Abstract
Epigenetic changes play significant roles in cancer development. UHRF1, an epigenetic regulator, has been shown to be overexpressed and to coordinate tumor suppressor gene (TSG) silencing in several cancers. In a previous study, we found that UHRF1 promoted gastric cancer (GC) invasion and metastasis. However, the role and underlying mechanism of UHRF1 in GC carcinogenesis remain largely unknown. In the present study, we investigated UHRF1 expression and function in GC proliferation and explored its downstream regulatory mechanism. The results demonstrated that UHRF1 overexpression was an independent and significant predictor of GC prognosis. Downregulation of UHRF1 suppressed GC proliferation and growth in vitro and in vivo, and UHRF1 upregulation showed opposite effects. Furthermore, downregulation of UHRF1 reactivated 7 TSGs, including CDX2, CDKN2A, RUNX3, FOXO4, PPARG, BRCA1 and PML, via promoter demethylation. These results provide insight into the GC proliferation process, and suggest that targeting UHRF1 represents a new therapeutic approach to block GC development.
Keywords: DNA methylation, gastric cancer, proliferation, tumor suppressor gene, UHRF1
Abbreviations
- BRCA
breast cancer
- CDX2
caudal type homeobox 2
- CDH4
cadherin 4
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- DNMT
DNA methyltransferase
- FOXO
forkhead box O
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GC
gastric cancer
- GO
gene ontology
- mRNA
messenger RNA
- MSP
methylation-specific PCR
- NC
negative control
- PBS
phosphate buffered saline
- PI
propidium iodide
- PLA
Chinese People's Liberation Army
- PML
promyelocytic leukemia
- PPARG
peroxisome proliferator-activated receptor gamma
- qRT-PCR
quantitative reverse transcription–polymerase chain reaction
- RB
retinoblastoma protein
- RUNX3
runt-related transcription factor 3
- shRNA
short hairpin RNA
- TSG
tumor suppressor gene
- UHRF1
ubiquitin-like containing PHD ring finger 1
Introduction
Gastric cancer (GC) is one of the most common malignancies worldwide. With approximately 723,100 deaths annually, GC is the second most common cause of cancer-related death1. High incidence rates of GC are found in Eastern Asian countries, particularly China and Japan, while much lower in Western Europe and the Unites States.1,2 Despite the improved prognosis of patients with GC resulting from earlier diagnosis, radical surgery, and the development of adjuvant therapy, the 5-year survival rate across all stages remains around 20% poorly.2 Therefore, understanding the molecular pathology of GC is critical for establishing novel therapeutic and diagnostic strategies against this fatal disease.
The epigenetic regulator UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1), also known as inverted CCAAT box-binding protein of 90 kDa (ICBP90), is a newly discovered gene reported to have a function in maintaining DNA methylation by helping recruit DNA methyltransferase 1 (DNMT1) to hemimethylated DNA.3,4 In recent years, many investigations have focused on UHRF1 functions in various physiological and pathological processes. UHRF1 is upregulated in multiple types of cancers, including breast,5 lung,6,7 colorectal,8,9 prostate,10 bladder,11,12 and liver cancer,13 and a consensus has been reached that UHRF1 may be an important biomarker for cancer diagnosis and prognosis. Moreover, emerging evidence has indicated that overexpression of UHRF1 is involved in tumorigenesis and cancer progression.14,15 UHRF1 increases the G1/S transition as the target of E2F transcription factor,16 and upregulation of UHRF1 promotes cell cycle progression, cell proliferation and cell migration.8-10,12,13 Our previous study demonstrated that UHRF1 is upregulated in GC tissues and that forced UHRF1 overexpression could promote GC invasion and metastasis.17 However, the function and mechanism of UHRF1 in GC proliferation and carcinogenesis remain largely unknown.
In this study, we investigated the potential involvement of UHRF1 in GC carcinogenesis. We examined the expression levels of UHRF1 in human GC tissues and cells and tested its effects on cell growth, cell-cycle distribution, and colony formation. In addition, we also investigated a potential role of UHRF1 in GC tumorigenesis in a murine model. Finally, we explored the underlying mechanism of UHRF1 functions in GC. Our study will provide a better understanding of GC pathogenesis.
Results
UHRF1 is significantly overexpressed in human GC tissues and cell lines, and indicates poor prognosis
In our previous study, we demonstrated that UHRF1 has higher expression levels in GC tissues than in adjacent nontumorous tissues from 106 GC patients, and correlates with a number of clinicopathological parameters including late stage, lymph node metastases, and distant metastasis to other organs.17 In the present study, to further explore the prognostic value of UHRF1 in GC, we assessed the association between UHRF1 expression and survival in tissue samples from 238 GC patients by immunohistochemistry (Table S3). Consistent with previous results,17 overexpression of UHRF1 in GC tissues was significantly related to depth of invasion (T3-T4), late TNM stage (stages III–IV), poor differentiation, lymph node metastasis, and distant tumor metastasis, while not related to gender, age, location or tumor size (Table 1). According to UHRF1 expression levels, the 3-year cumulative survival rates were 64% (–), 45% (+), 28% (++) and 11% (+++), respectively, and 5-year rates were 38% (–), 30% (+), 13% (++) and 6% (+++), respectively. The mean survival time for patients with different expression levels of UHRF1 were 52.26 (–), 43.37 (+), 28.33 (++) and 20.71 (+++) months. Univariate and Kaplan-Meier analyses clearly showed that GC patients with higher expression of UHRF1 had a poorer prognosis than those with lower expression (Table 2; Fig. 1A). Moreover, UHRF1 expression appeared to be more closely associated with GC prognosis in Stage I–II than Stage III–IV (Fig. 1B and C). To further evaluate the predictive roles of UHRF1 in GC prognosis, the expression and the clinicopathological parameters found to be significant by univariate analysis were included in the multivariate analysis. The covariates included in the Cox regression model were age, tumor size, depth of invasion, differentiation, TNM stage, lymph node metastasis, distant metastasis, and UHRF1 expression. After stepwise multivariate survival analysis, age, depth of invasion, lymph node metastasis, distant metastasis, and UHRF1 expression were found to be independent prognostic factors for patients with GC (Table 2). Thus, these findings confirm that UHRF1 is overexpressed in GC tissues and that UHRF1 expression is an independent and significant predictor of GC prognosis.
Table 3.
