Abstract
Background: Colorectal cancer (CRC) metastasectomy improves survival, however most patient develop recurrences. Circulating tumor cells (CTCs) are an independent prognostic marker in stage IV CRC. We hypothesized that CTCs can be enriched during metastasectomy applying different isolation techniques. Methods: 25 CRC patients undergoing liver (16 (64%)) or lung (9 (36%)) metastasectomy were prospectively enrolled (clinicaltrial.gov identifier: NCT01722903). Central venous (liver) or radial artery (lung) tumor outflow blood (7.5 ml) was collected at incision, during resection, 30 min after resection, and on postoperative day (POD) 1. CTCs were quantified with 1. EpCAM-based CellSearch® system and 2. size-based isolation with a novel filter device (FMSA). CTCs were immunohistochemically identified using CellSearch®‘s criteria (cytokeratin 8/18/19+, CD45- cells containing a nucleus (DAPI+)). CTCs were also enriched with a centrifugation technique (OncoQuick®). Results: CTC numbers peaked during the resection with the FMSA in contrast to CellSearch® (mean CTC number during resection: FMSA: 22.56 (SEM 7.48) (p = 0.0281), CellSearch®: 0.87 (SEM ± 0.44) (p = 0.3018)). Comparing the 2 techniques, CTC quantity was significantly higher with the FMSA device (range 0–101) than CellSearch® (range 0–9) at each of the 4 time points examined (P < 0.05). Immunofluorescence staining of cultured CTCs revealed that CTCs have a combined epithelial (CK8/18/19) and macrophage (CD45/CD14) phenotype. Conclusions: Blood sampling during CRC metastasis resection is an opportunity to increase CTC capture efficiency. CTC isolation with the FMSA yields more CTCs than the CellSearch® system. Future studies should focus on characterization of single CTCs to identify targets for molecular therapy and immune escape mechanisms of cancer cells.
Keywords: circulating tumor cells, colorectal cancer, liver metastasis, lung metastases, stage IV
Abbreviations
- CTCs
Circulating Tumor Cells
- CRC
Colorectal Cancer
- EpCAM
Epithelial Cell Adhesion Molecule
- FMSA
Flexible Micro Spring Array
Introduction
Colorectal cancer (CRC) is the 2nd leading cause of cancer mortality occurring in both genders in the United States.1 Liver and lung metastases are the most frequent sites of CRC spread, and complete metastasectomy in conjunction with multidisciplinary management leads to a 5-year survival of 30%, but the majority of patients develops a recurrence after surgery.2 The current challenge is to develop objective criteria of selection for surgery and principles of its integration with adjuvant systemic treatments to achieve better outcomes.
The ability to isolate circulating tumor cells (CTCs) is a powerful tool for monitoring cancer patients with minimal morbidity. Detection and characterization of CTCs has potential high-impact implications for prognostication and therapy. Significant improvements have been accomplished to analyze peripheral blood for the presence of CTCs.3 The CellSearch® system by Veridex utilizes several molecular parameters to isolate CTCs: immunomagnetic enrichment for epithelial cell adhesion molecule (EpCAM), nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI), and immunofluorescence detection of cytokeratins (CKs) and CD45.4 Due to its reliability and prognostic impact, the CellSearch® system is the only system approved by the Food and Drug Administration (FDA) for the enumeration of CTCs in metastatic colorectal, prostate, and breast cancers. More than 3 CTCs in 7.5 ml of blood are an independent prognostic marker for stage IV CRC patients.5 However, only a few of them have the potential to eventually initiate metastatic growth.6,7 Investigators have shown that CTCs can be isolated more frequently in the tumor outflow blood than in the peripheral blood during surgery of primary CRC and liver metastases.8-10 Size-based exclusion with the FMSA device has a higher CTC capture efficiency than CellSearch® since it is not limited to the detection of EpCAM expressing cells.11 In most carcinomas, tumor progression implicates a shift toward a mesenchymal phenotype, a process referred to as the epithelial-mesenchymal transition (EMT) and considered to be crucial for metastasis.12-14 In a previous study we were able to isolate CTCs from CRC patients with a mesenchymal phenotype expressing vimentin.15 Fitting into the concept of EMT, tumor cell fusion with white blood cells (‘hybrids’) has been a long-standing theory of metastasis.16 In a previous study we detected large CTCs that were enriched with a simple porous membrane gradient centrifugation device (OncoQuick®). OncoQuick® is based on CTCs having a lighter buoyant density than peripheral blood mononuclear cells and CTCs can be enriched ∼400–500× from whole blood. These enriched and cultured CTCs from CRC patients showed dual-staining for CK/CD45 and also expressed the pan-macrophage marker CD14.17 These hybrids might escape the immune system by inducing immune tolerance.
We hypothesized that perioperative CTC isolation is a unique opportunity to increase CTC yield with innovative detection methods. In a prospective pilot study, 25 patients with resectable CRC liver and lung metastases were recruited for perioperative CTC isolation with different techniques. The study demonstrates that detection of CTCs during CRC metastasectomy is highest during the resection phase, and size-based isolation is much more efficient than CellSearch®. Immunofluorescence staining of replicating CTCs in culture revealed that they have both epithelial (CK8/18/19) and immune cell/macrophage (CD45 and CD14) expression pattern, which could support a hypothesis that certain cancer cells fuse with macrophages. This would allow CTCs to induce T cell immune tolerance.
Materials and Methods
Patient selection
Institutional Review Board (IRB) approval for conductance of this prospective clinical study was obtained at Penn State Hershey Medical Center (IRB No. 29748EP). The trial was registered at ClinicalTrials.gov (Identifier: NCT01722903). Written consent was obtained and patients were enrolled for trial participation in the multidisciplinary CRC cancer outpatient clinics at the Penn State Hershey Cancer Institute. Twenty-five patients were recruited between August 2012 and May 2013. Inclusion criteria for subjects were: older than 18 years, all genders and ethnicities, the diagnosis of stage IV CRC with resectable metastases limited to liver and lungs, histopathology of the CRC primary tumor to be documented as adenocarcinoma. Liver and lung metastases were defined according to radiological criteria or proven by tissue biopsy. Exclusion criteria were as follows: pregnancy, concurrent diagnosis of an active second malignancy besides basal cell carcinoma of the skin if there was evidence of disease burden or if the patient was currently being treated with chemotherapy.
Clinicopathological data
Clinical and pathological data were collected by reviewing electronic records, including primary tumor characteristics (TNM) and carcinoembryonic antigen (CEA) serum levels. Imaging (CT, MRI and PET/CT scans) were reviewed in a multidisciplinary conference (including a radiologist) to determine metastatic organ involvement. Mutational status (KRAS, BRAF, PI3K, NRAS), microsatellite instability (MSI) and EGFR, ERCC1, TS (thymidylate synthetase) expression were determined within the Department of Pathology, or by sending out specimens to Response DX Colon® (Los Angeles, CA), or Quest Diagnostics® (Lyndhurst, NJ).
CTC detection
During metastasectomy blood was sampled for CTC analysis from sites close (central venous line/right atrium) to the liver metastases outflow, and from the lung metastases outflow (radial artery). At each time point 15 ml of blood were drawn directly before, during and after metastasectomy and then immediately split (7.5 ml each) at the bedside into a CellSave® tube (Veridex, Raritan, NJ) for CellSearch® analysis and an EDTA (K2) tube (Becton Dickinson, Vacutainer) for FMSA analysis.
