Abstract
VAMP-associated proteins (VAPs) are highly conserved among eukaryotes. Here, we report a functional analysis of one of the VAPs, PVA31, and demonstrate its novel function on leaf senescence in Arabidopsis. The expression of PVA31 is highly induced in senescence leaves, and localizes to the plasma membrane as well as the ARA7-positive endosomes. Yeast two-hybrid analysis demonstrates that PVA31 is interacted with the plasma membrane localized-VAMP proteins, VAMP721/722/724 but not with the endosome-localized VAMPs, VAMP711 and VAMP727, indicating that PVA31 is associated with VAMP721/722/724 on the plasma membrane. Strong constitutive expression of PVA31 under the control of the Cauliflower mosaic virus 35S promoter induces the typical symptom of leaf senescence earlier than WT in normal growth and an artificially induced senescence conditions. In addition, the marker genes for the SA-mediated signaling pathways, PR-1, is promptly expressed with elicitor application. These data indicate that PVA31-overexpressing plants exhibit the early senescence phenotype in their leaves, and suggest that PVA31 is involved in the SA-mediated programmed cell death process during leaf senescence and PR-protein secretion during pathogen infection in Arabidopsis.
Keywords: Arabidopsis thaliana, cell death, intracellular trafficking, leaf senescence, membrane trafficking, VAMP-associated protein
Introduction
In the final stage of leaf development, leaf tissue undergoes an age-dependent deterioration process that leads to cell death. This process is called leaf senescence. Leaf senescence is a highly regulated degradative process that involves age-dependent programmed cell death (PCD), which is important for plant reproduction as it relocates nutrients from leaves to growing seeds.1 PCD also plays a critical role in various developmental and physiological processes, including xylogenesis, embryogenesis, seed development, and seed germination.2,3
VAMPs (also referred as synaptobrevins) are the transport vesicle–residing soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins that are involved in the membrane fusion process between transport vesicles and target membranes.4 VAMP-associated proteins (VAPs) are highly conserved type-II integral membrane proteins in eukaryotes, which are involved in diverse cellular functions, including regulation of lipid transport, membrane trafficking, neurotransmitter release, stabilization of presynaptic microtubules, and unfolded protein response.5 First VAP protein, VAP-33 was isolated as a VAMP-associated protein with a yeast 2-hybrid screen of a sea hare, Aplysia californica; it is involved in the VAMP-mediated neurotransmitter release at the plasma membrane (PM).6 A mammalian VAP-related protein, VAP-A has been reported to bind not only VAMP protein but also other SNAREs and SNARE-binding proteins, including syntaxin 1A, rbet1, rsec22, αSNAP, and NSF.7 A rat VAP-33-related protein, ERG-30, has been reported to function in COPI-mediated ER-Golgi trafficking, although no direct interaction between ERG-30 and SNARE proteins was detected.8
An alternate function of VAPs that has been reported is lipid transfer from ER to a wide range of intracellular membranes via membrane contact sites (MCSs) coordinated with the FFAT (double phenylalanine in an acidic tract) motif containing lipid transfer proteins.5 The FFAT motif consists of the consensus EFFDAxE motif, which binds to the major sperm protein (MSP) domain of VAPs forming a 2:2 complex.9 VAPs have been reported to interact with various FFAT motif-containing lipid binding proteins, including oxysterol-binding proteins (OSBPs),10-12 ceramide transport proteins (CERTs),13 a glycolipid transfer protein (GLTP),14 and a PI/PC-transfer protein.15 In Arabidopsis, 10 VAP-33 family proteins, which are referred to as plant VAP homologs (PVAs), are found in the genome.16 Although several VAPs have been reported to interact with various proteins,17-20 the physiological functions of VAPs in plants remain largely unknown.
In this study, we found that one of the PVA-family proteins, PVA31, was highly expressed in senescent leaves in Arabidopsis. GFP-PVA31 localized to the PM and was interacted with the PM-resided VAMP proteins. The strong constitutive expression of PVA31 under the control of the Cauliflower mosaic virus 35S promoter induced the early senescence phenotypes in rosette leaves. In PVA31 overexpressing plants, the SA-mediated signaling pathway was promptly activated with elicitor application. These findings suggest that PVA31 may be involved in the PCD during leaf senescence and PR-protein secretion during pathogen infection to regulate the SNARE-mediated membrane trafficking to the PM in Arabidopsis.
Results
Identification of one of the VAP-family proteins, PVA31, as a senescence-induced VAP in Arabidopsis
It has been reported that 10 VAP33-related molecules designated as PVAs (plant VAPs) were found in the Arabidopsis genome.16 Typical VAP homologues among eukaryotes consist of an N-terminal major sperm protein (MSP) and a C-terminal transmembrane (TM) domains.5 According to the molecular structures and similarities of VAP proteins, we categorized 10 Arabidopsis PVA proteins into 5 subgroups; 3 PVAs in group-A (PVA11, PVA12 and PVA13) have typical N-terminal MSP and a C-terminal TM domains, group-B PVA consists of only one group protein, PVA14, which has a unique protein structure with 2 conserved MSP domains and a Toll/interleukin-1 receptor homology (TIR) domain in the middle of the molecule with no C-terminal TM domain. Two Group-C PVAs (PVA21 and PVA22) possess typical VAP protein structure with the N-terminal MSP, the C-terminal TM domains, and the coiled-coil domain adjacent to the TM. PVA31, which belongs to group D, has no TMD in its C-terminus, with the smallest molecular weight (17-kDa) among Arabidopsis PVAs, and 3 group-E (PVA41, PVA42 and PVA43) PVAs have an N-terminal extension sequence with no C-terminal TMD (Fig. 1).
