activity and DNA methylation. Among them, Zhang et al. reported a trend of higher levels of LINE-1 methylation with higher levels of physical activity.25 Nitert and colleagues also demonstrated that exercise for 6 months is associated with epigenetic changes.26
Our data shows that LINE-1 DNA methylation was associated with the risk of metabolic status worsening. Several models were analyzed including those confounder variables, and it was observed that levels of global DNA methylation at baseline were associated with the risk of developing some metabolic disorders after one year. This association was independent of other classic risk factors, such as age, gender and physical activity. The inclusion of lifestyle variables (smoking habits, diet, and alcohol) did not modify this association; in fact, DNA methylation, together with physical activity, was statistically significant in all models. Our study showed that this association was also independent of the intervention program. When either “group” or “dietary pattern” was added in the models, the association of DNA methylation level with carbohydrate metabolism remained significant.
DNA methylation pattern could be used as a biomarker to identify subjects susceptible to developing complex diseases, such as those presenting obesity or type 2 diabetes mellitus. Recently, Pearce et al. found an association between global LINE-1 DNA methylation and blood glycemic and lipid profiles in a sample of 228 individuals aged 49–51 from the Newcastle Thousand Families Study. Their data suggest that that LINE-1 DNA methylation might be useful in predicting the risk of common complex diseases such as Type 2 diabetes and CVD.17 On the other hand, other authors have also proposed a role for epigenetic modifications as a biomarker of CVD. For example, Baccarelli and colleagues showed that individual differences in repetitive element DNA methylation predicted the risk of developing ischemic heart disease and stroke in elderly men, independently of established risk factors.10 Another study performed by Smulders et al. found that individuals with metabolic syndrome showed relative DNA hypomethylation compared to participants without the syndrome. In the same way, the study showed that people with T2DM or impaired glucose metabolism showed DNA hypomethylation compared to normoglycemic individuals.27 Cash et al., reported that global DNA hypomethylation (measured in LINE-1 elements) was associated with altered levels of LDL and HDL, both risk factors of CVD, in peripheral blood samples from Samoan islanders.28 Most of these studies were cross-sectional and they were not able to determine the causal direction of this relationship.
Although several studies showed a link between epigenetic modifications and diabetes in animal models,29 reported studies of DNA methylation patterns of tissues derived from diabetic patients are limited.14,20 For example, Ling et al. found DNA hypermethylation at the peroxisome proliferator-activated receptor γ coactivator 1α (PPARGC1A) promoter and concomitant transcriptional repression in pancreatic islets isolated from patients with T2DM compared to healthy control subjects.16 Recently, McCarthy et al.30 published a review showing the different DNA methylation candidate genes and epigenome-wide association studies for T2DM. These projects were performed in pancreatic islets and whole blood. Taken together, there is considerable evidence that an altered DNA methylation pattern plays a role in the development of T2DM.30
Our study has shown that LINE-1 DNA methylation could be used to improve the predictive capacity of other risk factors, but these results should be interpreted with caution and replicated in other populations. The area under curve is not significant to consider it a good biomarker, and more studies are needed to confirm the role of global DNA methylation as a possible biomarker of T2DM risk.
One of the main strengths of our study is its prospective nature. Although we cannot determine the causal direction of this finding, the longitudinal design provides valuable information to advance in this exciting field. Additionally, we measured LINE-1 methylation levels, which are considered good markers of global DNA methylation, by pyrosequencing technology, a highly reproducible and accurate method to quantify DNA methylation. Our study also has some limitations. The main weakness is that the differences found are small. Recently, several studies13,31 have evaluated the association between levels of LINE-1 methylation and different characteristics of healthy subjects in blood DNA. The significantly differences found were also within the range of values of our study. Although greater differences in LINE-1 methylation have been shown in people with cancer compared to healthy subjects, studies that focused on common human diseases showed very subtle differences. Another limitation could be the sample size. Currently, very little is known about actual differences in the methylation spectra at epigenetic variants implicated in disease; therefore, there is no consensus about the recommended sample size.32 Further studies in larger populations are needed to confirm these data.
In summary, our study has shown that lower LINE-1 methylation levels were associated with a higher risk of metabolic status worsening, independently of other classic risk factors. Furthermore, we highlight the potential role of DNA methylation as a possible biomarker for the risk of metabolic diseases, such as Type 2 diabetes mellitus.
Material and Methods
Study population and experimental design
A prospective intervention study was undertaken with a control group. The intervention group comprised subjects from Cabra (Cordoba, Spain) and the control group subjects from Pizarra (Malaga, Spain). Both towns are located in the south of Spain, 80 km apart. The study was conducted in 2010, and the first year follow-up ended at the beginning of 2012. A total of 195 subjects completed the baseline study in each group. The cohort was evaluated one year after, and a total of 155 individuals completed the follow-up.