The 7.5 mL blood samples in CellSave® tubes were processed according to the manufacturer's instructions by a technician trained and certified by the manufacturer to operate the FDA-approved CellSearch® instrument. CTC detection was performed with CellSearch® system. Samples were analyzed within 3 d using the standard CellSearch® protocol and the CTC Epithelial Cell Kit. CellSearch® qualifies a cell as a CTC if it has an evident nucleus (by DAPI) and is EpCAM+, CK 8,18/19+, and CD45-. Analysis and enumeration of CTCs was conducted by a blinded, certified assay operator (D.D.). To increase capture quantity CTCs, blood was analyzed using the FMSA device that is a parylene-based filter that operates by size-based exclusion for CTC enrichment. The FMSA device was microfabricated from parylene polymer with alterations to a previously described process18. The device allows isolation of CTCs using the CellSearch® system's definition of CTCs (Pan-CK+/CD45−/DAPI+), however we chose an EpCAM-independent protocol. Blood (7.5 ml) was processed within 24 hours to facilitate optimal filtration conditions. Blood samples were passed through the FMSA device under precisely regulated pressures as described previously11. Filter devices were washed with Dulbecco's phosphate buffered saline (DPBS, Invitrogen) immediately after enrichment and then fixed with 4% paraformaldehyde (VWR) for 20 minutes. Cells were permeabilized in 0.3% Triton X-100 (VWR) for 10 minutes. One μg/mL of DAPI (Invitrogen) was added as a nucleic acid stain and then samples were blocked with 5% goat serum (Sigma-Aldrich). Cells were subsequently incubated with 1 μg/mL mouse monoclonal anti-cytokeratin 8/18/19 (Abcam, clone 2A4) and 10 μg/mL goat anti-mouse IgG conjugated to DyLight 488 (Thermo Scientific). Cells were blocked again with 100 μg/mL mouse IgG solution (Sigma-Aldrich) and incubated with 0.5 μg/mL monoclonal mouse anti-CD45 conjugated to Alexa Fluor 647 (Santa Cruz, clone 35-Z6). All blocking and antibody incubation steps were carried out for 8–12 hours at 4°C.
EpCAM immunostaining of resected metastases tissue
All resected CRC liver and lung metastases were immunohistochemically stained for EpCAM expression. Tissue specimens were fixed in buffered formalin, routinely processed and embedded in paraffin. Sections from representative tumor blocks of all cases were cut at 4-μm thickness, and hematoxylin and eosin stain was performed per routine histology protocol. Antigen retrieval was done with EDTA (pH 8.0), and immunohistochemical staining for EpCAM with a monoclonal mouse antihuman antibody (clone BerEP4) (Dako, Carpinteria, CA, USA) diluted 1:100 was performed on Dako Autostainer Plus® using the streptavidin-biotin-peroxidase system, and the signal was visualized with 3,3′-Diaminobenzidine (DAB) detection kit, applied according to the manufacturer's manual. The staining was visualized with Olympus microscope and the images were captured with Olympus DP26 digital camera (Tokyo, Japan). All slides were examined by a board-certified pathologist (Z. Y.). The staining intensity was graded in a scoring system ranging from 0 to 3 (0: no expression, 3: highest intensity). In addition, the percentages of EpCAM positive cancer cells were also determined.
CTC enrichment with OncoQuick and cell culture
Blood samples were collected and processed using OncoQuick® (Greiner Bio-One, Frickenhausen, Germany) columns as described previously.17 Briefly, blood samples were processed within 24 h after collection using OncoQuick® columns as per manufacturer's recommendations, using 7.5 ml blood + 7.5 ml wash buffer (PBS +0.5% BSA)17. OncoQuick® enrichment is based on the fact that CTCs have a lighter buoyant density than peripheral blood mononuclear cells (PBMCs), so that they remain on top of the liquid (of defined density) used for the separation. Cells were resuspended in RPMI-1640 medium and plated for 2 h or overnight, and the medium was then changed every other day throughout the culturing period (∼4 weeks). In some cases, medium was supplemented with M-CSF (50 ng/ml). For immunofluorescence staining, CTCs were plated in medium in an 8-well coated chamber slide (Lab-Tek II CC2) and incubated overnight at 37°C. Cells were blocked in 2.5% bovine serum albumin in PBS for 1 hour at RT. Primary antibodies (1 1 μ mu; Abs) were diluted to the desired concentrations in the same blocking solution and were incubated for 1–3 hours at RT or in some cases at 4°C overnight in a humidifying chamber. After a washing step with PBS buffer, all steps were performed in the dark. To counterstain nuclei, DAPI was diluted in PBS (1:30,000) and incubated for 5 min at RT in the dark. Coverslips were mounted with ProLong Gold Antifade mounting media and examined using fluorescence microscopy. Antibodies used were as follows: Pan-cytokeratin (pan-KRT) rabbit polyclonal (Santa Cruz, #SC-15367) was used with Alexafluor-labeled donkey anti-rabbit 488 antibody (Invitrogen, #A21206). This pan-CK antibody is broadly reactive with human cytokeratin family members. For detection of the common leukocyte antigen CD-45 (officially known as PTPRC, Gene ID#5788), a rat monoclonal antibody (Santa Cruz, #SC59071) was used with Alexafluor-labeled goat anti-rat 568 antibody (Invitrogen, #A11077). For detection of the monocyte differentiation antigen CD-14 (Gene ID#929) a mouse monoclonal antibody (BD PharMingen, #555396) was applied with Alexafluor-labeled goat anti-mouse 568 antibody (Invitrogen, #A11004).
Statistical analysis
Descriptive statistics were generated to summarize patients' characteristics. The main statistical tests performed were the non-parametric Wilcoxon signed-rank test for matched pairs, Mann-Whitney U test and repeated-measure ANOVA models. Statistical analyses were performed using statistical software SAS version 9.3 (SAS Institute, Cary, NC). Significance statements refer to a P-value of <0.05.
Results
Patients' characteristics
Twenty-five stage IV CRC patients were prospectively recruited in a multidisciplinary CRC clinic at a tertiary care center for intraoperative CTC detection with EpCAM-based CellSearch® and a novel size-based filter isolation technique (FMSA) (ClinicalTrials.gov Identifier: NCT01722903). Sixteen (64%) patients with liver and 9 (36%) with lung metastases were enrolled. Blood was isolated at incision, during resection, 30 min after resection, and on postoperative day 1 (POD 1). 7.5 ml of blood were analyzed with both CTC detection methods. All patients had stage IV CRC as determined by clinical history, radiologic imaging and tissue biopsies. 16/25 (64%) patients had chemotherapy treatments before metastasectomy.
Patients' characteristics are listed in Table 1. The mean age of all patients was 58.5 (SEM ± 2.26), with a median of 57 y (range 40–87). Ten (40%) patients were females and 15 (60%) males. The location of the primary tumors was predominantly rectosigmoid (16 (64%)), followed by the right (cecum and ascending) (7 (28%)) and descending (2 (8%)) colon. Ten (40%) patients presented with synchronous metastases at initial CRC diagnosis, and 15 (60%) had metachronous (>6 months after initial diagnosis) metastatic disease. TNM data of the primary tumors were available in 20/25 (80%) patients, as many patients had treatments for the primary CRC in outside institutions, and reports were not retrievable in 5 (20%) patients. Most CRC patients had initially a pT3 (10 (50%)) or pT4 (5 (25%))), whereas the minority had a pT1 (2 (10%)) or pT2 (3 (15%)) invasion depth. The lymph node status was negative in 8 (40%), and 12 (60%) primary CRC had metastatic local lymph spread associated with the primary colorectal tumor.