Figure 1.
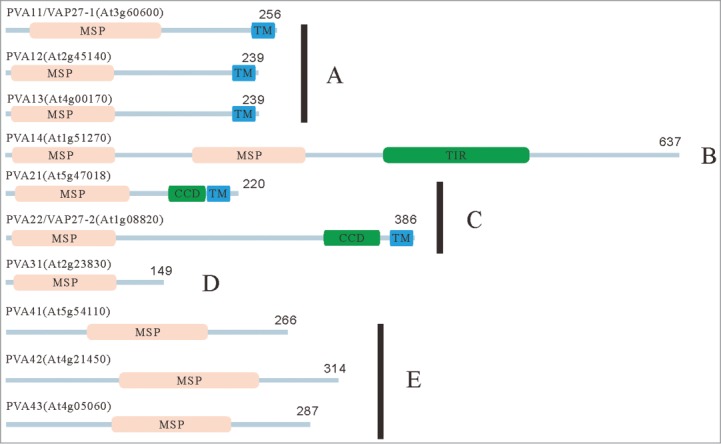
The domain structures of Arabidopsis VAP proteins. Domain structures of Arabidopsis VAP (PVA) proteins were predicted with the SMART program (http://smart.embl-heidelberg.de/). On the basis of the molecular structures and similarities of VAP proteins, we categorized 10 Arabidopsis VAP proteins into 5 subgroups. The abbreviations of domains used in this figure are as follows: CCD: coiled-coil domain, MSP: major sperm protein domain, TIR: Toll/interleukin-1 receptor homology domain TM: transmembrane domain. The numbers indicate the length of molecules in amino acid.
To identify VAP proteins that might be involved in the leaf senescence process, we searched genes encoding the VAP proteins in the publicly available Arabidopsis microarray database on the various developmental stages of leaves by using the Arabidopsis eFP Browser,21 and found that PVA31 is specifically expressed in the senescent leaves (Fig. S1).
PVA31 localizes to the plasma membrane and the endosomes
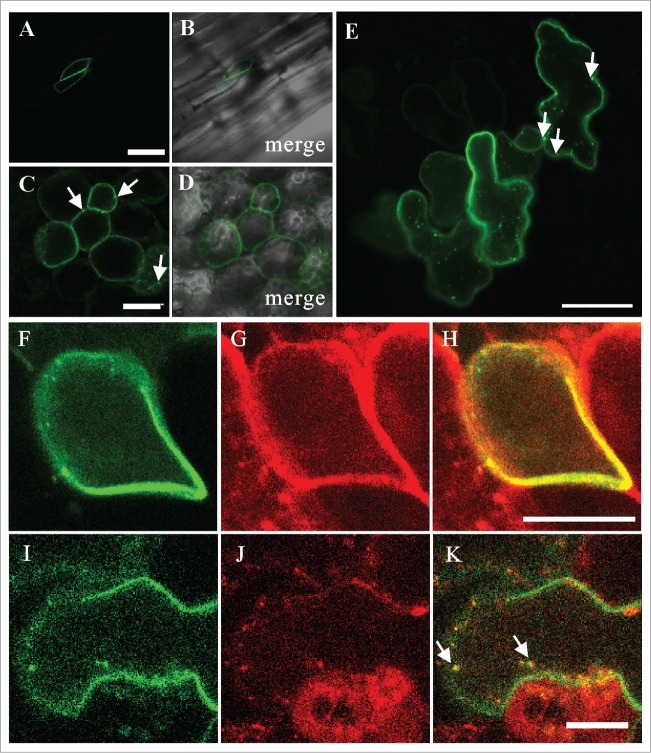
Next, we transiently expressed GFP-fused PVA31 (GFP-PVA31) under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter by Agrobacterium-mediated transient transformation, and observed the subcellular localization of GFP-PVA31 with confocal laser scanning microscopy. The GFP-PVA31 protein predominantly localized to the plasma membrane (PM) of the epidermal cells of hypocotyl (Fig. 2A and B), leaf mesophyll cells as well as leaf epidermal cells (Fig. 2C-E). The GFP-fluorescence was also observed on the intracellular mobile punctate structures near the PM (arrowheads in Figure 2C and E; Movie S1). To determine the subcellular localization of the punctate structures of GFP-PVA31, we co-expressed GFP-PVA31 with a PM-localized VAMP protein, mRFP-VAMP72222 and an endosomal RAB protein, mRFP-ARA7.23 The fluorescence of PVA31 and VAMP722 was completely merged on the PM (Fig. 2F-H), and the fluorescent punctate structures of GFP-PVA31 were merged with mRFP-ARA7 (arrowheads in Figure 2I-K), indicating that PVA31 localizes to the PM and the ARA7-positive endosomes.