The inclusion age was 40–65 y, and persons were excluded from the study if they were institutionalized for any reason, were pregnant, or had some severe clinical or psychological disorder that impeded their attendance.
Briefly, after OGTT, those subjects with impaired fasting glucose, impaired glucose tolerance or unknown diabetes mellitus were selected in both populations according to the criteria of the World Health Organization (1998).33 Subjects from the control group received standard recommendations on lifestyle changes and subjects in the intervention group were included in a lifestyle program. We collected phenotypic, anthropometric, biochemical, and nutritional information from all the subjects, both at baseline and one year later.
All the participants were informed of the nature of the study and gave their written consent. Likewise, the participants and their family doctors were informed of the most relevant clinical results, whether or not they were abnormal. The study was approved by the Ethics and Clinical Research Committee of Carlos Haya Regional University Hospital, Malaga.
Procedures
Anthropometric and clinical measurements
The study was started at the same time in both groups and the same methodology was used for both the baseline and the follow-up measurements. All the participants underwent an interview and a standardized clinical examination. Standardized measurements were made of weight, height, and body mass index (BMI). Persons were considered to be obese if their BMI was ≥30 kg/m2. A blood sample was taken after overnight fasting and after OGTT, both at baseline and one year after starting the nutritional intervention. Serum was stored at −70°C for later analysis.
Intervention program
The intervention program consisted of 2 levels. Level 1: All the subjects from the control group received general dietary recommendations and guidelines about physical activity. Level 2: The intervention program consisted of a program with regular controls to achieve goals for dietary habits, exercise and weight within the Mediterranean dietary pattern. Subjects from the intervention group underwent this program, whereas the control subjects only underwent Level 1. Subjects from the level 2 programs were monitored at visits every 4 months over the year.
A nutritional evaluation was carried out for all the participants using a 14-item screening questionnaire about adherence to the traditional Mediterranean diet. We used a questionnaire previously validated.34 The surveys were done by experienced dieticians previously trained for this project. We considered as a high adherence to Mediterranean diet pattern a score greater than 10.35 Physical activity was classified as: slight (sit down or stand almost all day without walking), moderate (frequent walking with light loads) or intense (strenuous physical effort).
Classification criteria
Subjects who had IFG or IGT at baseline and had OGTT-N one year later were considered to have improved their carbohydrate metabolism. Otherwise, subjects with T2DM at baseline and who had IFG, IGT, or OGTT-N after the follow-up were also included in the same group.
The rest of the subjects were considered to have maintained or worsened their metabolic status. Those subjects who were taking oral antidiabetic drugs were excluded from the study.
DNA isolation
Venous blood samples were collected at the beginning of the study and one year after starting the nutritional intervention. Genomic DNA was isolated from peripheral blood using the QIAamp DNA Blood kit (Qiagen, Hilden, Germany) in a QIACUBE instrument (Qiagen) according to the manufacturer's recommended protocols. DNA samples were stored at −20°C.
DNA methylation analysis by pyrosequencing
DNA methylation analyses were performed on bisulfite-treated DNA using highly quantitative analysis based on PCR-pyrosequencing. The bisulfite conversion was performed with 500 ng genomic DNA isolated from peripheral blood using the EpiTect bisulfite kit (QIAGEN) as recommended by the manufacturer. The PyroMark™Q96 ID Pyrosequencing System (QIAGEN) was used to determine the methylation status of the CpG island region of the LINE-1 element.
For the methylation analysis of the LINE-1 element we used the primer sequences previously published by Daskalos et al.,36 including 6 CpG sites. Supplementary Table 1 shows the primer sequences for the assay.
Briefly, the PCR was performed in a 25 μl total volume. The final primer concentrations were 0.2 μM. One of the primers was biotinylated in order to purify the final PCR product using sepharose beads. The biotinylated PCR products were purified using the pyrosequencing Vacuum PrepTool (Qiagen). Finally, 15 μl of the PCR products underwent pyrosequencing using the PyroMark™Q96 ID Pyrosequencing System using a 0.4 μΜ sequencing primer.
The degree of methylation was expressed for each DNA locus as the percentage of methylated cytosine (%5 mC) over the sum of methylated and unmethylated cytosines. Non-CpG cytosine residues were used as built-in controls to verify efficient sodium bisulfite DNA conversion. The control peak for the bisulphite conversion passed the quality assessment for all the samples. Correlation between methylation at all 6 CpG sites was high (P < 0.001), therefore the mean for all the sites was expressed as%5 mC. Unmethylated (Jurkat Genomic DNA, cat. N4001S), and methylated DNA (CpG Methylated Jurkat Genomic DNA, cat. N4002S) from New England BioLabs, were also included as controls in each run. We measured LINE-1 methylation in 32 randomly selected duplicate samples and the intra-assay coefficient variation was <1.0%. To calculate inter-assay variability we ran a group of 50 samples in 2 plates on different days, and the coefficient variation was <2.5%.