Table 1.
Patients’ characteristics
| Total number of patients | 25 |
| • Liver metastasectomy | 16 (64%) |
| • Lung metastasectomy | 9 (36%) |
| Age | |
| • Mean (SEM) | 58.5 (±2.26) |
| • Median (range) | 57 (40-87) |
| Gender | Females: 10 (40%), males: 15 (60%) |
| Location of primary tumor | |
| • rectosigmoid | 16 (64%) |
| • descending colon | 2 (8%) |
| • right colon/cecum | 7 (28%) |
| Synchronous vs. metachronous metastases | 10 (40%) vs. 15 (60%) |
| Neoadjuvant chemotherapy before metastasectomy | 16/25 (64%) |
| pT stage of primary tumor (data available in 20/25 (80%) patients) | |
| • pT1 | 2 (10%) |
| • pT2 | 3 (15%) |
| • pT3 | 10 (50%) |
| • pT4 | 5 (25%) |
| Lymph node status of the primary colorectal tumor N- vs. N+ (data available in 20/25 (80%) patients) | 8 (40%) vs. 12 (60%) |
| CEA serum level (ng/ml) (data available in 24/25 (96%) patients) | |
| • Mean (SEM) | 132.4 (±75.8) |
| • Median (range) | 5.6 (1.0-1437.0) |
| CA19.9 serum level (U/ml) (data available in 7/25 (28%) patients) | |
| • Mean (SEM) | 126.2 (±101.5) |
| • Median (range) | 40.0 (1.0-734.0) |
| Microsatellite instability (MSI-H and MSI-L) | 2/9 (22.2%) |
| KRAS mutation (codon 12 or 13) | 6/16 (37.5%) |
| BRAF mutation (codon 600) | 0/8 (0%) |
| Liver metastasectomy | 16 |
| • Resection | 10 (62.5%) |
| • Radiofrequency ablation (RFA) | 1 (6.3%) |
| • Resection & RFA | 5 (31.2%) |
| Lung metastasectomy | 9 |
| • Resection | 9 (100%) |
| EpCAM protein expression of resected metastases (n) | |
| Staining intensity (score 0-3) | 25 |
| • Mean (SEM) | 2.48 (±0.13) |
| • Median (range) | 3 (1-3) |
| Percentage (%) of positive cancer cells | |
| • Mean (SEM) | 69.6% (±23.89) |
| • Median (range) | 70.0% (20-100%) |
Baseline CEA serum levels at the time of metastases diagnoses were available in 24/25 (96%) patients. The mean CEA serum level at the time of CTC detection was 132.4 ng/ml (SEM ± 75.8), with a median of 5.6 ng/ml (range 1.0–1437.0). CA19.9 serum levels at the time of baseline CTC detection were available in 7/25 (28%). The mean CA19.9 serum level was 126.2 U/ml (SEM ± 101.5), with a median of 40.0 U/ml (range 1.0–734.0). Mutational statuses (KRAS, BRAF) and microsatellite instability (MSI) was also reviewed. MSI (MSI-H and MSI-L) was present in 2/9 (22.2%). Mutational analysis showed that 6/16 (37.5%) tumors had a KRAS mutation (codon 12 or 13), and 0/8 (0%) had a BRAF mutation (codon 600).
Among the 16 patients that underwent liver metastasectomy, 10 (62.5%) underwent resection, 1 (6.3%) underwent radiofrequency ablation (RFA), and 5 (31.2%) simultaneous resection and RFA in the same surgery. All 9 patients with lung metastases underwent resection. Complete resection (R0) of the metastases was confirmed by microscopically negative pathologic margins in all 25 participating patients.
Kinetics of perioperative CTC dissemination with CellSearch® and FMSA
CellSearch® data were available in 92 (92%) out of 100 possible detection time points, 8 (8%) samples were not analyzed (mostly due to clotting) (Table 2). FMSA data were available in 65 (65%) out of 100 possible perioperative blood sample analyses. 35 (35%) were excluded from analysis (mostly due to clotting).
Table 2.
Comparison of CTC numbers quantified with CellSearch® and filter-based CTC isolation (FMSA)
| Incision | During resection | 30 min after resection | Postoperative day 1 | Comparison of timepoints: P value2 | |
|---|---|---|---|---|---|
| CellSearch® | |||||
| Samples analyzed (total 92/100 possible) | 24 | 23 | 22 | 23 | |
| CTCs present (%) | 3 (12.5%) | 6 (26.1%) | 8 (36.4%) | 2 (8.7%) | |
| Mean (SEM) | 0.42 (±0.33) | 0.87 (±0.44) | 0.41 (±0.13) | 0.09 (±0.06) | 0.3018 |
| Median (Range) | 0 (0–8) | 0 (0–9) | 0 (0–2) | 0 (0–1) | |
| Filter-based CTC isolation (FMSA) | |||||
| Samples analyzed (total 65/100 possible) | 16 | 16 | 16 | 17 | |
| CTCs present (%) | 15 (93.4%) | 15 (93.4%) | 11 (68.8%) | 15 (88.2%) | |
| Mean (SEM) | 5.06 (±1.47) | 22.56 (±7.48) | 7.00 (±3.23) | 10.65 (±2.77) | 0.0281 |
| Median (Range) | 3 (0–19) | 8 (0–101) | 4 (0–53) | 6 (0–38) | |
| Comparison of CellSearch® vs. FMSA: P value1 | 0.0002 | 0.0001 | 0.0078 | 0.0001 |
SEM: standard error of the mean; FMSA: flexible micro spring array.
Wilcoxon Signed-rank test for matched pairs (non-parametric).
Repeated-measures ANOVA model.
Central venous (liver) or radial artery (lung) tumor outflow blood (7.5 ml) was collected at incision, during resection, 30 min after resection, and on postoperative day (POD) 1. Before comparing the 2 isolation techniques we analyzed each of them independently looking at CTC numbers at different time points with a non-parametric repeated-measures ANOVA model (Table 2). CTC numbers with CellSearch® peaked during the resection of the metastases, although not reaching level of significance comparing the 4 different timepoints (p = 0.3018) (Fig. 1). At incision (baseline level) CTCs were present with CellSearch® detection in 3/24 (12%) patients with a mean of 0.42 (SEM ± 0.33). Mean CTC numbers during resection were 0.87 (SEM ± 0.44), and after resection CTC mean was 0.41 (SEM ± 0.13). On postoperative day 1, CTC values with CellSearch® were at comparable levels as at incision and during and after resection (mean 0.09 (SEM ± 0.06). With the FMSA device, 15/16 (93.4%) of patients had CTCs present at incision (baseline) which was at a much higher incidence than with CellSearch® (12.5%). FMSA CTC numbers were significantly highest during resection in contrast to before and after (p = 0.0281) (Fig. 1). The mean baseline CTC number with the FMSA was at incision 5.06 (SEM ± 1.47), and significantly higher during resection 22.56 (SEM ± 7.48). After resection the mean was 7.00 (SEM ± 3.23). On postoperative day 1 the CTC mean was 10.65 (SEM ± 2.77). With the FMSA the CTC numbers were not significantly different preoperatively at baseline (incision) and postoperatively (POD 1) (p = 0.2024).
Figure 1.