Proliferation related genes upregulated after UHRF1 downregulation in MKN45 cells.
| Accession | Gene name | Full name | value in MKN45-NC | value in MKN45-shUHRF1 |
|---|---|---|---|---|
| NM_001265 | CDX2 | caudal type homeobox 2 | 0.6994 | 0.2876 |
| NM_000077 | CDKN2A(p16 INK4) | cyclin-dependent kinase inhibitor 2A | 0.1387 | 0.0643 |
| NM_001031680 | RUNX3 | runt-related transcription factor 3 | 0.2603 | 0.1421 |
| NM_00117093 | FOXO4 | forkhead box O 4 | 0.1216 | 0.0705 |
| NM_005037 | PPARG(PPARγ) | peroxisome proliferator-activated receptor gamma | 0.0178 | 0.0114 |
| NM_007294 | BRCA1 | breast cancer 1, early onset | 0.1166 | 0.0761 |
| NM_002675 | PML | promyelocytic leukemia | 0.8048 | 0.6693 |
Table 1.
Association of UHRF1 expression in the tumor tissues with demographic and clinicopathologic characteristics in 238 patients with GC
| Category | n | Expression of UHRF1 (n) |
P valuea | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| 238 | 42 | 83 | 78 | 35 | ||
| Gender | 0.475 | |||||
| Male | 151 | 24 | 58 | 48 | 21 | |
| Female | 87 | 18 | 25 | 30 | 14 | |
| Age | 0.096 | |||||
| < 60 | 131 | 23 | 54 | 39 | 15 | |
| ≥ 60 | 107 | 19 | 29 | 39 | 20 | |
| Location of tumor | 0.345 | |||||
| Cardia | 13 | 4 | 4 | 2 | 3 | |
| Body | 77 | 11 | 22 | 30 | 13 | |
| Antrum | 148 | 27 | 57 | 46 | 19 | |
| Tumor size, cm | 0.134 | |||||
| <5 | 139 | 30 | 51 | 40 | 18 | |
| ≥5 | 99 | 12 | 32 | 38 | 17 | |
| Depth of invasion | 0.016 | |||||
| T1-T2 | 40 | 11 | 19 | 8 | 2 | |
| T3-T4 | 198 | 31 | 64 | 70 | 33 | |
| Stages | 0.001 | |||||
| I–II | 76 | 20 | 34 | 16 | 6 | |
| III–IV | 162 | 22 | 49 | 62 | 29 | |
| Differentiation | 0.049 | |||||
| Well and moderate | 121 | 24 | 50 | 34 | 13 | |
| Poorly and not | 117 | 18 | 33 | 44 | 22 | |
| Lymph node metastases | 0.013 | |||||
| 0 | 43 | 10 | 22 | 9 | 2 | |
| ≥ 1 | 195 | 32 | 61 | 69 | 33 | |
| Metastases to other organs | 0.001 | |||||
| Present | 15 | 0 | 1 | 8 | 6 | |
| Absent | 223 | 42 | 82 | 70 | 29 | |
P values were calculated through the Kruskal–Wallis H-test and the Mann–Whitney U test to analyze the relationship between UHRF1 expression and clinicopathologic characteristics.
Table 2.
Univariate Analysis and Multivariate analysis of the Correlation Between Clinicopathological Parameters and Survival of Patients With Gastric Cancer
| Clinicopathological Parameters | Cumulative Survival Rates, % |
Mean Survival Time, mo | Univariate Analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|---|---|---|
| 3-Year | 5-Year | HR | 95% CI | P | HR | 95% CI | P | ||
| Gender | |||||||||
| Male | 38 | 22 | 37.49 | 1.000 | reference | ||||
| Female | 37 | 23 | 35.28 | 1.106 | 0.836–1.463 | 0.479 | |||
| Age | |||||||||
| < 60 | 50 | 29 | 45.15 | 1.000 | reference | 1.000 | reference | ||
| ≥ 60 | 23 | 14 | 26.32 | 1.91 | 1.458–2.523 | < 0.001 | 1.791 | 1.327–2.417 | < 0.001 |
| Location of tumor | |||||||||
| Cardia | 69 | 15 | 48.23 | 1.000 | reference | 1.000 | reference | ||
| Body | 26 | 13 | 26.53 | 1.949 | 1.032–3.680 | < 0.05 | 1.665 | 0.847–3.275 | 0.140 |
| Antrum | 41 | 28 | 40.95 | 1.189 | 0.640–2.207 | 0.584 | 1.185 | 0.617–2.277 | 0.610 |
| Tumor size, cm | |||||||||
| < 5 | 43 | 27 | 40.70 | 1.000 | reference | 1.000 | reference | ||
| ≥ 5 | 30 | 16 | 31.04 | 1.382 | 1.049–1.820 | < 0.05 | 0.821 | 0.606–1.111 | 0.202 |
| Depth of invasion | |||||||||
| T1-T2 | 80 | 65 | 70.03 | 1.000 | reference | 1.000 | reference | ||
| T3-T4 | 29 | 14 | 29.94 | 3.824 | 2.438–5.998 | < 0.001 | 1.803 | 1.100–2.953 | < 0.05 |
| Stages | |||||||||
| I–II | 49 | 34 | 47.33 | 1.000 | reference | 1.000 | reference | ||
| III–IV | 33 | 17 | 31.69 | 1.941 | 1.219–3.088 | < 0.01 | 1.130 | 0.765–1.669 | 0.539 |
| Differentiation | |||||||||
| Well and moderate | 42 | 27 | 41.15 | 1.000 | reference | 1.000 | reference | ||
| Poorly and not | 33 | 17 | 32.06 | 1.358 | 1.033–1.785 | < 0.05 | 0.957 | 0.662–1.382 | 0.814 |
| Lymph node metastases | |||||||||
| 0 | 77 | 67 | 70.67 | 1.000 | reference | 1.000 | reference | ||
| ≥1 | 29 | 12 | 29.18 | 4.285 | 2.725–6.738 | < 0.001 | 2.965 | 1.777–4.948 | < 0.001 |
| Metastases to other organs | |||||||||
| Present | 7 | 7 | 18.93 | 1.000 | reference | 1.000 | reference | ||
| Absent | 40 | 23 | 37.87 | 2.245 | 1.297–3.884 | < 0.01 | 2.277 | 1.223–4.240 | < 0.01 |
| UHRF1 expression | |||||||||
| − | 64 | 38 | 52.26 | 1.000 | reference | 1.000 | reference | ||
| + | 45 | 30 | 43.37 | 1.287 | 0.849–1.951 | 0.235 | 1.421 | 0.920–2.194 | 0.113 |
| ++ | 28 | 13 | 28.33 | 2.164 | 1.431–3.273 | < 0.001 | 1.711 | 1.093–2.680 | < 0.05 |
| +++ | 11 | 6 | 20.71 | 3.085 | 1.8905.035 | < 0.001 | 2.553 | 1.506–4.327 | < 0.001 |
CI indicates confidence interval; HR, hazard ratio.
Figure 1.