Statistical analysis
The continuous variables are shown as the mean and standard deviation and the classification variables as proportions. Calculation of the statistical difference between the means of the continuous variables was done by one-way ANOVA and the qualitative variables by the χ2 test. The statistical differences before vs. after the intervention were determined using the Wilcoxon test.
The strength of association between variables was measured by calculating the odds ratio (OR) and 95% confidence intervals by logistic regression. The multivariate logistic regression model was controlled for potential confounders such as age, gender, and BMI at baseline.
To evaluate whether epigenetic markers have some clinical utility and can help to improve the prediction of T2DM, the receiver operator characteristic (ROC) curves 37 was applied on the test set and the area under the curve (AUC).
All analyses were performed using R statistical software, version 2.8.1 (Department of Statistics, University of Auckland, Auckland, NZ; http://www.r-project.org/).38
Supplementary Material
Disclosure of Potential Conflicts of Interest
No conflicts of interest were disclosed.
Funding
This study was supported by the Fondo de Investigación Sanitaria (PI09/2117, PI08/1592), Consejería de Innovación CTS-5125/2009 and II Ayudas SED (Sociedad Española de Diabetes) a Proyectos de Investigación en Diabetes Clínica y Básica.
References
- 1.Bird A. Perceptions of epigenetics. Nature 2007; 447:396-8; PMID:17522671; http://dx.doi.org/ 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- 2.Rakyan VK, Beck S. Epigenetic variation and inheritance in mammals. Curr Opin Genet Dev 2006; 16:573-7; PMID:17005390; http://dx.doi.org/ 10.1016/j.gde.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 3.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 2010; 38:3909-22; PMID:20215437; http://dx.doi.org/ 10.1093/nar/gkq132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol 2002; 14:343-50; PMID:12067657; http://dx.doi.org/ 10.1016/S0955-0674(02)00338-1 [DOI] [PubMed] [Google Scholar]
- 5.Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 2003; 13:651-8; PMID:14638329; http://dx.doi.org/ 10.1016/j.gde.2003.10.013 [DOI] [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33: 6823-36; PMID:16326863; http://dx.doi.org/ 10.1093/nar/gki987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2007; 16:108-14; PMID:17220338; http://dx.doi.org/ 10.1158/1055-9965.EPI-06-0636 [DOI] [PubMed] [Google Scholar]
- 8.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol 2008; 9:359-66; PMID:18339581; http://dx.doi.org/ 10.1016/S1470-2045(08)70038-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 2010; 5:e9692; PMID:20300621; http://dx.doi.org/ 10.1371/journal.pone.0009692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 2010; 21:819-28; PMID:20805753; http://dx.doi.org/ 10.1097/EDE.0b013e3181f20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol 2008; 27:357-65; PMID:18613790; http://dx.doi.org/ 10.1089/dna.2007.0694 [DOI] [PubMed] [Google Scholar]
- 12.Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, Wainstein J, Friedlander Y, Levy-Lahad E, Glaser B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet 2012; 21:371-83; PMID:21994764; http://dx.doi.org/ 10.1093/hmg/ddr472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tajuddin SM, Amaral AF, Fernandez AF, Rodriguez-Rodero S, Rodriguez RM, Moore LE, Tardon A, Carrato A, Garcia-Closas M, Silverman DT, et al. Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect 2013; 121:650-6; PMID:23552396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fradin D, Bougneres P. T2DM: why epigenetics? J Nutr Metab 2011; 2011:647514; PMID:22132323; http://dx.doi.org/ 10.1155/2011/647514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol 2010; 299:F14-25; PMID:20462972; http://dx.doi.org/ 10.1152/ajprenal.00200.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008; 51:615-22; PMID:18270681; http://dx.doi.org/ 10.1007/s00125-007-0916-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce MS, McConnell JC, Potter C, Barrett LM, Parker L, Mathers JC, Relton CL. Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles. Int J Epidemiol 2012; 41:210-7; PMID:22422454; http://dx.doi.org/ 10.1093/ije/dys020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol 2006; 310:211-50; PMID:16909913; (0070-217X (Print). 0070-217X (Linking)) [DOI] [PubMed] [Google Scholar]