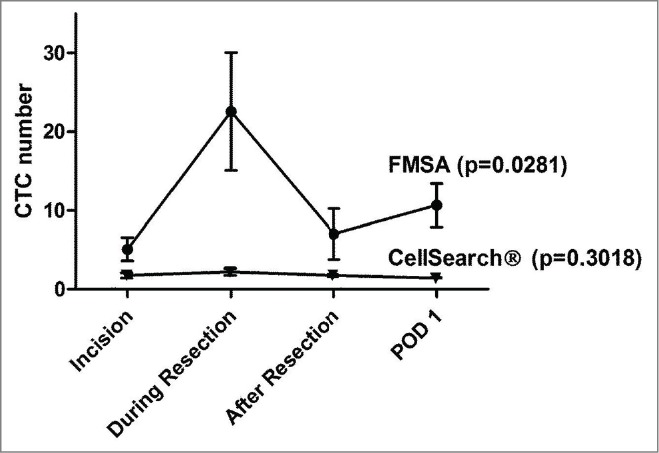
CTC dissemination kinetics during CRC liver and lung metastasectomy. Blood (7.5 ml) was drawn at incision, during resection, 30 min after resection and on postoperative day 1 (POD 1) during CRC liver (n = 16) and lung (n = 9) metastasectomy. CTC isolation and enumeration was performed with FDA-approved CellSearch® and a novel filter isolation technique (FMSA) that enables enrichment by size. CTCs peaked statistically significant during resection with the FMSA device in comparison to the other 3 time points tested with the FMSA. With the FMSA, CTC numbers were not significantly different between incision (baseline) and POD 1 (p = 0.2024, Mann-Whitney U test). However, CTC numbers were not significantly different with the CellSearch® system comparing the 4 time points. Shown are mean CTC values, and bars represent ± standard error of the mean (SEM). P values were calculated with repeated-measures ANOVA model.
Taken together, overall CTC quantity and incidence of detection was significantly higher with the FMSA device in comparison to CellSearch®. CTC numbers peaked during the resection of the metastases in comparison to before (baseline) and after resection with the FMSA in contrast to CellSearch®.
Comparison of CellSearch® with size-based filter isolation (FMSA)
Comparing the 2 techniques CTC overall incidence and quantity was significantly higher with the FMSA device (range 0–101) than CellSearch® (range 0–9) at each of the 4 perioperative timepoints (P < 0.01) (Table 2 and Fig. 2). A matched paired t test (Wilcoxon) was applied to compare CellSearch® and the FMSA CTC numbers (Table 2). At incision, during resection, 30 min after resection and on POD 1 mean baseline CTC numbers were significantly higher with FMSA in comparison to CellSearch® 0 (p = 0.0002, p = 0.0001, p = 0.0078, p = 0.0001 at the 4 time points, respectively). The most striking difference between CellSearch® and the FMSA device was determined during resection, as listed above and in Table 2. The FMSA device consistently detected more CTCs with higher overall incidence at any timepoint analyzed.
Figure 2.
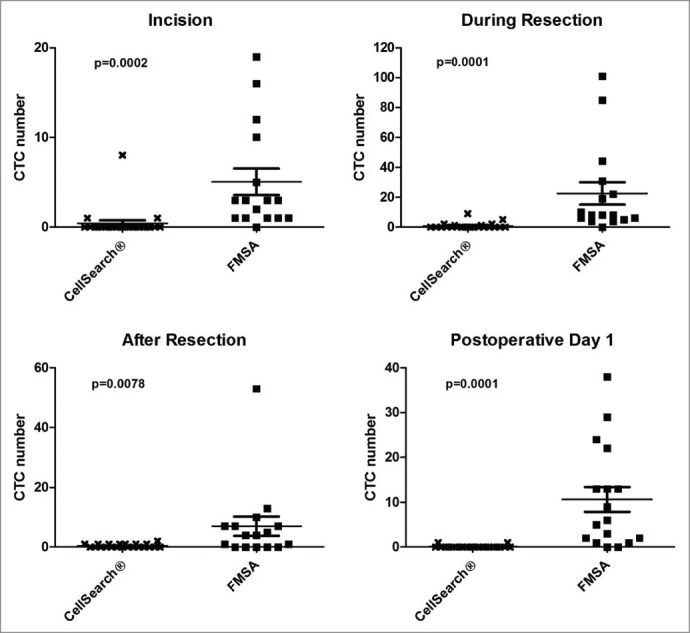
Comparison of CTC isolation with CellSearch® and a novel filter-based isolation technique (FMSA) during CRC metastasectomy. Blood (7.5 ml) was drawn at incision, during resection, 30 min after resection and on postoperative day 1 (POD1). CTCs numbers isolated were significantly higher with the FMSA device at all 4 time points tested, with the most striking difference during the resection phase. Shown are mean CTC values, and bars represent ± standard error of the mean (SEM). P values shown were calculated with Wilcoxon Signed-rank test for matched pairs (non-parametric).
EpCAM protein expression analysis of resected liver and lung metastasis
As the CellSearch® detection system is based on immunomagnetic bead selection of exclusively EpCAM+ CTCs, peroxidase immunostaining of the resected liver and lung metastases with a monoclonal antibody against EpCAM (BerEP4) was performed (Table 1 and Figs. 3–5). EpCAM expression was graded by a staining intensity score (score 0–3). The mean staining intensity score was high with a mean of 2.48 (SEM ± 0.13) and a median of 3 (range 1–3). In addition, the percentage of EpCAM+ cancer cells was determined in the resected liver and lung metastases specimens, and the mean was 69.9% (SEM ± 23.89) and the median was 70% (range 20–100). Patients with no CTCs detectable by CellSearch® had also consistently high EpCAM expression intensity scores (0: no expression, 3: highest intensity) (Fig. 3). Non-parametric ANOVA analysis revealed no statistically significant difference between the maximum CTC numbers and EpCAM intensity score (p = 0.2855). Fig. 4 illustrates 2 examples of stage IV CRC patients that underwent liver and lung metastasectomy that includes EpCAM expression and CTC analysis with CellSearch® and FMSA. In summary, resected liver and lung metastases consistently expressed EpCAM with high intensity and percentages, demonstrating that it was not downregulated.
Figure 3.
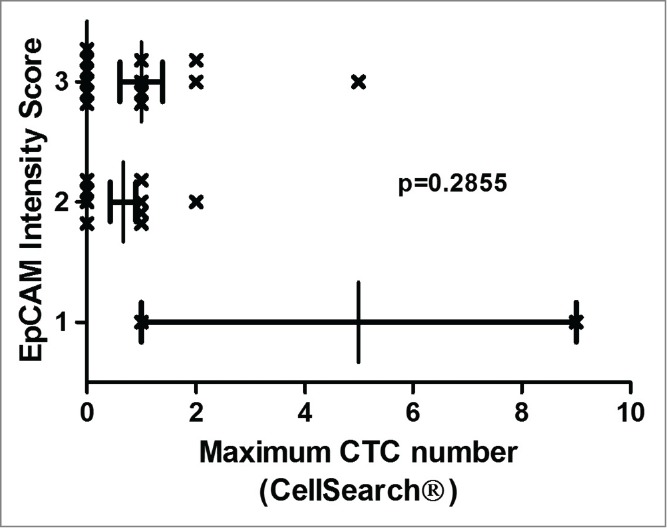
EpCAM expression intensity and CTC numbers. EpCAM expression was analyzed immunohistochemically with monoclonal BerEP4 antibody in resected CRC liver and lung metastases tissue. EpCAM expression was graded by a staining intensity score (score 0–3 (0: no expression, 3: highest intensity)). The mean EpCAM intensity score was high with a mean of 2.48 (SEM ±0.13) and a median of 3 (range 1–3). Patients with no CTCs detectable by CellSearch had consistently high EpCAM expression intensity scores. Non-parametric ANOVA analysis revealed no statistically significant association between the maximum CTC numbers and EpCAM intensity scores (p = 0.2855).