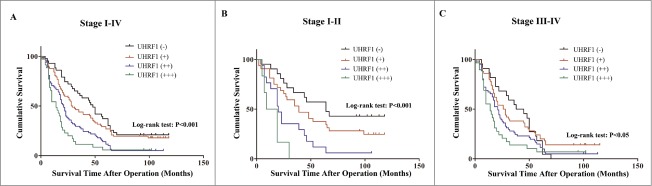
Kaplan–Meier survival curves of GC patients with different level of UHRF1 expression stratified by the TNM stage of the tumor (log-rank test). (A) Correlation of UHRF1 expression with overall survival (cum survival) in all stages. (B) Correlation of UHRF1 expression with overall survival in Stage I–II. (C) Correlation of UHRF1 expression with overall survival in Stage III–IV.
To further characterize the role of UHRF1 in GC development, we measured UHRF1 expression in a panel of human GC cell lines (SGC7901, AGS, and MKN45) and in one immortalized normal gastric mucosal epithelial cell line (GES). As shown in Figure 2A, results from qRT-PCR analysis indicated that UHRF1 mRNA levels were markedly higher in all the GC cell lines, compared with that in GES cells. The MKN45 cell line had the highest UHRF1 expression level, while SGC7901 had the lowest expression level among the GC cell lines. This result was consistent with the protein expression levels from the western blot analysis (Fig. 2B) and suggested that UHRF1 expression could correlate with GC tumorigenesis.
Figure 2.
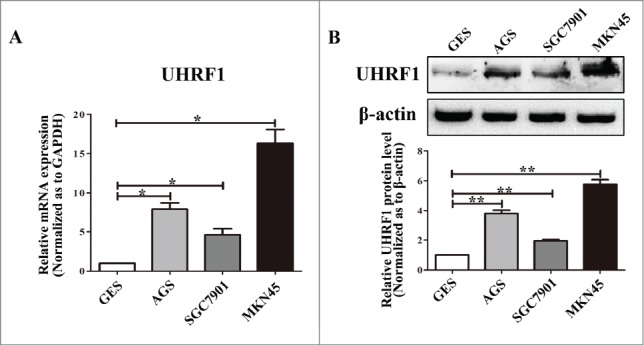
Expression of UHRF1 in GC cell lines. (A) The expression level of UHRF1 mRNA in 3 GC cell lines and one immortalized normal gastric mucosal epithelial cell line (GES) was measured using qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (B) The expression of UHRF1 in 3 GC cell lines and GES cell line was examined through western blot analysis. β-actin was used as an internal control.
Downregulation of UHRF1 expression inhibits proliferation and induces apoptosis of GC cells in vitro
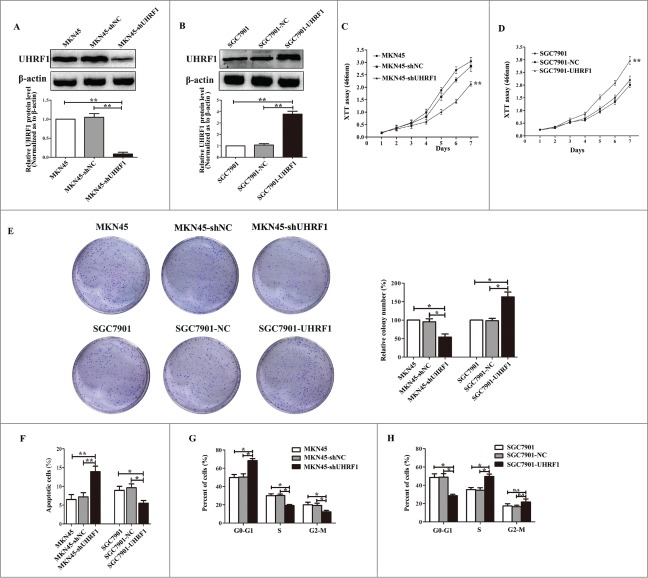
To investigate whether UHRF1 is involved in the increased growth of GC cells, lentiviral vectors encoding shUHRF1 or negative control (shNC) were transfected into MKN45 cells. After cell transfection and fluorescence screening, extracts from MKN45-shUHRF1 and MKN45-shNC cells were compared by protein gel blot (Fig. 3A). shUHRF1 significantly downregulated UHRF1 expression by more than 85% in MKN45 cells (Fig. 3A). XTT assays showed that cell growth was significantly decreased by the downregulation of UHRF1 in MKN45 cells compared with MKN45 or MKN45-shNC cells (Fig. 3C). Colony formation assay results indicated that the number of colonies from MKN45 cells transfected with shUHRF1 was less than half of that from the control and parental groups (Fig. 3E). To further explore the induced effects of UHRF1 on GC cell growth, we performed apoptosis and cell cycle analyses by flow cytometry. Apoptosis assays demonstrated that the percentage of apoptotic MKN45-shUHRF1 cells increased compared with MKN45 or MKN45-shNC cells (Fig. 3F). Moreover, cell cycle assays showed that downregulation of UHRF1 in MKN45 cells increased the G0/G1 population and decreased the S and G2/M population compared with MKN45 or MKN45-shNC cells (Fig. 3G). In contrast, overexpression of UHRF1 in SGC7901 cells increased cell growth and colony formation, and reduced apoptosis and the G0/G1 population (Fig. 3B, D, F and H). Taken together, these results suggest that UHRF1 correlates with gastric carcinogenesis and that downregulation of UHRF1 may inhibit GC cells growth by blocking cell cycle progression and inducing apoptosis.
Figure 3.
UHRF1 promotes GC cell proliferation in vitro. (A) Western blot analysis of UHRF1 expression in MKN45 cells infected with UHRF1 shRNA (shUHRF1) or negative control (shNC) and parental cells. β-actin was used as an internal control. (B) Western blot analysis of UHRF1 in SGC7901 cells transfected with UHRF1 plasmid or vector control (NC) and parental cells. (C) XTT assay of MKN45, MKN45-shNC and MKN45-shUHRF1 cells. (D) XTT assay of SGC7901, SGC7901-NC and SGC7901-UHRF1 cells. (E) Colony formation assay of MKN45 cells infected with shUHRF1 and SGC7901 cells transfected with UHRF1 plasmid. Colonies were evaluated and values were reported as the ratio. (F) Apoptosis assay of MKN45 cells infected with shUHRF1 and SGC7901 cells transfected with UHRF1 plasmid. (G) Flow cytometry cell cycle analysis of MKN45, MKN45-shNC and MKN45-shUHRF1 cells. (H) Flow cytometry cell cycle analysis of SGC7901, SGC7901-NC and SGC7901-UHRF1 cells. Data are shown as mean ± SEM (n = 3) of one representative experiment. Similar results were obtained in 3 independent experiments.