Figure 2.
Localization of GFP-PVA31 in Arabidopsis. The confocal images of GFP fluorescence (A, C) and merged DIC and GFP-fluorescence (B, D) were captured in epidermal cells of hypocotyls (A, B) and mesophyll cells of cotyledon (C, D) of Arabidopsis plants transiently expressing GFP-PVA31. The images of GFP fluorescence in epidermal cells (E). The confocal images of GFP-PVA31 (F), mRFP-VAMP722 (G) and merged image of F and G (H) in epidermal cells of hypocotyl. Arrows indicate the mobile punctate structures of GFP-PVA31. Bars = 50 μm (A) , 20 μm (C) and 10 μm (H, K).
PVA31 is interacted with the plasma membrane localized-VAMPs but not with endosome-localized VAMPs
VAP-33 was firstly isolated as a VAMP-associated protein in a yeast 2-hybrid screen of a sea hare, Aplysia californica, and is involved in the VAMP-mediated neurotransmitter release at the PM,17 suggesting that PVA31 is also interacted with particular VAMP proteins in Arabidopsis. To investigate whether PVA31 is interacted with particular Arabidopsis VAMP proteins, we tested an interaction of PVA31 with several Arabidopsis VAMPs by using a yeast 2-hybrid interaction analysis. As shown in Figure 3, PVA31 is specifically interacted with VAMP721/722/724 but not with VAMP711 and VAMP727. VAMP721/722/724 reportedly localized to the PM in Arabidopsis cells, while VAMP711 and VAMP727 localized to the vacuolar membrane and the endosomes, respectively.27,28 These results suggest that PVA31 is specifically associated with VAMP721/722/724 at the PM.
Figure 3.
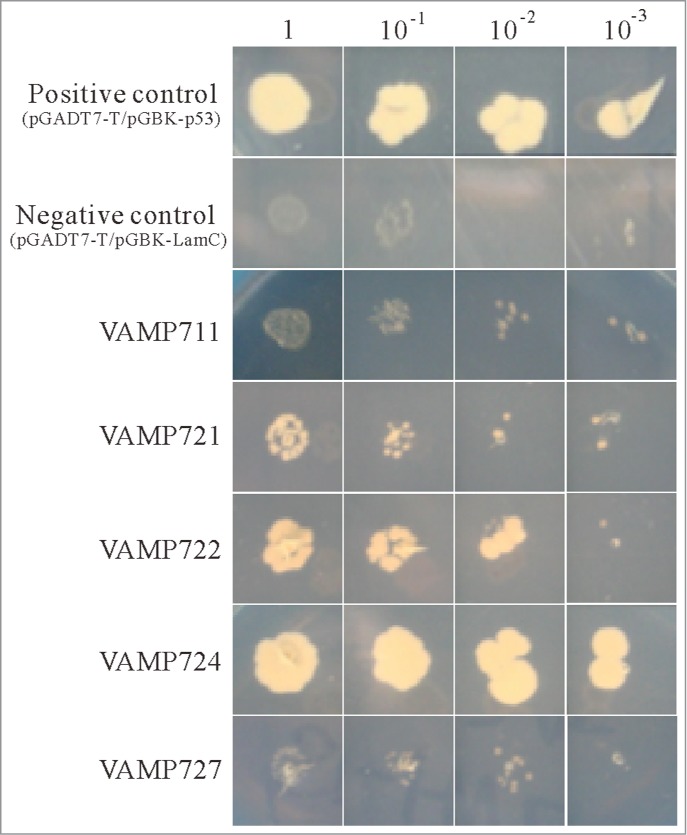
Interaction of PVA31 and VAMP proteins. Yeast two-hybrid assay were performed to confirm between PVA31 and Arabidopsis VAMP proteins. PVA31 and VAMP/711/721/722/724/727 were fused with activation domain (AD) and DNA binding domain (BD) of the transcription factor GAL4, respectively. The yeast strains co-transformed with PVA31 and indicated VAMPs plasmids were grown on selective media lacking leucine, tryptophan and histidine and adding 3-AT. Positive control: pGADT7-T-antigen and pGBKT7-p53. Negative control: pGADT7-T-antigen and pGBKT7-Lamin C.