Figure 4.
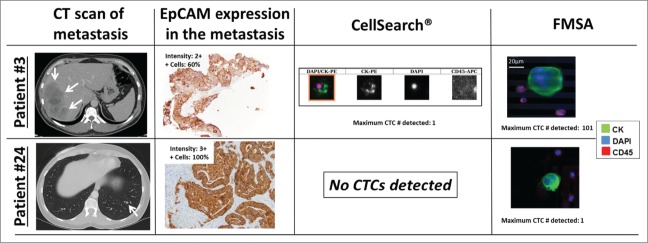
Illustrated are CTCs isolated with CellSearch®, the FMSA, CT scans and EpCAM immunohistochemistry of resected metastases of 2 patients that underwent CRC liver (patients #3) and lung (patient #24) metastasectomy. CTC immunofluorescence staining images from the CellSearch® system and the FMSA, CT scans of liver and lung metastases, and EpCAM immunostaining (peroxidase method) and the respective expression intensity scores (0–3) and percentages of EpCAM+ tumor cells in resected and paraffin-embedded metastases are shown (from left to right column). EpCAM was consistently expressed in CRC liver and lung metastases at high levels, also in patients that had no CTCs detectable by EpCAM-based CellSearch® system (patient #24).
Figure 5.
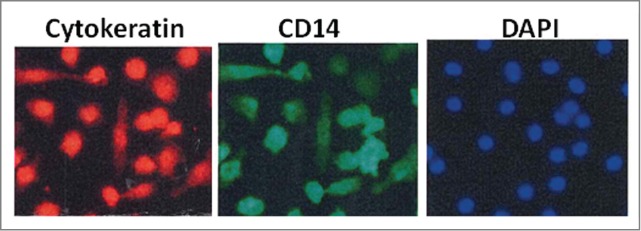
CTCs enriched from a CRC patient undergoing metastasectomy with a simple centrifugation technique (OncoQuick®) were cultured directly. Further staining revealed large CTCs that were pan-cytokeratin+ (left image), DAPI+ (right image) and also expressed macrophage marker CD14 (middle image) in addition to leucocyte marker CD45 (not shown). We hypothesize that CTCs capable to spread and replicate in other organs induce immune tolerance by fusing with tumor-associated macrophages.
Isolation of CTCs with OncoQuick® and phenotype analysis of CTCs
We have previously reported on CRC CK/CD45+ CTCs expressing the pan-macrophage marker CD14 in CRC patients17. Isolation of viable CTCs for culturing was performed with a simple centrifugation technique (OncoQuick®) on samples from 3 trial patients. CTCs were consistently cultured from all 3 CRC patients undergoing metastasectomy over multiple passages. No cells could be cultured from blood from healthy controls individuals. Essentially all of the cells dual-stained for epithelial and immune cell markers throughout the culture period. Immunofluorescence staining after 3 weeks of culturing revealed large CK8/18/19+, DAPI+ CTCs that also expressed macrophage marker CD14 and leucocyte marker CD45 by immunofluorescence (Fig. 5). This finding could support the hypothesis that certain epithelial cancer cells can fuse with CD45/CD14+ macrophages that would allow CTCs to induce T cell immune tolerance.
Discussion
Liver and lungs are the most frequent sites for CRC spread, and complete surgical removal of oligometastatic disease can potentially cure patients with a 5 y survival of over 30%.2,19,20 However, most patients develop cancer recurrence and the current challenge is to develop objective criteria of selection for surgery and establish indications and principles of its integration with adjuvant systemic treatments. CTCs might be a tool to detect occult disease that currently escapes with routine staging methods.
Several studies have demonstrated the clinical significance of CTC numbers as a prognostic marker when enumerated with the CellSearch® system in metastatic CRC, breast and prostate cancer.5,21-24 In general, a major limitation with CTC detection is that CTC numbers are low in the peripheral blood.6,7 The FDA-approved CellSearch® technology does not detect CTCs in many patients with widely metastatic disease, while some of them have exceedingly high CTC numbers in the peripheral blood.24 In recent clinical trials we showed that CTCs correlate with response to chemotherapy in metastatic CRC.25 Furthermore we demonstrated that CTCs can be exceedingly high in patients with high radiologic tumor burden in the liver and CTC quantity also varies among different stage IV CRC patients, and are lowest in patients with metastases limited to the liver or lung in contrast to diffuse spread involving multiple sites.26,27 Hence we hypothesized that intraoperative CTC isolation is a unique opportunity to increase CTC yield in patients with limited metastases that usually have low CTC numbers. Intraoperative CTC isolation could be an exceptional opportunity to isolate more CTCs due to surgical manipulation and access to blood in proximity to the tumor outflow. Other studies with few patients demonstrated that CTCs can be isolated more frequently in tumor outflow than in the peripheral blood during surgery of primary CRC.10,28 A recent trial revealed that CTCs are elevated in the outflow (mesenteric vein) of primary CRC, and comparison with peripheral blood demonstrated lower numbers of CTCs, suggesting that the liver captures CTCs before they enter the peripheral circulation.9 In the present trial the filter effect of the liver was not important as we investigated patients with liver and lung metastases.
Due to CellSearch® system's limited capture efficiency to detect rare CTCs, we chose to compare this established, FDA-approved technology with a novel, innovative size-based filter CTC isolation (FMSA).29 In previous studies we have shown that the FMSA device has a higher CTC capture efficiency than CellSearch®.11,30 Microfiltration allows rapid processing of blood and these devices have been widely tested for cell enrichment.18,31-35 The FMSA allows isolation of CTCs using the CellSearch® system's definition of CTCs (Pan-CK+/CD45-/DAPI+) and gives option to add additional markers and culture CTCs.11,17 In addition, usage is quick, cheap and can be performed at the bedside with no requirement for pre-processing. Rapid enrichment is desirable to minimize disruption to cells and to preserve cell phenotypes. Of note in one of our previous studies the FMSA device detected CTC microclusters consisting of up to 20 cells from clinical blood samples of CRC patients that may be preserved due to the gentle handling and processing of the blood samples.11 Aggregate microclusters of CTCs have been reported in other cancer types though their clinical significance is not fully understood.35-41 Their exact biologic role in tumor spread applying novel technologies has still to be investigated. In our investigation we observed several clotted samples that were not suitable for filter analysis. Although we processed samples quickly within a day, the exact reason remains to be solved. Clotting of blood samples could most likely be due to hypercoagulable states in cancer patients and risk for thrombotic events is particularly high during cancer surgery.42,43 Future studies will have to test other conditions to reduce rates of clotted samples for perioperative CTC analysis.