Tumor suppressors contribute to UHRF1-induced growth inhibition
In a previous study, we confirmed that UHRF1 reactivates Slit3, CDH4 and RUNX3 genes via promoter hypermethylation to induce GC invasion and metastasis in the previous study17. To determine the mechanisms by which UHRF1 promotes proliferation and growth in GC cells, we compared the gene methylation status in MKN45-shNC cells and MKN45-shUHRF1 cells using methylation arrays and focused on the genes involved in proliferation. Subsequent to the Gene Ontology (GO) analysis, several proliferation-related genes were selected for further analysis (Table 3). We were particularly interested in CDX2, CDKN2A, RUNX3, FOXO4, PPARG, BRCA1 and PML because they had been shown to play negative roles in GC proliferation and growth.18-24
To verify the effects of methylation changes on the expression of these genes, we examined the methylation status of the promoter regions using methylation-specific PCR in MKN45 cells transfected with shUHRF1, SGC7901 cells with UHRF1 and their negative controls. As shown in Figure 4A, UHRF1 inhibition reversed the promoter methylation of all these genes and upregulation of UHRF1 enhanced promoter methylation. qRT-PCR analysis showed that the mRNA levels of these genes increased after UHRF1 silencing (Fig. 4C). By contrast, UHRF1 forced expression produced opposite results (Fig. 4B and D). Collectively, these results suggested that UHRF1 promotes GC cell proliferation by enhancing tumor suppressor gene (TSG) methylation.
Figure 4.
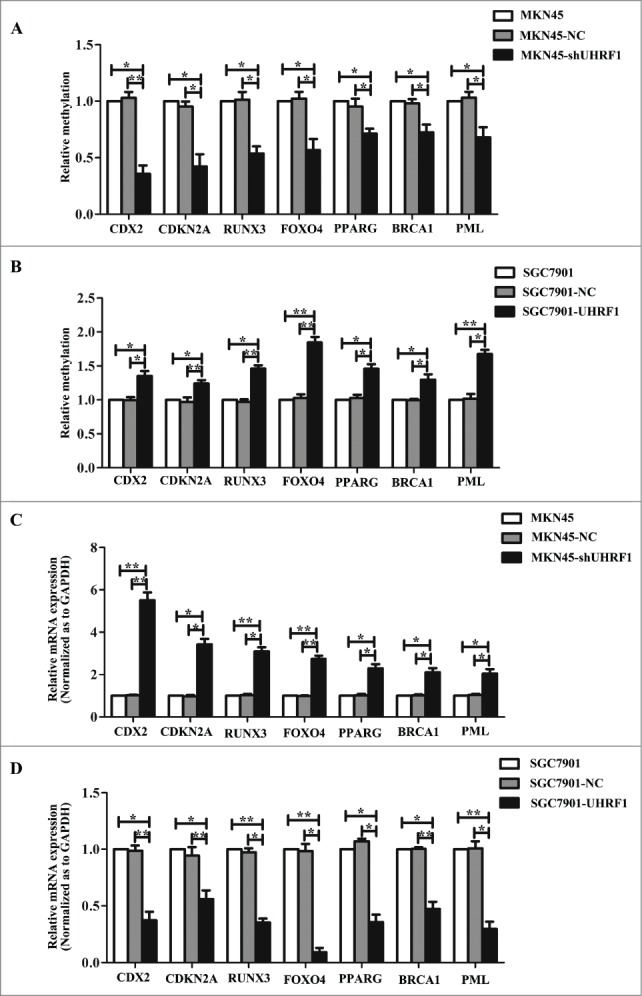
UHRF1 reduces tumor suppressor expression via enhancing methylation. (A) Effects of UHRF1 silencing on gene methylation of 7 tumor suppressor genes in MKN45 cells infected with UHRF1 shRNA (shUHRF1) assayed by Methylation-specific PCR (MSP). (B) Effects of UHRF1 upregulation on gene methylation of 7 tumor suppressor genes in SGC7901 cells transfected with UHRF1 plasmid assayed by MSP. (C) Effects of UHRF1 silencing on mRNA expression of 7 tumor suppressor genes in MKN45 cells infected with shUHRF1 assayed by qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (D) Effects of UHRF1 upregulation on mRNA expression of 7 tumor suppressor genes in SGC7901 cells transfected with UHRF1 plasmid assayed by qRT-PCR.
Downregulation of UHRF1 suppresses tumor growth of GC cells in vivo
To evaluate the functional role of UHRF1 in vivo, we injected both MKN45-shUHRF1 and MKN45-shNC cells into athymic nude mice and measured the tumor volume over time. As early as 10 d post-implantation, the growth of transplanted tumors between the 2 groups became statistically significant (P < 0.05). At 2 weeks after implantation, those mice injected with MKN45-shNC carried larger tumor burdens. Compared with control group, the average tumor volume of the shUHRF1–treated group was markedly reduced by more than 60% (Fig. 5A and B; P < 0.05). After 4 weeks, the mice were euthanized, and the average tumor weight was significantly reduced in the shUHRF1 treated group (0.48 ± 0 .18 g vs. One.07 ± 0 .26 g; P < 0.01). The low expression of UHRF1 in excised tumors was further confirmed by qRT-PCR (Fig. 5D; P < 0.01). In addition, immunohistochemical analysis of Ki-67 antigen revealed that the number of hyperproliferative Ki-67+ tumor cells significantly decreased compared with control group (Fig. 5E; P < 0.01).
Figure 5.
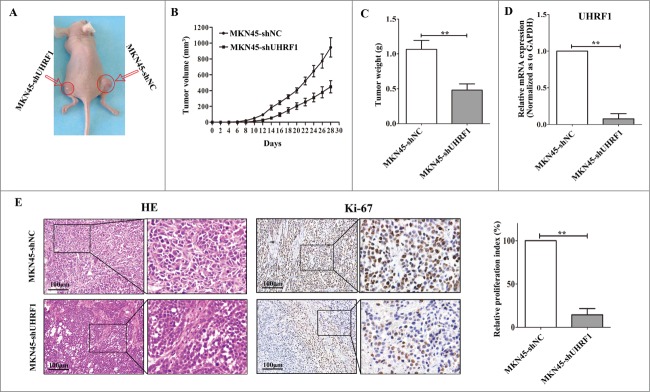
Down-regulation of UHRF1 inhibits GC growth in vivo. (A) MKN45-shUHRF1 and MKN45-shNC cells were injected subcutaneously into nude mice. At 4 weeks after implantation, MKN45-shUHRF1 cells produced smaller tumors than control cells. (B) Growth curve of tumor volumes. Each data point represents the mean ±SEM of 5 mice. (C) Average weight of tumors in nude mice. (D) The mRNA expression of UHRF1 in transplanted tumors formed by MKN45-shUHRF1 and MKN45-shNC cells was measured using qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (E) Representative photographs of H&E staining and immunohistochemical analysis of Ki-67 antigen in tumors of nude mice (left). Comparison of proliferation index is shown on the right.