Overexpression of PVA31 confers the early senescence phenotype in Arabidopsis
Next, we attempted to generate transgenic Arabidopsis plants overexpressing PVA31 under the control of the CaMV 35S promoter to investigate the physiological role of PVA31 on leaf senescence. From several hygromycin-resistant lines of the transgenic plants, we selected 2 overexpressing (OE) lines with about fold8- (line #7) and fold2- (line #13) higher expression levels compared with that in WT. The growth of root and rosette leaf was inhibited in the OE lines, especially in line #7 (Fig. S2 A-C). The relative expression levels of a senescent marker, SAG12 of rosette leaves of 4-weeks old plants was enhanced in the overexpression lines (Fig. 4A), suggesting that leaf senescence is enhanced dependent on the PVA31 expression level. Both OE-lines exhibited the early senescence phenotype in the rosette leaves of 8-week-old plants (Fig. 4B). To measure the rate of senescent rosette leaves in 8-week-old plants, we counted the numbers of leaves revealing the feature of leaf senescence in which the edge to leaves have yellowish color, and found that about half of the rosette leaves of the both VAP31-OE lines (58% in #7 and 59% in #13) exhibited typical feature of senescence, whereas the rate of senescent leaves was 12% in WT (Fig. 4C and D; Fig. S2D; Table S1). We also characterized the leaf senescence phenotype by the artificially induced senescence experiment. After incubated the detached leaves on 3 mM MES under the dark condition for 6 d, the rosette leaves of the PVA31-OE lines, #7 and #13, became more yellowish than WT (Fig. S3). Especially, the chlorophyll contents of rosette leaves in the dark condition was significantly lower in lines #7 than that in WT (Fig. 4E), suggesting that the degree of the senescence is consistent with the expression dosage of PVA31. Taken together, these results suggest that PVA31 was involved in the leaf senescence process in Arabidopsis.
Figure 4.
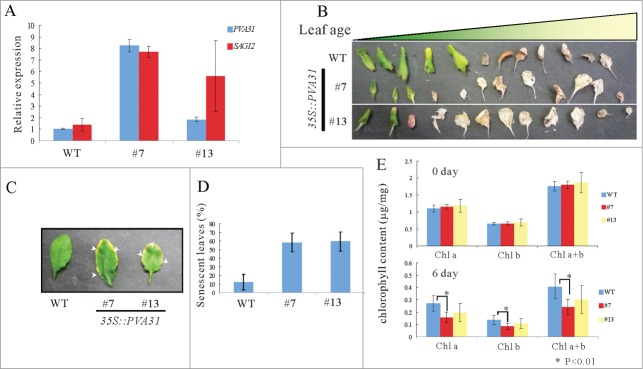
Age-dependent senescence symptoms in PVA31-overexpressing plants. A quantitative RT-PCR analysis of SAG12 transcripts isolated green rosette leaves of WT and PVA31-overexpressing plants growing for 4-weeks. UBQ10 was used as an internal control. Each vertical bars represented the mean ± standard error (3 leaves from each plant were used for the analysis) (A). Senescence phenotype of the rosette leaves of WT and PVA31-overexpressing plants (lines #7 and #13) grown for 8 weeks. The whole rosette leaves of 8-weeks-old plants were detached and aligned in leaf age order (B). The senescence symptom of rosette leaves. Arrowheads indicate the senescence symptom in representative leaves (C). The percentages of senescent leaves of each plant (WT or PVA31-overexpressing lines #7 and #13) with respect to total leaf numbers are presented. Error bars indicate standard deviations. Ten plants from each line were evaluated according to the senescence symptom shown in C (D). The chlorophyll content was measured before and after dark treatment in detached leaves. The difference between WT and PVA31-overexpressing line was indicated by an asterisk (P < 0.01) (E).
Salicylic acid–dependent biotic stress responsive pathway is highly activated in PVA31 overexpressing plant
A search of the publicly available Arabidopsis microarray expression data of PVA31 for various stress treatments revealed that the expression of PVA31 is strongly induced by Pseudomonas syringae infection (Fig. S4). We therefore measured the expression level of #7 line and the expression of marker genes, PR-1 for the SA-mediated defense pathway after a bacterial elicitor, flg22 treatment. As depicted in Fig. 5, the expression of PR-1 was gradually induced over 24 h after the flg22 treatment in WT, whereas in PVA31-OE plants, the PR-1 gene was expressed earlier than WT after the flg22 treatment. These data indicate that the constitutive overexpression of PVA31 promptly induced the SA-mediated signaling pathway with the elicitor stimulation in Arabidopsis.
Figure 5.
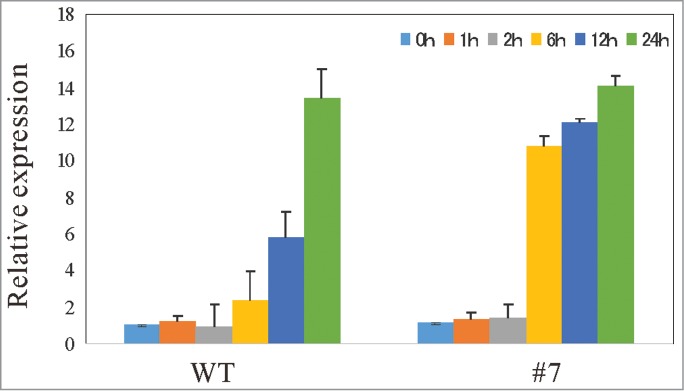
Expression of marker genes for SA-mediated biotic stress responsive pathway are upregulated in PVA31-overexpressing plants with elicitor application A quantitative RT-PCR analysis of PR-1 gene was performed with total RNA isolated from a whole 14-day-old WT plant at various time points (0, 1, 6, 12, and 24 h) following flg22 treatment. UBQ10 was used as an internal control. Each vertical bars represented the mean ± standard error (3 leaves from each plant were used for the analysis).