In most carcinomas, tumor progression is associated a loss of epithelial features and a transition toward a mesenchymal phenotype.12 This process, referred to as the epithelial-mesenchymal transition (EMT), is considered to be crucial for metastasis.13,14 Mesenchymal-like CTCs present in the blood of carcinoma patients are likely to be missed with CellSearch® as CTC detection depends on EpCAM expression. However, CTCs without EpCAM expression may have a crucial role in developing distant metastasis.44,45 CRC patients with metastases limited to the lungs have rarely CTCs detected by CellSearch®, but in the present study the EpCAM-independent CTC FMSA isolation technique detected many more CTCs in these patients even at baseline.27 Independently of CTC detection quantity all resected liver and lung metastases expressed EpCAM at high levels as determined by immunohistochemistry, suggesting that EpCAM downregulation might not have contributed to absence of CTCs by CellSearch® detection technique. Though a subset of CTCs may still down-regulate EpCAM and escape the CellSearch® system, and that could also explain the higher CTC numbers detected with the EpCAM independent FMSA.
Another advantage of filter isolation is that it allows analysis of additional markers and isolation of viable CTCs for culture.11,17 Recently we observed that the majority of FMSA enriched CTCs from CRC stained positively for both epithelial markers (CKs) and the mesenchymal cell marker vimentin (CK+/vimentin+/CD45-).11 Expression of vimentin intermediate filaments and downregulation of epithelial cell markers is implicated in EMT, which is considered a pre-requisite for CTC dissemination.39,46-48 Other molecules involved in cell adhesion, migration and chemotaxis have also been described to have prognostic impact and potential key roles in metastatic dissemination of single cancer cells to the lymph nodes and bone marrow.49,50
Different studies suggested macrophage–tumor cell fusions (´hybrids´) in different cancer types, including CRC.16 Tumor-associated macrophages may fuse with epithelial cancer cells at the site of the primary tumor or metastasis. These hybrids might then induce the EMT transition in some cancer cells, allowing them to escape into the blood and lymphatic system (along with the hybrid cells) and colonize distant organs.16,17 Like other authors we hypothesize that CTCs undergo EMT and acquire the skill to induce immune tolerance. In a previous study we detected large CK/CD45/CD14+ CTCs enriched by an established and simple centrifugation technique (OncoQuick®) and cultured CTCs from melanoma, pancreatic and CRC patients.17 In the present trial were also able to culture CTCs from peripheral blood of CRC patients (but not healthy controls) through multiple passages. Further analysis of viable and replicating CTCs in culture revealed again CK positive CTCs expressing leucocyte marker CD45 and macrophage marker CD14 supporting the hypothesis that certain CTCs fuse with macrophages. This would allow CTCs to induce T cell immune tolerance. A recent study on CTCs in CRC included a gene expression analysis that revealed both a pronounced upregulation of CD47 as a potential immune-escape mechanism and a significant downregulation of several other pathways, suggesting a dormant state of viable CTCs.51 Results suggested mutational heterogeneity between tumor tissue and CTCs and upregulation of immune-escape pathways that may be responsible for survival of CTCs in CRC patients. Further emphasis on analysis of cultured CTCs that are capable to replicate could explain what kind of metastases cause CTC dissemination, and these studies can lead to the detection of novel molecular therapy targets.
This prospective single institution trial on stage IV CRC patients has a potential for selection bias. Patients with stage IV CRC are heterogeneous and have previously received various systemic and surgical treatments. However, our study population had consistently oligometastatic disease that was completely resected. Furthermore, it is a low sample size analysis and patient recruitment needs to be increased in future studies. Finally, no survival analysis was performed as it would not be statistically relevant due to low sample size, but further longitudinal follow-up of more patients will further clarify what spread patterns are associated with presence of certain CTCs. Some patients had low numbers of CTCs detectable, and it could indicate that tumor cell dissemination occurs in a pulsatile, and not in a continuous pattern. However, by determining CTC numbers at multiple time points during surgery, it is unlikely that pulsatile shed CTCs were missed. Intraoperative CTC sampling is a unique setting that is not applicable to all cancer patients, but CTC sampling in direct proximity to the metastases could be easily added in a routine clinical setting, as transjugular hepatic vein (outflow of liver) and radial artery (outflow of lung) blood samplings are both often applied clinical diagnostic procedures.
The efficiency of Phase I actually raises a potential concern, since there are now a number of reports which describe physical translocation of CTCs into the circulation (blood or lymph) during surgical procedures.52-57 In some instances, their forced release appears to have prognostic relevance for patients; for example, hematogenous dissemination of cancer cells during surgery for lung cancer has been reported and was related to clinical prognosis, and similar observations have been reported for hematogenous and lymphatic spread in esophageal, stomach and colorectal cancers.55,56,58-60 In other instances (e. g, pancreatic cancer), the forced dissemination does not affect prognosis.61 In a related vein, there are also reports of direct physical seeding of cancer cells during biopsies.62,63
In general, CTC isolation during CRC liver and lung metastasectomy increases understanding of CTC shedding during surgery. Future studies will need to improve techniques for isolation, culturing and characterization of relevant CTCs to further understand cancer spread, and improve management of patients undergoing cancer-related surgery.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank and our Clinical Program Coordinator Teresa Smink, BSN, OCN, for assisting in identifying the patients, and all participating individuals for their involvement in this study.
Funding
J.T.K. and S.Y. were supported by the Commonwealth Universal Research Enhancement Program (CURE) of the Tobacco Funds of the State of Pennsylvania and an American Cancer Society Pilot Award. W.S.E-D. is an American Cancer Society Research Professor.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60:277-300; PMID:20610543; http://dx.doi.org/ 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- 2.Headrick JR, Miller DL, Nagorney DM, Allen MS, Deschamps C, Trastek VF, Pairolero PC. Surgical treatment of hepatic and pulmonary metastases from colon cancer. Ann Thorac Surg 2001; 71:975-9; discussion 9–80; PMID:11269484; http://dx.doi.org/ 10.1016/S0003-4975(00)02522-4 [DOI] [PubMed] [Google Scholar]
- 3.Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, Rao SB, Eng-Wong J, Seillier-Moiseiwitsch F, Noone AM, Isaacs C. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol 2009; 27:5153-9; PMID:19752342; http://dx.doi.org/ 10.1200/JCO.2008.20.6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes DF, Smerage JB. Circulating tumor cells. Prog Mol Biol Transl Sci 2010; 95:95-112; PMID:21075330; http://dx.doi.org/ 10.1016/B978-0-12-385071-3.00005-8 [DOI] [PubMed] [Google Scholar]