Discussion
DNA methylation, which is associated with transcriptional silencing and TSG function loss, plays an important role during carcinogenesis.25-27 The epigenetic regulator UHRF1 is known to maintain genomic DNA methylation by recruiting the DNA methyltransferase DNMT1 to DNA replication forks, which is involved in a large number of physiological and pathological phenomena, from embryogenesis to cell migration and cancer development and progression.3,4 As a putative oncogenic factor, UHRF1 is a significant biomarker for differentially diagnosing pancreatic adenocarcinoma, chronic pancreatitis and normal pancreas.28 UHRF1 overexpression is associated with late tumor stages and predicts poor prognoses in lung and bladder cancers.7,11 Yang et al. showed that UHRF1 overexpression is an independent prognostic factor for tumor recurrence in nonmuscle-invasive bladder cancer and proposed that UHRF1 might be a molecular marker that could be used to predict the recurrence of this type of cancer.29 In our previous study, UHRF1 expression was shown to be much higher in GC tissues compared with adjacent tissues and to be closely associated with depth of invasion (T3-T4), late TNM stage, poor differentiation, lymph node metastasis, and distant metastasis.17 In the present study, we further demonstrated that UHRF1 was overexpressed in GC tissues and that its expression was an independent and significant predictor of GC prognosis. Moreover, UHRF1 expression appeared to be more closely associated with GC prognosis in early stage than late stage. Multivariate analysis indicated that UHRF1 expression was still an independent predictor after stratifying by early and late stage (Table S4). Taken together, these results suggest that UHRF1 could be used as a novel molecular marker for GC diagnosis and prognosis.
High UHRF1expression is a key factor for several tumor features that can maintain cells in a proliferative state and prevent their differentiation.14,15,30 UHRF1 is not expressed in highly differentiated tissues, whereas it is particularly overexpressed in proliferative tissue.31 Moreover, UHRF1 promotes proliferation and progression in many cancers, including colorectal,8,9 bladder cancer32 and liver cancer.13,33 These results indicate that overexpression of UHRF1 is closely associated with tumor proliferation. Thus, we speculate that UHRF1 may enhance GC cell proliferation and tumorigenesis. Our results demonstrated that forced UHRF1 expression markedly enhances GC cell proliferation, whereas UHRF1 knockdown significantly inhibits cell proliferation in vitro and in vivo. These data suggest that UHRF1 overexpression contributes to GC cell proliferation and growth.
Emerging evidence has indicated that UHRF1 represses the transcription of several TSGs via promoter methylation, including p16INK4A, hMLH1, p21 and RB, thus promoting cancer growth and metastasis.6,8,34,35 In this study, we found that UHRF1 promoted GC proliferation via methylation of CDX2, CDKN2A, RUNX3, FOXO4, PPARG, BRCA1 and PML.
Of the genes examined in this study, some have been identified as targets of UHRF1 in other cancers; these genes include CDKN2A, RUNX3, PPARG, BRCA1 and PML. Many studies have shown the regulatory role of UHRF1 in the expression of CDKN2A (also known as p16INK4A), which is one of the most commonly studied gene candidates in the pathogenesis of human cancers.9,36,37 In 2012, Wang et al. demonstrated that UHRF1 expression inversely correlates with p16INK4A expression in colorectal cancer cell tissues and that the transfection of DLD1 and SW620 cells with lenti-shUHRF1 markedly increases the expression of p16INK4A. RUNX3 was found to be regulated by UHRF1 during GC invasion and metastasis in our previous study.17 Moreover, loss of RUNX3 expression can enhance the Akt1-mediated signaling pathway and promote the tumorigenesis process in human GC.20 PPARG, which plays an important role in GC tumorigenesis serving as suppressor gene, has been proved to be negatively regulated by UHRF1 in colorectal cancer.8 According to Sabatino et al.'s study8, UHRF1 stimulates the migration and invasion capacities of tumor cells through silencing PPARγ and increasing of cyclin A and cyclin D1 expression. BRCA1 is a well-known TSG implicated in the predisposition to early-onset breast and ovarian cancers.38 It was first described in sporadic breast cancer that UHRF1 is responsible for regulating BRCA1 transcription by inducing DNA methylation.39 PML has been shown to be a tumor suppressor protein that regulates cell cycle progression, gene transcription, transformation suppression, and apoptosis.40 Recently it was reported that loss of PML expression contributes to the enhancement of lymphocyte infiltration into gastric cancer tissue via regulation of IP-10 expression.24 Guan et al. found that UHRF1 promotes ubiquitination-mediated degradation of PML to regulate endothelial cell migration and capillary tube formation.41 The present study further demonstrated that UHRF1 represses CDKN2A, RUNX3, PPARG, BRCA1 and PML expression via promoter hypermethylation, promoting GC proliferation and carcinogenesis.
CDX2 and FOXO4 are newly discovered downstream genes of UHRF1. The transcription factor, CDX2, is a member of the caudal-related homeobox gene family, and is mainly expressed in the intestine. CDX2 has been reported to be associated with intestinal metaplasia of the stomach, in which ectopic CDX2 expression is speculated to induce the trans-differentiation of gastric epithelial cells to an intestinal phenotype.42 In addition, CDX2 transgenic mice have been shown to present intestinal metaplasia and a high incidence of gastric carcinoma.18 FOXO4, a member of the FOXO family, is an ubiquitously expressed transcription factor that functions as a tumor suppressor protein via its ability to repress the expression of genes encoding proliferative, survival or anti-differentiation functions.43,44 Loss of FOXO4 expression contributes to GC growth and metastasis, and it may serve as a potential therapeutic target for GC.21 Taken together, these findings indicate that UHRF1 regulates a large panel of TSGs and thus may be a major effector of malignant transformation or malignant status maintenance. Therefore, UHRF1 expression could be targeted to prevent cell transformation via the repression of TSGs.
In summary, the results obtained in the present study reveal that UHRF1 expression is an independent and significant predictor of GC prognosis and that UHRF1 overexpression promotes GC proliferation and carcinogenesis by inhibiting apoptosis and increasing the G1/S transition, as well as inducing the hypermethylation of 7 TSGs. These findings shed new insight regarding the function of UHRF1 and suggest that the downregulation of UHRF1 may be another possible approach for the management of human GC.