Discussion
Leaf senescence includes the PCD, which is a sequential deterioration process that results in the massive degradation of subcellular components, and finally leads to the cell death. During the PCD process in leaf senescence, a highly dynamic disintegration of host cell organelles is induced; the grana structure of chloroplast is disrupted and chlorophyll and chloroplast proteins are massively degraded. Concomitantly, chromatin condensation and DNA laddering are detected.25-27 In the final stage of PCD, loss of the integrity of the plasma and vacuolar membranes appears to disrupt cellular homeostasis, leading to cell death.1,28 PCD is also induced in the defense response to pathogens in plants. At the site of infection, localized PCD known as a hypersensitive response (HR) occurs in order to prevent the spread of biotrophic pathogens.9,10 HR is known to be regulated by the salicylic acid (SA)-mediated defense signaling pathway.11,12 Intriguingly, SA also plays a critical role in age-dependent leaf senescence.13,14 The concentration of endogenous SA is considerably higher in senescent leaves of Arabidopsis, and age-dependent leaf senescence is delayed in NahG overexpressing transgenic plants with reduced levels of endogenous SA.13 Thus, part of the regulatory pathway of PCD is shared among leaf senescence and HR.
In this study, we found that one of the Arabidopsis VAP orthologs, PVA31, was highly expressed during leaf senescence and that the constitutive overexpression of PVA31 induced the early senescence phenotype in rosette leaves (Fig. 4; S2D). The results indicate that PVA31 might be involved in the senescence-associated PCD process in Arabidopsis. Although PVA31 protein lacks the C-terminal transmembrane domain and coiled-coil domain typically found in the conventional VAP proteins,5 PVA31 localizes to the PM and the ARA7-resided endosomes (Fig. 2). Additionally, the yeast-2 hybrid assay showed that PVA31 is interacted with the PM-VAMPs, VAMP721/722/724 (Fig. 3). These data suggest that PVA31 is associated with the PM and the endosome membranes via the VAMP protein interaction, and is involved in the senescence-associated PCD process in Arabidopsis.
Although a wide variety of function of VAP proteins have been reported in various eukaryotes, one possible fundamental function of the VAP proteins is thought to be an ER-resident tethering factor that forms a complex with lipid transfer proteins and SNAREs to facilitate lipids transport and protein secretion from the ER to various membranous compartments, thereby mediating the lipid transfer and the membrane trafficking in eukaryotes.5 Taking into the fundamental function of VAP proteins, what kinds of function of PVA31 might be considered in the leaf senescence process? One of the Arabidopsis VAP proteins, PVA11/VAP27–1, was isolated as one of the interactors of a shingosine transfer protein, ACD11, in a yeast 2-hybrid screen.19 The loss-of-function acd11 mutant is highly expressed in several genes related to the SA-mediated disease resistance process, HR,29 and exhibited an accelerated cell death phenotype even under normal growth conditions.30 Given that similar PCD phenotype between acd11 mutant and PVA31 overexpressing plants, the overexpression of PVA31 may interfere the ACD11 function during leaf senescence in a dose dependent manner. To improve this hypothesis, a confirmation of the interaction between PVA31 and ACD11 is needed in the future study.
Avirulent and virulent P. syringae infection induced PVA31 expression within 24 h (Fig. S4). Furthermore, the expression of the pathogenesis-related (PR) gene was promptly activated with the elicitor application in the PVA31-overexpressing plants (Fig. 5). These data suggest that increased expression of PVA31 strongly induces PR gene expression. Intriguingly, Not only the expression of PR genes, but also the expression of the genes of the ER-resident secretory proteins is also up-regulated by the control of NPR1-regulated SA-signaling pathway, and the mutations in the secretory proteins result in diminishing in the secretion of PR proteins, thereby reducing the resistance to pathogen, suggesting that the enhanced expression of secretory proteins is necessary for the secretion of PR proteins for the defense reaction.31 We found that PVA31 resides on the PM and the ARA7-positive endosomes, and interacts with the PM-resident VAMPs, VAMP721/722/724. VAMP721/722 is proposed to be involved in the defense pathway against a wide variety of pathogens including a virulent oomycete Hyaloperonospora arabidopsidis, non-adapted Blumeria graminis f. sp. hordei, Erysiphe pisi and host-adapted Golovinomyces orontii powdery mildew fungi.32,33 Recently, it was reported that VAMP721/722 is stabilized by the application of flg22 and forms SNARE complex with the plasma membrane syntaxin SYP132 that is required for plant growth and immunity.34 Since SYP132 is reported to be involved in the secretion of PR proteins, the SYP132-VAMP721/722 SNARE complex might mediate the PR protein secretion.35 Although VAMP721/722 is known to form SNARE complexes with distinct PM syntaxins including PEN1, SYP122 and SYP132 to perform multiple functions in plant growth and defense reaction,33,35,36 how VAMP721/722 could execute various functions to form SNARE complexes with distinct syntaxins is largely unknown. Therefore, it is likely that the specific vesicle fusion is mediated not only by SNARE molecules but also requiring additional regulatory proteins including Rab, SM and synaptotagmin proteins.37 For instance, changes in the VAMP727 interaction with PEN1 and SYP22 is regulated by the plant-specific Rab protein Ara6 for balancing membrane fusion of intracellular compartments.22 If this is the case, PVA31 might be involved in the secretion process of PR proteins and the changes in the membrane integrity of the PM to regulate the formation of the PM-localized SNARE complex composed of VAMP721/722 and syntaxins as a SNARE accessory protein in response to pathogen infection or leaf senescence process. Of course, we cannot exclude the possibility that PVA31 could be interacted with the PM and endosomal proteins other than VAMP721/722. Future studies need to identify the interacting proteins of PVA31 in Arabidopsis.