- 5.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al.. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26:3213-21; PMID:18591556; http://dx.doi.org/ 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 6.Lianidou ES, Mavroudis D, Sotiropoulou G, Agelaki S, Pantel K. What's new on circulating tumor cells? A meeting report. Breast Cancer Res 2010; 12:307; PMID:20727231; http://dx.doi.org/ 10.1186/bcr2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011; 192:373-82; PMID:21300848; http://dx.doi.org/ 10.1083/jcb.201010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papavasiliou P, Fisher T, Kuhn J, Nemunaitis J, Lamont J. Circulating tumor cells in patients undergoing surgery for hepatic metastases from colorectal cancer. Proc (Bayl Univ Med Cent) 2010; 23:11-4; PMID:20157496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneve E, Riethdorf S, Ramos J, Nocca D, Coffy A, Daures JP, Maudelonde T, Fabre JM, Pantel K, Alix-Panabieres C. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem 2013; 59:1384-92; PMID:23695297; http://dx.doi.org/ 10.1373/clinchem.2013.202846 [DOI] [PubMed] [Google Scholar]
- 10.Jiao LR, Apostolopoulos C, Jacob J, Szydlo R, Johnson N, Tsim N, Habib NA, Coombes RC, Stebbing J. Unique localization of circulating tumor cells in patients with hepatic metastases. J Clin Oncol 2009; 27:6160-5; PMID:19884529; http://dx.doi.org/ 10.1200/JCO.2009.24.5837 [DOI] [PubMed] [Google Scholar]
- 11.Harouaka RA, Zhou MD, Yeh YT, Khan WJ, Das A, Liu X, Christ CC, Dicker DT, Baney TS, Kaifi JT, et al.. Flexible micro spring array device for high-throughput enrichment of viable circulating tumor cells. Clin Chem 2013; PMID:24132944 [DOI] [PubMed] [Google Scholar]
- 12.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9:265-73; PMID:19262571; http://dx.doi.org/ 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- 13.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 2011; 105:1338-41; PMID:21970878; http://dx.doi.org/ 10.1038/bjc.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al.. EMT and dissemination precede pancreatic tumor formation. Cell 2012; 148:349-61; PMID:22265420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harouaka RC, Zhou MD, Yeh YT, Khan WJ, Das A, Liu X, Christ CC, Dicker DT, Baney TS, Kaifi JT, et al.. Flexible micro spring array device for high throughput enrichment of viable circulating tumor cells. Clin Chem. 2014. Feb;60(2):323–33. doi: 10.1373/clinchem.2013.206805. Epub 2013 Oct 16. PMID:24132944; http://dx.doi.org/ 10.1373/clinchem.2013.206805. [DOI] [PubMed] [Google Scholar]
- 16.Clawson GA. Cancer. Fusion for moving. Science 2013; 342:699-700; PMID:24202164; http://dx.doi.org/ 10.1126/science.1244270 [DOI] [PubMed] [Google Scholar]
- 17.Clawson GA, Kimchi E, Patrick SD, Xin P, Harouaka R, Zheng S, Berg A, Schell T, Staveley-O'Carroll KF, Neves RI, et al.. Circulating tumor cells in melanoma patients. PloS One 2012; 7:e41052; PMID:22829910; http://dx.doi.org/ 10.1371/journal.pone.0041052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng S, Lin HK, Liu J-Q, Balic M, Datar R, Cote RJ, Tai Y-C. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A 2007; 1162:154-61; PMID:17561026; http://dx.doi.org/ 10.1016/j.chroma.2007.05.064 [DOI] [PubMed] [Google Scholar]
- 19.Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J, Ganiats T, Levin T, Woolf S, Johnson D, et al.. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 2003; 124:544-60; PMID:12557158; http://dx.doi.org/ 10.1053/gast.2003.50044 [DOI] [PubMed] [Google Scholar]
- 20.Kaifi JT, Gusani NJ, Deshaies I, Kimchi ET, Reed MF, Mahraj RP, Staveley-O'Carroll KF. Indications and approach to surgical resection of lung metastases. J Surg Oncol 2010; 102:187-95; PMID:20648593; http://dx.doi.org/ 10.1002/jso.21596 [DOI] [PubMed] [Google Scholar]
- 21.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al.. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004; 351:781-91; PMID:15317891; http://dx.doi.org/ 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 22.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008; 14:6302-9; PMID:18829513; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 23.Groot Koerkamp B, Rahbari NN, Buchler MW, Koch M, Weitz J. Circulating tumor cells and prognosis of patients with resectable colorectal liver metastases or widespread metastatic colorectal cancer: a meta-analysis. Ann Surg Oncol 2013; 20:2156-65; PMID:23456317 [DOI] [PubMed] [Google Scholar]
- 24.Joosse SA, Pantel K. Biologic challenges in the detection of circulating tumor cells. Cancer Res 2013; 73:8-11; PMID:23271724; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3422 [DOI] [PubMed] [Google Scholar]
- 25.Das A, Kunkel M, Dicker DT, Joudeh J, Scicchitano A, Allen JE, Sarwani NE, Yang Z, Kaifi JT, Zhu J, et al.. Clinico-pathological significance of serial measurement of circulating tumor cells in 24 metastatic colorectal cancer patients treated with combination chemotherapy. Cancer Biol Ther 2015; In press; PMID:25785486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaifi JT, Kunkel M, Dicker DT, Joude J, Allen J, Zhu J, Yang Z, Sarwani NE, Staveley-O'Carroll KF, El-Deiry ES. Circulating tumor cell levels are elevated in colorectal cancer patients with high tumor burden in the liver. Cancer Biol Ther 2015; [Epub ahead of print]; PMID:25785486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaifi JT, Kunkel M, Zhu J, Dicker DT, Gusani NJ, Yang Z, Sarwani NE, Li G, Kimchi ET, Staveley-O'Carroll KF, et al.. Circulating tumor cells are associated with diffuse spread in stage IV colorectal cancer patients. Cancer Biol Ther 2013; 14:1174-81; PMID:24153154; http://dx.doi.org/ 10.4161/cbt.26884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahbari NN, Bork U, Kircher A, Nimitz T, Scholch S, Kahlert C, Schmidt T, Steinert G, Ulrich AB, Reissfelder C, et al.. Compartmental differences of circulating tumor cells in colorectal cancer. Ann Surg Oncol 2012; 19:2195-202; PMID:22230943; http://dx.doi.org/ 10.1245/s10434-011-2178-1 [DOI] [PubMed] [Google Scholar]
- 29.Williams A, Balic M, Datar R, Cote R. Size-based enrichment technologies for CTC detection and characterization. Recent Results Cancer Res 2012; 195:87-95; PMID:22527497; http://dx.doi.org/ 10.1007/978-3-642-28160-0_8 [DOI] [PubMed] [Google Scholar]
- 30.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, et al.. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res 2010; 16(20):5011-8; PMID:20876796; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleischer RL, Price PB, Symes EM. Novel filter for biological materials. Science 1964; 143:249-50; PMID:17753151; http://dx.doi.org/ 10.1126/science.143.3603.249 [DOI] [PubMed] [Google Scholar]
- 32.Seal SH. A sieve for the isolation of cancer cells and other large cells from the blood. Cancer 1964; 17:637-42; PMID:14159810; http://dx.doi.org/ 10.1002/1097-0142(196405)17:5%3c637::AID-CNCR2820170512%3e3.0.CO;2-I [DOI] [PubMed] [Google Scholar]
- 33.Fleischer R, Alter H, Furman S, Price P, Walker R. Particle track etching diverse technological uses range from virus identification to uranium exploration. Science 1972; 178:255-63; PMID:5078248; http://dx.doi.org/ 10.1126/science.178.4058.255 [DOI] [PubMed] [Google Scholar]
- 34.Vona G, Estepa L, Beroud C, Damotte D, Capron F, Nalpas B, Mineur A, Franco D, Lacour B, Pol S, et al.. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology 2004; 39:792-7; PMID:14999698; http://dx.doi.org/ 10.1002/hep.20091 [DOI] [PubMed] [Google Scholar]