Materials and Methods
Cell culture
Human normal gastric epithelial cell line, GES and human gastric cancer cell lines, including AGS, SGC7901 and MKN45 cells were obtained from Beijing Institute of Oncology. Cells were maintained in RPMI-1640 medium (Thermo Scientific HyClone, Beijing, China) supplemented with 10% fetal calf serum. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Immunohistochemistry
The GC tissue microarray protocol was described in detail in our previous study.45 All research protocols were approved by the Ethics Committee of Xijing Hospital. Each array included 238 GC tissues and 72 non-cancerous gastric mucosae, with exact follow-up data. Immunohistochemistry staining and evaluation were performed as described previously17 using monoclonal anti-UHRF1 antibody (ab57083; Abcam, Cambridge, UK; 1:100 dilution) incubating for overnight. The staining was evaluated by scanning the entire tissue specimen under low magnification (×40) and then confirming under high magnification (×200 and ×400). Protein expression was visualized and classified based on the percentage of positive cells and the intensity of staining. The percentage of positive cells was divided into 5 grades (percentage scores): <1% (0), 1–25% (1), 26–50% (2), 51–75% (3) and >75% (4). The intensity of staining was divided into 4 grades (intensity scores): negative (0), weak (1), moderate (2) and strong (3). The histological score (H-score) was determined using the following formula: overall scores = percentage score × intensity score. An overall score of 0–12 was calculated and graded as negative (–, score: 0), weak (+, score: 1–4), moderate (++, score: 5–8) or strong (+++, score: 9–12).
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The PCR primers for all the genes are shown in Table S1. First-strand cDNA was synthesized using a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). Real-time PCR was performed using SYBR Premix Ex Taq II (TaKaRa) and measured using a LightCycler 480 system (Roche, Basel, Switzerland). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control, and an internal control was used to verify that sample loading was equal. The fold change was calculated by 2−ΔΔCt. All reactions were performed in triplicate.
Western blot
Total proteins were prepared from cultured cell samples by complete cell lysis (Roche) with protease and phosphatase inhibitors. Denatured proteins (20–50 μg) were separated on SDS-PAGE gels and transferred to PVDF membranes. The primary antibodies used were anti-UHRF1 (Abcam; 1:150 dilution), and β-actin (Sigma, St Louis, MO, USA; 1:2000 dilution), both of which were monoclonal antibodies and incubated for 12 hours at 4°C. The bands were scanned using a ChemiDoc XRS+ Imaging System (Bio-Rad, Hercules, CA, USA) and quantified using Quantity One v4.6.2 software (Bio-Rad).
Plasmid construction and transfection
The full-length open reading frame sequence of UHRF1 was obtained by RT-PCR amplification (primers were shown in Supplementary Table 1) of normal human stomach cDNA. The PCR aliquots were subcloned into the mammalian expression vector pcDNA3.1 containing an HA-tag (Invitrogen) at N terminus and then verified by DNA sequencing. The plasmids were transiently transfected into cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol.
Lentivirus infection
The short hairpin RNA (shRNA) vector pGCSIL-GFP-shUHRF1 was purchased from GeneChem (Shanghai, China). The UHRF1 shRNA sequence was: 5′-TGCAGTATCCAGAAGGCTA-3′. The target cells (1 × 105) were infected with 1 × 107 lentivirus-transducing units in the presence of 10-μg/ml polybrene. As a control, we also obtained a lentiviral vector that expressed green fluorescent protein alone (LV-GFP) from GeneChem.
XTT assays
XTT assays were conducted to determine cell growth according to the manufacturer's instructions (Cell Proliferation Kit II (XTT), Roche). Target cells (1000/well) were seeded in 96-well plates, and the volume of culture medium was set to 100 μl. XTT (sodium 3´-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate) labeling reagent (final concentration of 0.3 mg/ml) and electron-coupling reagent (PMS, N-methyl dibenzopyrazine methyl sulfate) were mixed, and 50 μl of the mixture was added to the wells. The cells were incubated with the reagents for 6 hours. A Varioskan Flash Multimode Reader (Thermo Fisher, Waltham, MA, USA) was used to read the absorbance at 466 nm with a reference wavelength at 650 nm.
Plate colony formation assay
Log phase cells were trypsinized into single cell suspensions and plated in 90 mm2 plates at a density of 1 × 103 cells/well. The colonies were stained with Giemsa, and the total number of colonies was counted. Each assay was performed in triplicate.
Cell cycle and apoptosis assays
For cell cycle analysis, target cells were fixed in 75% ethanol and stained with PI (propidium iodide) supplemented with RNase A (Roche) for 30 minutes at 22°C. An Annexin V-FITC Apoptosis Detection Kit was used for apoptosis assays. The cells (1 × 104) were stained according to the manufacturer's protocol and sorted using a FACS sorter (BD Biosciences, Maryland, USA), and the data were analyzed using Modfit software (BD Biosciences).
In vivo tumor growth assay
Female BALB/c nude mice aged 4 to 5 weeks were purchased from Laboratory Animal Services Center of the Fourth Military Medical University (FMMU). Animal handling and experimental procedures were approved by the Animal Experimental Ethics Committee of FMMU. For the tumor growth assay, 2 × 106 MKN45-shUHRF1 and MKN45-shNC cells were suspended in 200 μl PBS and injected subcutaneously into the left and right dorsal flanks of nude mice, respectively. The experiment contained 5 mice and was repeated 3 times. Tumor size was measured every 2 d After 4 weeks, the mice were sacrificed, and the tumors were dissected. Tumor volumes were calculated as follows: volume = (D×d2)/2, where D meant the longest diameter and d meant the shortest diameter. After the transplanted tumor tissues from mice were formalin-fixed and paraffin-embedded, they were sectioned at 4-mm thickness and analyzed for hematoxylin and eosin staining, and for Ki-67 (ab833; Abcam; 1:50 dilution) expression. The proliferative index score was measured by the mean percentage of nuclei staining positive for Ki-67 antigen in 1000 cells under a microscope.
DNA extraction
Genomic DNA from MKN45, MKN45-shNC, MKN45-shUHRF1, SGC7901, SGC7901-NC and SGC7901-UHRF1 cells was isolated from tumor cells using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) after the manufacturer's protocol.
Methylation arrays and data analysis
A total of 500 ng of DNA extracted from MKN45-shNC or MKN45-shUHRF1 cells was bisulfite converted using a EZ DNA Methylation Kit (Zymo Research) and subsequently processed for hybridization onto an Infinium HumanMethylation450 BeadArray (Illumina, California, USA) according to the manufacturer's instructions. This array interrogates more than 450,000 methylation sites of >99 % RefSeq genes.. Briefly, the DNA was treated with bisulfite, converting non-methylated C nucleotides to U (T), whereas methylated C nucleotides remained unaffected. The bisulfite-treated DNA was subsequently amplified, fragmented, and hybridized to locus-specific oligonucleotides on the BeadArray. C or T nucleotides were detected through the fluorescence signal obtained from the single-nucleotide extension of the DNA fragments. The results were interpreted as a ratio (β value) of methylated signal (C) compared with the sum of methylated and unmethylated signal (C + T) for each locus, where a β-value of 0 represents fully unmethylated DNA and a value of 1 fully methylated DNA.