- 19.Ostertag EM, Kazazian HH Jr. Biology of mammalian L1 retrotransposons. Annu Rev Genet 2001; 35:501-38; PMID:11700292 [DOI] [PubMed] [Google Scholar]
- 20.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 2009; 58:2718-25; PMID:19940235; http://dx.doi.org/ 10.2337/db09-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turan N, Katari S, Coutifaris C, Sapienza C. Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics 2010; 5:16-9; PMID:20083905; http://dx.doi.org/ 10.4161/epi.5.1.10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langevin SM, Kelsey KT. The fate is not always written in the genes: epigenomics in epidemiologic studies. Environ Mol Mutagen 2013; 54:533-41; PMID:23444110; http://dx.doi.org/ 10.1002/em.21762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA 2008; 299:2877-83; PMID:18577732; http://dx.doi.org/ 10.1001/jama.299.24.2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Maarri O, Walier M, Behne F, van Uum J, Singer H, Diaz-Lacava A, Nusgen N, Niemann B, Watzka M, Reinsberg J, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One 2011; 6:e16252; PMID:21311577; http://dx.doi.org/ 10.1371/journal.pone.0016252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, Vishwanatha JK, Morabia A, Santella RM. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011; 6:293-9; PMID:21178401; http://dx.doi.org/ 10.4161/epi.6.3.14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexeeff SE, Baccarelli AA, Halonen J, Coull BA, Wright RO, Tarantini L, Bollati V, Sparrow D, Vokonas P, Schwartz J. Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes. Int J Epidemiol 2013; 42:270-80; PMID:23508416; http://dx.doi.org/ 10.1093/ije/dys220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luttmer R, Spijkerman AM, Kok RM, Jakobs C, Blom HJ, Serne EH, Dekker JM, Smulders YM. Metabolic syndrome components are associated with DNA hypomethylation. Obes Res Clin Pract 2013; 7:e106-e115; PMID:24331772; http://dx.doi.org/ 10.1016/j.orcp.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 28.Cash HL, McGarvey ST, Houseman EA, Marsit CJ, Hawley NL, Lambert-Messerlian GM, Viali S, Tuitele J, Kelsey KT. Cardiovascular disease risk factors and DNA methylation at the LINE-1 repeat region in peripheral blood from samoan islanders. Epigenetics 2011; 6:1257-64; PMID:21937883; http://dx.doi.org/ 10.4161/epi.6.10.17728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang M, Zhang Y, Liu M, Lan MS, Fei J, Fan W, Gao X, Lu D. Hypermethylation of hepatic glucokinase and L-type pyruvate kinase promoters in high-fat diet-induced obese rats. Endocrinology 2011; 152:1284-9; PMID:21239437; http://dx.doi.org/ 10.1210/en.2010-1162; (1945-7170 (Electronic). 0013-7227 (Linking)) [DOI] [PubMed] [Google Scholar]
- 30.Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther 2012; 92:707-15; PMID:23047653; http://dx.doi.org/ 10.1038/clpt.2012.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2012; 41:126-39; PMID:20846947; http://dx.doi.org/ 10.1093/ije/dyq154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet 2011; 12:529-41; PMID:21747404; http://dx.doi.org/ 10.1038/nrg3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998; 15:539-53; PMID:9686693; http://dx.doi.org/ 10.1002/(SICI)1096-9136(199807)15:7%3c539::AID-DIA668%3e3.0.CO;2-S [DOI] [PubMed] [Google Scholar]
- 34.Schroder H, Fito M, Estruch R, Martinez-Gonzalez MA, Corella D, Salas-Salvado J, Lamuela-Raventos R, Ros E, Salaverria I, Fiol M, et al. A short screener is valid for assessing Mediterranean diet adherence among older Spanish men and women. J Nutr 2011; 141:1140-5; PMID:21508208; http://dx.doi.org/ 10.3945/jn.110.135566 [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Gonzalez MA, Garcia-Arellano A, Toledo E, Salas-Salvado J, Buil-Cosiales P, Corella D, Covas MI, Schroder H, Aros F, Gomez-Gracia E, et al. A 14-item Mediterranean diet assessment tool and obesity indexes among high-risk subjects: the PREDIMED trial. PLoS One 2012; 7:e43134; PMID:22905215; http://dx.doi.org/ 10.1371/journal.pone.0043134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 2009; 124:81-7; PMID:18823011; http://dx.doi.org/ 10.1002/ijc.23849 [DOI] [PubMed] [Google Scholar]
- 37.Pencina MJ, D’Agostino RB Sr, D’Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008; 27:157-72; discussion 207-12; PMID:17569110; http://dx.doi.org/ 10.1002/sim.2929 [DOI] [PubMed] [Google Scholar]
- 38.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2012. http://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