- 35.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, et al.. Isolation by size of epithelial tumor cells – a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol 2000; 156:57-63; PMID:10623654; http://dx.doi.org/ 10.1016/S0002-9440(10)64706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, et al.. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol 2012; 9:016003; PMID:22306768; http://dx.doi.org/ 10.1088/1478-3975/9/1/016003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci 2010; 107:18392-7; http://dx.doi.org/ 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS, Lim CT. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 2013; 3:1259; PMID:23405273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013; 339:580-4; PMID:23372014; http://dx.doi.org/ 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur J Cancer 1973; 9:223-7; PMID:4787857; http://dx.doi.org/ 10.1016/S0014-2964(73)80022-2 [DOI] [PubMed] [Google Scholar]
- 41.Liotta LA, Kleinerman J, Saldel GM. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res 1976; 36:889-94; PMID:1253177 [PubMed] [Google Scholar]
- 42.Lip GY, Chin BS, Blann AD. Cancer and the prothrombotic state. Lancet Oncol 2002; 3:27-34; PMID:11908507; http://dx.doi.org/ 10.1016/S1470-2045(01)00619-2 [DOI] [PubMed] [Google Scholar]
- 43.De Martino RR, Goodney PP, Spangler EL, Wallaert JB, Corriere MA, Rzucidlo EM, Walsh DB, Stone DH. Variation in thromboembolic complications among patients undergoing commonly performed cancer operations. J Vasc Surg 2012; 55:1035-40 e4; PMID:22409858; http://dx.doi.org/ 10.1016/j.jvs.2011.10.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med 2013; 5:180ra48; PMID:23576814; http://dx.doi.org/ 10.1126/scitranslmed.3005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mego M, De Giorgi U, Dawood S, Wang X, Valero V, Andreopoulou E, Handy B, Ueno NT, Reuben JM, Cristofanilli M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int J Cancer 2011; 129:417-23; PMID:20857493; http://dx.doi.org/ 10.1002/ijc.25690 [DOI] [PubMed] [Google Scholar]
- 46.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420-8; PMID:19487818; http://dx.doi.org/ 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, Garcia-Blanco MA. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 2011; 9:997-1007; PMID:21665936; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria J, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 2011; 105:1338-41; PMID:21970878; http://dx.doi.org/ 10.1038/bjc.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaifi JT, Reichelt U, Quaas A, Schurr PG, Wachowiak R, Yekebas EF, Strate T, Schneider C, Pantel K, Schachner M, et al.. L1 is associated with micrometastatic spread and poor outcome in colorectal cancer. Mod Pathol 2007; 20:1183-90; PMID:17873897; http://dx.doi.org/ 10.1038/modpathol.3800955 [DOI] [PubMed] [Google Scholar]
- 50.Kaifi JT, Yekebas EF, Schurr P, Obonyo D, Wachowiak R, Busch P, Heinecke A, Pantel K, Izbicki JR. Tumor-cell homing to lymph nodes and bone marrow and CXCR4 expression in esophageal cancer. J Natl Cancer Inst 2005; 97:1840-7; PMID:16368946; http://dx.doi.org/ 10.1093/jnci/dji431 [DOI] [PubMed] [Google Scholar]
- 51.Steinert G, Scholch S, Niemietz T, Iwata N, Garcia SA, Behrens B, Voigt A, Kloor M, Benner A, Bork U, et al.. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res 2014; 74:1694-704; PMID:24599131; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-1885 [DOI] [PubMed] [Google Scholar]
- 52.Camara O, Kavallaris A, Noschel H, Rengsberger M, Jorke C, Pachmann K. Seeding of epithelial cells into circulation during surgery for breast cancer: the fate of malignant and benign mobilized cells. World J Surg Oncol 2006; 4:67; PMID:17002789; http://dx.doi.org/ 10.1186/1477-7819-4-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camara O, Kavallaris A, Noschel H, Rengsberger M, Jorke C, Pachmann K. Seeding of epithelial cells into circulation during surgery for breast cancer: the fate of malignant and benign mobilized cells. World J Surg Oncol 2006; 4:67-75; PMID:17002789; http://dx.doi.org/ 10.1186/1477-7819-4-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawabata N, Okumura M, Utsumi T, Inoue M, Shiono H, Minami M, Nishida T, Sawa Y. Circulating tumor cells in peripheral blood caused by surgical manipulation of non-small-cell lung cancer: pilot study using an immunocytology method. Gen Thorac Cardiovasc Surg 2007; 55:189-92; PMID:17554991; http://dx.doi.org/ 10.1007/s11748-007-0101-2 [DOI] [PubMed] [Google Scholar]
- 55.Ge MJ, Shi D, Wu OC, Wang M, Li LB. Observation of circulating tumour cells in patients with non-small cell lung cancer by real-time fluorescent quantitative reverse transcriptase-polymerase chain reaction in perioperative period. J Cancer Res Clin Oncol 2006; 132:248-56; PMID:16320073; http://dx.doi.org/ 10.1007/s00432-005-0059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong Q, Huang J, Zhou Y, Li L, Bao G, Feng J, Sha H. Hematogenous dissemination of lung cancer cells during surgery: quantitative deetection by flow cytometry and prognostic significance. Lung Cancer 2002; 37:293-301; PMID:12234699; http://dx.doi.org/ 10.1016/S0169-5002(02)00102-2 [DOI] [PubMed] [Google Scholar]
- 57.Engilbertsson H, Aaltonen KE, Bjornsson S, Kristmundsson T, Patschan O, Ryden L, Gudjonsson S. TURBT can cause seeding of cancer cells into the bloodstream. J Urol 2014; Epub July 1; PMID:24996129 [DOI] [PubMed] [Google Scholar]
- 58.Broll R, Lembcke K, Stock C, Zingler M, Duchrow M, Schimmelpenning H, Strik M, Muller G, Kujath P, Bruch HP. Tumor cell dissemination in bone marrow and peritoneal cavity. An immunocytochemical study of patients with stomach or colorectal carcinoma (German). Langenbecks Arch Chir 1996; 381:51-8; PMID:8717176; http://dx.doi.org/ 10.1007/BF00184256 [DOI] [PubMed] [Google Scholar]
- 59.Guller U, Zajac P, Schnider A, Bosch B, Vorburger S, Zuber M, Spagnoli GC, Oertli D, Maurer R, Metzger U, et al.. Disseminated single tumor cells as detected by real-time quantitative plymerase chain reaction represent a prognostic factor in patients undergoing surgery for colorectal cancer. Ann Surg 2002; 236:768-75; PMID:12454515; http://dx.doi.org/ 10.1097/00000658-200212000-00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Z, Jiang M, Zhao J, Ju H. Circulating tumor cells in perioperative esophageal cancer patients: quantitative assay system and potential clinical utility. Clin Cancer Res 2007; 13:2992-7; PMID:17505001; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-2072 [DOI] [PubMed] [Google Scholar]
- 61.Sergeant G, Roskams T, van Pelt J, Houtmeyers F, Aerts R, Topal B. Perioperative cancer cell dissemination detected with a real-time RT-PCR assay for EpCAM is not associated with worse prognosis in pancreatic ductal adenocarcinoma. BMC Cancer 2011; 11:47; PMID:21281486; http://dx.doi.org/ 10.1186/1471-2407-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishizuna K, Ota D, Okamoto J, Fukuuchi A, Tanaka R, Fujii A, Mori M, Nishi T. A case of mucinous carcinoma of the breast in which needle tract seeding was diagnosed by preoperative diagnostic imaging. Breast Cancer 2011; 18:324-7; PMID:19701680; http://dx.doi.org/ 10.1007/s12282-009-0151-7 [DOI] [PubMed] [Google Scholar]
- 63.Shyamala K, Girish HC, Murgod S. Risk of tumor cell seeding through biopsy and aspiration cytology. J Int Soc Prev Community Dent 2014; 4:5-11; PMID:24818087; http://dx.doi.org/ 10.4103/2231-0762.129446 [DOI] [PMC free article] [PubMed] [Google Scholar]