GO analysis of the genes obtained from methylation arrays was performed using DAVID online analysis tool (http://david.abcc.ncifcrf.gov/) with Biological Process FAT data set [25,26]. The threshold of the Expression Analysis Systematic Explorer (EASE) score, a modified Fisher exact P value, for gene-enrichment analysis was set at 0.05 (P ≤0 .05 is considered strongly enriched in the annotation categories). The GO biological processes, including cell proliferation, were selected and the differentially expressed genes involved in these biological processes were screened out for further study.
Methylation-specific PCR analysis
DNA was treated with an EZ DNA Methylation-Gold KitTM (Zymo Research Co., Orange, CA) according to the manufacturer's instructions. Methylation-specific PCR (MSP) was performed using TaKaRa TaqTM Hot Start Version (TaKaRa) with primers specific for methylated and unmethylated sequences of the genes. Methylation-specific PCR primers for each gene, as verified in previous reports are available in Table S2. The PCR products were electrophoresed in 2.5% agarose gels. The bands were scanned using a ChemiDoc XRS+ Imaging System (Bio-Rad) and quantified using Quantity One v4.6.2 software (Bio-Rad). β value of methylated signal (C) compared with the sum of methylated and unmethylated signal (C + T) was used to evaluate the methylation status.
Statistical analysis
SPSS software (version 12.0, SPSS Inc.., Chicago, IL, USA) was used for the statistical analyses. The continuous data were presented as the means ± s.e.m. and compared between 2 groups using Student's unpaired t-test. The Kruskal–Wallis H-test and the Mann–Whitney U test were used to analyze the relationship between UHRF1 expression and clinicopathological factors of gastric cancer samples. Univariate survival analysis was performed using the Kaplan-Meier method, and the significance of differences between groups was analyzed using the log-rank test. Stepwise multivariate survival analysis was performed using the Cox proportional hazards model. Significant variables in the univariate analysis were included in the model using the Backward Wald method. P < 0.05 was considered statistically significant (*P < 0.05, **P < 0.01 and ***P < 0.001).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This study was partially sponsored by the National Natural Science Foundation of China (No.81402337 to Dr. Lin Zhou, No.81402387 to Dr. Yulong Shang, No.30900551 and No. 81322037 to Dr. Jie Liang) and the National Excellent Doctoral dissertation of China (No. 201182 to Dr. Jie Liang).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87-108; PMID:25651787; http://dx.doi.org/ 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet 2009; 374:477-90; PMID:19625077; http://dx.doi.org/ 10.1016/S0140-6736(09)60617-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al.. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450:908-12; PMID:17994007; http://dx.doi.org/ 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- 4.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317:1760-4; PMID:17673620; http://dx.doi.org/ 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 5.Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-α by pRb family proteins: a novel mechanism in breast cancer. Cancer Res 2007; 67:7731-7; PMID:17699777; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1476 [DOI] [PubMed] [Google Scholar]
- 6.Daskalos A, Oleksiewicz U, Filia A, Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK, Liloglou T. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer 2011; 117:1027-37; PMID:21351083; http://dx.doi.org/ 10.1002/cncr.25531 [DOI] [PubMed] [Google Scholar]
- 7.Unoki M, Daigo Y, Koinuma J, Tsuchiya E, Hamamoto R, Nakamura Y. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer 2010; 103:217-22; PMID:20517312; http://dx.doi.org/ 10.1038/sj.bjc.6605717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabatino L, Fucci A, Pancione M, Carafa V, Nebbioso A, Pistore C, Babbio F, Votino C, Laudanna C, Ceccarelli M, et al.. UHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progression. Oncogene 2012; 31:5061-72; PMID:22286757; http://dx.doi.org/ 10.1038/onc.2012.3 [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol 2012; 19:2753-62; PMID:22219067; http://dx.doi.org/ 10.1245/s10434-011-2194-1 [DOI] [PubMed] [Google Scholar]
- 10.Babbio F, Pistore C, Curti L, Castiglioni I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero E, et al.. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene 2012; 31:4878-87; PMID:22330138; http://dx.doi.org/ 10.1038/onc.2011.641 [DOI] [PubMed] [Google Scholar]
- 11.Unoki M, Kelly JD, Neal DE, Castiglioni I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero E. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer 2009; 101:98-105; PMID:19491893; http://dx.doi.org/ 10.1038/sj.bjc.6605123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Huang Z, Zhu Z, Castiglioni I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero E. Upregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1. PLoS One 2014; 9:e104252; PMID:25272010; http://dx.doi.org/ 10.1371/journal.pone.0104252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al.. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 2014; 25:196-209; PMID:24486181; http://dx.doi.org/ 10.1016/j.ccr.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronner C, Krifa M, Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem Pharmacol 2013; 86:1643-9; PMID:24134914; http://dx.doi.org/ 10.1016/j.bcp.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther 2007; 115:419-34; PMID:17658611; http://dx.doi.org/ 10.1016/j.pharmthera.2007.06.003 [DOI] [PubMed] [Google Scholar]
- 16.Mousli M, Hopfner R, Abbady AQ, Monté D, Jeanblanc M, Oudet P, Louis B, Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer 2003; 89:120-7; PMID:12838312; http://dx.doi.org/ 10.1038/sj.bjc.6601068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Zhao X, Han Y, Monté D, Jeanblanc M, Oudet P, Louis B, Bronner C. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J 2013; 27:4929-39; PMID:23982143; http://dx.doi.org/ 10.1096/fj.13-233387 [DOI] [PubMed] [Google Scholar]
- 18.Mutoh H, Sakurai S, Satoh K, Tamada K, Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N, et al.. Development of gastric carcinoma from intestinal metaplasia in Cdx2-transgenic mice. Cancer Res 2004; 64:7740-7; PMID:15520178; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1617 [DOI] [PubMed] [Google Scholar]