Materials and Methods
Plant materials
Arabidopsis thaliana ecotype Columbia (Col-0) was used for all experiments in this study. Plants were grown on soil or 1/2 Murashige and Skoog (MS) salts at 22°C under a 16-h light, 8-h dark photoperiod. Transgenic Arabidopsis plants expressing the full-length PVA31 cDNA were constructed as described as follows. The full-length cDNA of PVA31 was amplified from total RNA of rosette leaves of 5-weeks-old WT Arabidopsis by RT-PCR. The amplified cDNA was subcloned into pENTR/D-TOPO vector (Invitrogen), and then the sequence of the clone was confirmed. The DNA construct was introduced into pGWB50238 by the LR reaction. Transgenic lines were screened on 1/2 MS plates containing 50μg/ml hygromycin. The candidate plants were selected by reverse transcription polymerase chain reaction (RT-PCR) and 2 independent homozygous lines expressing PVA31 under the control of the 35S CaMV promoter were used for further analysis.
Generation of PVA31-overexpressing plants
Full-length cDNAs corresponding to PVA31 were amplified by PCR, cloned into pENTR/D-TOPO vectors (Invitrogen), and subcloned into Gateway binary vectors by LR recombination (Invitrogen). For analysis of subcellular localization of PVA31, PVA31 cDNA was subcloned into pGWB402 destination vectors to generate N-terminal fusions with GFP. For gain-of-function analysis, PVA31 full-length cDNA was inserted into pGWB502 destination vectors. The constructs were transformed into Arabidopsis plants by Agrobacterium-mediated transformation using the floral dip method.39 Screening of transgenic plants was performed on 1/2MS plates containing 50 μg/ml hygromycin. Homozygous T2 transgenic lines were used for further experiments.
Transient expression of GFP-PVA31 by agrobacterium-based transformation
To transiently express GFP-PVA31 in Arabidopsis seedlings, we employed the fast Agro-mediated seedling transformation (FAST) method.39 Briefly, WT and mRFP-ARA739 and mRFP-VAMP722 expressing transgenic Arabidopsis seedlings were grown in 1/2 MS plate at 22°C under a 16-h light, 8-h dark photoperiod for 5 d and transformed by co-cultivation with A. tumefaciens GV3100 harboring the GFP-PVA31 construct in the dark for 36 hr.
Confocal microscopy
GFP fluorescence signals and differential interference contrast (DIC) images were obtained using a Nikon ECLIPSE E600 laser scanning microscope equipped with a C1 si-ready confocal system (Nikon, Tokyo, Japan) and excited with a blue argon ion laser (488 nm) for GFP-PVA31 and with a Green HeNe laser (543 nm) for mRFP-ARA7. The collected images were processed using Nikon EZ-C1 software.
Yeast two-hybrid assay
The full-length cDNAs of PVA31 and VAMPs (VAMP721/722/724/727/711) were introduced into pGADT7 and pGBKT7 vectors, respectively. The pGADT7 and pGBKT7 constructs were co-transformed into yeast AH109 strain as described in the Yeast Protocols Handbook (Clonetech). The yeasts were incubated on the solid SD medium lacking leucine and tryptophan at 30 ºC for 5 d For each 2-hybrid interaction tested, each colony was suspended in 100 μl of DW then diluted 10-, 100- and 1000-fold. Each dilution series was spotted on a solid SD medium lacking leucine, tryptophan and histidine but adding 5 mM 3-amino-1, 2, 4-triazole (3-AT). Plates were incubated for 7 d at 30 ºC.
RT-PCR and quantitative RT-PCR analyzes
For qRT-PCR, total RNA was isolated WT and PVA31-overexpressing plant with the Nucleo Spin RNA kit (TaKaRa). cDNAs were synthesized from 500 ng (for amplifying SAG12) and 100 ng (for amplifying PR-1) of total RNA of each plant using ReverTra Ace qPCR RT Master Mix (TOYOBO). Quantitative real-time PCR was performed using the THUNDERBIRD SYBR qPCR mix (TOYOBO) and Eco Real-Time PCR System (illumine) according to the manual. The expression level was normalized to an internal control gene (UBQ10, At4g05320). The primers used are listed in Supplementary Table S2.