- 19.Kim JK, Noh JH, Eun JW, Jung KH, Bae HJ, Shen Q, Kim MG, Chang YG, Kim SJ, Park WS, et al.. Targeted inactivation of HDAC2 restores p16INK4a activity and exerts antitumor effects on human gastric cancer. Mol Cancer Res 2013; 11:62-73; PMID:23175521; http://dx.doi.org/ 10.1158/1541-7786.MCR-12-0332 [DOI] [PubMed] [Google Scholar]
- 20.Lin FC, Liu YP, Lai CH, Shan YS, Cheng HC, Hsu PI, Lee CH, Lee YC, Wang HY, Wang CH, et al.. RUNX3-mediated transcriptional inhibition of Akt suppresses tumorigenesis of human gastric cancer cells. Oncogene 2012; 31:4302-16; PMID:22231444; http://dx.doi.org/ 10.1038/onc.2011.596 [DOI] [PubMed] [Google Scholar]
- 21.Su L, Liu X, Chai N, Lv L, Wang R, Li X, Nie Y, Shi Y, Fan D. The transcription factor FOXO4 is down-regulated and inhibits tumor proliferation and metastasis in gastric cancer. BMC Cancer 2014; 14:378; PMID:24886657; http://dx.doi.org/ 10.1186/1471-2407-14-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JM, Kim SS, Cho YS. The Role of PPARgamma in Helicobacter pylori Infection and Gastric Carcinogenesis. PPAR Res 2012; 2012:687570; PMID:22936949; http://dx.doi.org/ 10.1155/2012/687570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H, Zhang Y, Zou M, Yang S, Liang XQ. Expression of TRF1, TRF2, TIN2, TERT, KU70, and BRCA1 proteins is associated with telomere shortening and may contribute to multistage carcinogenesis of gastric cancer. J Cancer Res Clin Oncol 2010; 136:1407-14; PMID:20127252; http://dx.doi.org/ 10.1007/s00432-010-0795-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HJ, Song DE, Lim SY, Lee SH, Kang JL, Lee SJ, Benveniste EN, Choi YH. Loss of the promyelocytic leukemia protein in gastric cancer: implications for IP-10 expression and tumor-infiltrating lymphocytes. PLoS One 2011; 6:e26264; PMID:22022583; http://dx.doi.org/ 10.1371/journal.pone.0026264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong KY, Huang X, Chim CS. DNA methylation of microRNA genes in multiple myeloma. Carcinogenesis 2012; 33:1629-38; PMID:22715154; http://dx.doi.org/ 10.1093/carcin/bgs212 [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012; 31:1609-22; PMID:21860412; http://dx.doi.org/ 10.1038/onc.2011.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012; 143:550-63; PMID:22796521; http://dx.doi.org/ 10.1053/j.gastro.2012.07.009 [DOI] [PubMed] [Google Scholar]
- 28.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, et al.. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology 2005; 129:1454-63; PMID:16285947; http://dx.doi.org/ 10.1053/j.gastro.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 29.Yang GL, Zhang LH, Bo JJ, Chen HG, Cao M, Liu DM, Huang YR. UHRF1 is associated with tumor recurrence in non-muscle-invasive bladder cancer. Med Oncol 2012; 29:842-7; PMID:21611839; http://dx.doi.org/ 10.1007/s12032-011-9983-z [DOI] [PubMed] [Google Scholar]
- 30.Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, et al.. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol 2012; 14:753-63; PMID:22729083; http://dx.doi.org/ 10.1038/ncb2520 [DOI] [PubMed] [Google Scholar]
- 31.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res 2000; 60:121-8; PMID:10646863 [PubMed] [Google Scholar]
- 32.Ying L, Lin J, Qiu F, et al.. Epigenetic repression of RGS2 by UHRF1 promotes bladder cancer progression. FEBS J 2015; 282174-82; PMID:2532376621496237 [DOI] [PubMed] [Google Scholar]
- 33.Wu SM, Cheng WL, Liao CJ, et al.. Negative modulation of the epigenetic regulator, UHRF1, by thyroid hormone receptors suppresses liver cancer cell growth. Int J Cancer 2015; 137:37-49; PMID:2543063921496237 [DOI] [PubMed] [Google Scholar]
- 34.Alhosin M, Sharif T, Mousli M, Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB, Bronner C. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J Exp Clin Cancer Res 2011; 30:41; PMID:21496237; http://dx.doi.org/ 10.1186/1756-9966-30-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene 2005; 24:7337-45; PMID:16007129; http://dx.doi.org/ 10.1038/sj.onc.1208878 [DOI] [PubMed] [Google Scholar]
- 36.Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al.. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 2008; 27:2187-97; PMID:17934516; http://dx.doi.org/ 10.1038/sj.onc.1210855 [DOI] [PubMed] [Google Scholar]
- 37.Shamma A, Suzuki M, Hayashi N, Kobayashi M, Sasaki N, Nishiuchi T, Doki Y, Okamoto T, Kohno S, Muranaka H, et al.. ATM mediates pRB function to control DNMT1 protein stability and DNA methylation. Mol Cell Biol 2013; 33:3113-24; PMID:23754744; http://dx.doi.org/ 10.1128/MCB.01597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al.. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266:66-71; PMID:7545954; http://dx.doi.org/ 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 39.Jin W, Chen L, Chen Y, Xu SG, Di GH, Yin WJ, Wu J, Shao ZM. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat 2010; 123:359-73; PMID:19943104; http://dx.doi.org/ 10.1007/s10549-009-0652-2 [DOI] [PubMed] [Google Scholar]
- 40.Salomoni P, Ferguson BJ, Wyllie AH, Rich T. New insights into the role of PML in tumour suppression. Cell Res 2008; 18:622-40; PMID:18504460; http://dx.doi.org/ 10.1038/cr.2008.58 [DOI] [PubMed] [Google Scholar]
- 41.Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene 2013; 32:3819-28; PMID:22945642; http://dx.doi.org/ 10.1038/onc.2012.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiotani A, Kamada T, Yamanaka Y, Manabe N, Kusunoki H, Hata J, Haruma K. Sonic hedgehog and CDX2 expression in the stomach. J Gastroenterol Hepatol 2008; 23 (Suppl 2):S161-6; PMID:19120891; http://dx.doi.org/ 10.1111/j.1440-1746.2008.05406.x [DOI] [PubMed] [Google Scholar]
- 43.van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal 2011; 14:579-92; PMID:20673124; http://dx.doi.org/ 10.1089/ars.2010.3419 [DOI] [PubMed] [Google Scholar]
- 44.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013; 13:482-95; PMID:23792361; http://dx.doi.org/ 10.1038/nrc3539 [DOI] [PubMed] [Google Scholar]
- 45.Zhou L, Shang Y, Liu C, Li J, Hu H, Liang C, Han Y, Zhang W, Liang J, Wu K. Overexpression of PrPc, combined with MGr1-Ag/37LRP, is predictive of poor prognosis in gastric cancer. Int J Cancer 2014; 135:2329-37; PMID:24706505; http://dx.doi.org/ 10.1002/ijc.28883 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.