Artificial induction of leaf senescence
Detached rosette leaves of 4-weeks-old plants were floated adaxial side up on 2 ml of 3 mM MES (pH 5.8) in 12-well Petri dishes. Leaves were incubated at 22 ºC in the dark for 6 d. Rosette leaves were collected from WT and PVA31-overexpressing plants and detached rosette leaves. The rosette leaves were extracted with 95% (v/v) EtOH at 80°C. The absorbance was measured at 649 nm, 665 nm and 750 nm. Total chlorophyll and each chlorophyll (a and b) concentrations were calculated according to.41
Supplementary Material
Author Contributions
M. H. S. conceived the study. M. I., Y. N., K.A. and S. N. carried out most of the genetic, biochemical, and physiological experiments. T. H. performed the quantitative RT-PCR analysis. M. I. performed the confocal laser scanning microscopy experiment. M. H. S. supervised the study and wrote the paper.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank T. Nakagawa (Shimane University) and T. Ueda (University of Tokyo) for generously providing the pGWB vector, and the transgenic Arabidopsis expressing mRFP-ARA7, respectively. We also thank K. Yamazaki (Kyoto Prefectural University) for technical assistances.
Funding
This work was supported by a Grant-in-Aid for Basic Science Research (C) and a Grant-in-Aid for Scientific Research on Innovative Areas (No. 25119720) from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and the Strategic Research Funds of Kyoto Prefectural University to M.H.S.
References
- 1. Lim PO, Kim HJ, Nam HG. Leaf senescence. Ann Rev Plant Biol 2007. 58:115-136; PMID:17177638; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- 2. Pennell R, Lamb C. Programmed Cell Death in Plants. Plant Cell 1997. 9:1157-1168; PMID:12237381; http://dx.doi.org/ 10.1105/tpc.9.7.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones AM. Programmed cell death in development and defense. Plant Physiol 2001. 125:94-97; PMID:11154305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2001. 2:98-106; PMID:11252968; http://dx.doi.org/ 10.1038/35052017 [DOI] [PubMed] [Google Scholar]
- 5. Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol 2008. 18:282-290; PMID:18468439; http://dx.doi.org/ 10.1016/j.tcb.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 6. Skehel PA, Martin KC, Kandel ER, Bartsch D A. VAMP-Binding Protein from Aplysia Required for Neurotransmitter Release. Science 1995. 269:1580-1583; PMID:7667638; http://dx.doi.org/ 10.1126/science.7667638 [DOI] [PubMed] [Google Scholar]
- 7. Weir ML, Xie H, Klip A, Trimble WS. VAP-A binds promiscuously to both v- and tSNAREs. Biochem Biophys Res Commun 2001. 286:616-621; PMID:11511104; http://dx.doi.org/ 10.1006/bbrc.2001.5437 [DOI] [PubMed] [Google Scholar]
- 8. Soussan L, Burakov D, Daniels MP, Toister-Achituv M, Porat A, Elazar Z. ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J Cell Biol 1999. 146:301-311; http://dx.doi.org/ 10.1083/jcb.146.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure 2005. 13:1035-1045; http://dx.doi.org/ 10.1016/j.str.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 10. Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A VAP-A interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem 2002. 277:29908-29918; PMID:12023275; http://dx.doi.org/ 10.1074/jbc.M201191200 [DOI] [PubMed] [Google Scholar]
- 11. Wyles JP, Ridgway ND. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp Cell Res 2004. 297:533-547; PMID:15212954; http://dx.doi.org/ 10.1016/j.yexcr.2004.03.052 [DOI] [PubMed] [Google Scholar]
- 12. Lehto M, Hynynen R, Karjalainen K, Kuismanen E, Hyvärinen K, Olkkonen VM. Targeting of OSBP-related protein 3 ORP3 to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp Cell Res 2005. 310:445-462; PMID:16143324; http://dx.doi.org/ 10.1016/j.yexcr.2005.08.003 [DOI] [PubMed] [Google Scholar]
- 13. Kawano M, Kumagai K, Nishijima M, Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem 2006. 281:30279-30288; PMID:16895911; http://dx.doi.org/ 10.1074/jbc.M605032200 [DOI] [PubMed] [Google Scholar]
- 14. Tuuf J, Wistbacka L, Mattjus P. The glycolipid transfer protein interacts with the vesicle-associated membrane protein-associated protein VAP-A. Biochem Biophys Res Commun 2009. 388:395-399; PMID:19665998; http://dx.doi.org/ 10.1016/j.bbrc.2009.08.023 [DOI] [PubMed] [Google Scholar]
- 15. Amarilio R, Ramachandran S, Sabanay H, Lev S. Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem 2005. 280:5934-5944; PMID:15545272; http://dx.doi.org/ 10.1074/jbc.M409566200 [DOI] [PubMed] [Google Scholar]
- 16. Sutter J, Campanoni P. Setting SNAREs in a Different Wood. Traffic 2006. 7:627-638; PMID:16683913; http://dx.doi.org/ 10.1111/j.1600-0854.2006.00414.x [DOI] [PubMed] [Google Scholar]
- 17. Laurent F, Labesse G, De Wit P. Molecular cloning and partial characterization of a plant VAP33 homologue with a major sperm protein domain. Biochem Biophys Res Commun 2000. 270:286-292; PMID:10733941; http://dx.doi.org/ 10.1006/bbrc.2000.2387 [DOI] [PubMed] [Google Scholar]
- 18. Saravanan RS, Slabaugh E, Singh VR, Lapidus LJ, Haas T, Brandizzi F. The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J 2009. 58:817-830; PMID:19207211; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03815.x [DOI] [PubMed] [Google Scholar]
- 19. Petersen NHT, Joensen J, McKinney L V, Brodersen P, Petersen M, Hofius D, Mundy J. Identification of proteins interacting with Arabidopsis ACD11. Plant Physiol 2009. 166:661-666; http://dx.doi.org/ 10.1016/j.jplph.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 20. Oufattole M, Park H, Poxleitner M, Jiang L, Rogers JC. Selective Membrane Protein Internalization Accompanies Movement from the Endoplasmic Reticulum to the Protein Storage Vacuole Pathway in Arabidopsis. Plant Cell 2005. 17:3066-3080; PMID:16227454; http://dx.doi.org/ 10.1105/tpc.105.035212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PloS One 2007. 2:e718; PMID:17684564; http://dx.doi.org/ 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nature Cell Biol 2011. 13:853-859; PMID:21666683; http://dx.doi.org/ 10.1038/ncb2270 [DOI] [PubMed] [Google Scholar]
- 23. Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J 2004. 40:783-789; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02249.x [DOI] [PubMed] [Google Scholar]
- 24. Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct and Funct 2004. 29:49-65; http://dx.doi.org/ 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- 25. Simeonova E, Sikora A, Charzyńska M, Mostowska A. Aspects of programmed cell death during leaf senescence of mono- and dicotyledonous plants. Protoplasma 2000. 214:93-101; http://dx.doi.org/ 10.1007/BF02524266 [DOI] [Google Scholar]
- 26. Yen C-H, Yang C-H. Evidence for Programmed Cell Death during Leaf Senescence in Plants. Plant Cell Physiol 1998. 39:922-927; http://dx.doi.org/ 10.1093/oxfordjournals.pcp.a029455 [DOI] [Google Scholar]
- 27. Cao J, Jiang F, Sodmergen , Cui K. Time-course of programmed cell death during leaf senescence in Eucommia ulmoides. J Plant Res 2003. 116:7-12; PMID:12605294 [DOI] [PubMed] [Google Scholar]
- 28. Obara K. Direct Evidence of Active and Rapid Nuclear Degradation Triggered by Vacuole Rupture during Programmed Cell Death in Zinnia. Plant Physiol 2001. 125:615-626; PMID:11161019; http://dx.doi.org/ 10.1104/pp.125.2.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, et al. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res 2007. 17:1030-1040; PMID:18059378; http://dx.doi.org/ 10.1038/cr.2007.100 [DOI] [PubMed] [Google Scholar]
- 30. Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J. Knockout of Arabidopsis encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 2002. 16:490-502; PMID:11850411; http://dx.doi.org/ 10.1101/gad.218202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science 2005. 308:1036-1040; PMID:15890886; http://dx.doi.org/ 10.1126/science.1108791 [DOI] [PubMed] [Google Scholar]
- 32. Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. Co-option of a default secretory pathway for plant immune responses. Nature 2008. 451:835-840; PMID:18273019; http://dx.doi.org/ 10.1038/nature06545 [DOI] [PubMed] [Google Scholar]
- 33. Collins NC, Thordal-christensen H, Lipka V. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003. 312:310-312. [DOI] [PubMed] [Google Scholar]
- 34. Yun HS, Kwaaitaal M, Kato N, Yi C, Park S, Sato MH, Schulze-Lefert P, Kwon C, et al. Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol Cells 2013. 6:481-488; http://dx.doi.org/ 10.1007/s10059-013-2130-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalde M, Nu TS, Findlay K, Peck SC. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA 2007. 10:11850-11855; http://dx.doi.org/ 10.1073/pnas.0701083104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. El Kasmi F, Krause C, Hiller U, Stierhof Y-D, Mayer U, Conner L, Kong L, Reichardt I, Sanderfoot AA, Jürgens G.SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell 2013. 24:1593-1601; PMID:23515225; http://dx.doi.org/ 10.1091/mbc.E13-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong W. SNAREs and traffic. Biochim Biophys Acta 2005. 1744:120-144; PMID:15893389; http://dx.doi.org/ 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 38. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 2007. 104:34-41; PMID:17697981; http://dx.doi.org/ 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- 39. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998. 16:735-743; PMID:10069079; http://dx.doi.org/ 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 40. Li J-F, Park E, Von Arnim AG, Nebenführ A. The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 2009. 5:6; PMID:19457242; http://dx.doi.org/ 10.1186/1746-4811-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wintermans JFGM, De Mots A. Spectrophotometric characteristics of chlorophylls a and b and their phenophytins in ethanol. Biochim Biophys Acta 1965. 109:448-453; PMID:5867546; http://dx.doi.org/ 10.1016/0926-6585(65)90170-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.