Abstract
Background/Aims
Smooth muscle cells (SMCs) characteristically express serum response factor (SRF), which regulates their development. The role of SRF in SMC plasticity in the pathophysiological conditions of gastrointestinal (GI) tract is less characterized.
Methods
We generated SMC-specific Srf knockout mice and characterized the prenatally lethal phenotype using ultrasound biomicroscopy and histological analysis. We used small bowel partial obstruction surgeries and primary cell culture using cell-specific enhanced green fluorescent protein (EGFP) mouse lines to study phenotypic and molecular changes of SMCs by immunofluorescence, Western blotting, and quantitative polymerase chain reaction. Finally we examined SRF change in human rectal prolapse tissue by immunofluorescence.
Results
Congenital SMC-specific Srf knockout mice died before birth and displayed severe GI and cardiac defects. Partial obstruction resulted in an overall increase in SRF protein expression. However, individual SMCs appeared to gradually lose SRF in the hypertrophic muscle. Cells expressing low levels of SRF also expressed low levels of platelet-derived growth factor receptor alpha (PDGFRαlow) and Ki67. SMCs grown in culture recaptured the phenotypic switch from differentiated SMCs to proliferative PDGFRαlow cells. The immediate and dramatic reduction of Srf and Myh11 mRNA expression confirmed the phenotypic change. Human rectal prolapse tissue also demonstrated significant loss of SRF expression.
Conclusions
SRF expression in SMCs is essential for prenatal development of the GI tract and heart. Following partial obstruction, SMCs down-regulate SRF to transition into proliferative PDGFRαlow cells that may represent a phenotype responsible for their plasticity. These findings demonstrate that SRF also plays a critical role in the remodeling process following GI injury.
Keywords: Gastrointestinal tract, Myocyte, Platelet-derived growth factor receptor alpha, Rectal prolapse, Serum response factor, Smooth muscle cell
Introduction
Serum response factor (SRF) is a transcription factor that plays a critical role in embryonic development as well as the growth and maintenance of all differentiated muscle types.1–3 SRF belongs to the MADS (MCM1, Agamous, Deficiens, and SRF) superfamily of transcription factors, which recognize 10-nucleotide (CC [A/T]6 GG) sequences called CArG boxes through a DNA binding domain.4 In smooth muscle cells (SMCs), SRF drives expression of contractile and cytoskeletal proteins through recognition and binding of these CArG boxes, which are primarily located in the promoter and intronic regions of most SMC-specific genes.5,6 The role of SRF as a “master switch” for contractility is further demonstrated by the loss of contractile phenotype with loss of SRF expression in muscle cells and the differentiation of fibroblasts to myofibroblasts with SRF overexpression in cell culture.7
The importance of SRF in embryonic development has been previously demonstrated in a SM22α-Cre knockout (KO) system in mice that resulted in Srf deletion in the majority of cardiomyocytes and vascular smooth muscle cells.8 Interestingly, the SM22α-Cre Srf KO mice died at embryonic day (E) 11.5 and had severe structural defects in cardiomyocytes and SMCs.
In a similar transgenic mouse model that restricts deletion of Srf to primarily SMCs through the expression cassette Myh11-Cre-EGFP, we confirm the prenatal lethality of congenital Srf deletion in GI and cardiac muscle cells. We also provide new evidence that expression of SRF in SMCs is lost in a surgically induced hypertrophy model, in human rectal prolapse tissue, and in cell culture. Importantly, we present a new finding that the loss of SRF protein expression during dedifferentiation of SMCs may be accompanied by a gain of platelet-derived growth factor receptor alpha (PDGFRα) expression during proliferative expansion. Collectively, our data further supports the critical role of SRF in contractile tissues and its loss in pathologic states.
Materials and Methods
Generation of Congenital Srf Knockout Mice
The SMC-specific congenital Srf KO mouse line, Tg(Myh11-Cre-EGFP);Srflox/lox, was generated by cross-breeding a Srf lox/lox female homozygote mouse (The Jackson Laboratory, Bar Harbor, ME, USA) with a Tg(Myh11-Cre-EGFP) male mouse9 according to procedures approved by the Institutional Animal Care and Use Committee at the University of Nevada, Reno.
Ultrasound Biomicroscopy
Pregnant dams were sedated with isoflurane and scanned with a high frequency ultrasound system containing a 32-MHz linear array transducer (VisualSonics Vevo 2100, Toronto, ON, Canada) for diagnosis of congenital heart defects. Ultrasound scanning was performed according to the manufacturer’s instructions and recorded in real-time.
Partial Obstruction Surgery
One month old transgenic mice with the genotypes Tg(Myh11-Cre-EGFP), Tg(Pdgfra-EGFP), or Tg(Myh11-Cre-EGFP); Tg(Pdgfra-EGFP) were used for intestinal partial obstruction surgeries as previously described.10 Briefly, a silicon ring was surgically placed on the distal ileum just oral to the cecum. As a modification to the previously described protocol, a 5-0 polyglycolic acid suture was placed through the ends of the silicon ring opening to secure its closing. After ∼2 weeks, the obstructed mice were sacrificed along with the sham operated control mice for histological and molecular analyses. Of note, the Tg(Myh11-Cre-EGFP) mice expressed cytoplasmic EGFP (cEGFP) in SMCs, whereas Tg(Pdgfra-EGFP) expressed nuclear EGFP (nEGFP) in PDGFRα+ cells. The surgical procedure was approved by the Institutional Animal Care and Use Committee at the University of Nevada, Reno, USA.
Histological Analysis
Srf KO and WT fetuses from gestational days 18 to 19 were fixed in Bouin’s fixative for at least 24 hours followed by washing in 70% ethanol for several days for picric acid removal. Fixed embryos were dehydrated through an alcohol gradient, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Three sections from at least 2 different embryos were analyzed. Small intestine tissues from partially obstructed and sham control animals were fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) for 1 hour 20 minutes at 4°C prior to dehydration through an alcohol gradient and H&E staining. Paraffin embedded sections of human rectal prolapse and control colon tissues were obtained from Stanford University Medical Center, Palo Alto, California, USA, where Institutional Review Board approval was obtained. Sections were examined using the iScan Coreo scanner (Ventana Medical Systems, Tucson, AZ, USA).
Immunohistochemical Analysis
Immunofluorescence microscopy of the jejunum was performed as previously described with minor modifications.11 For immunohistochemistry, tissues were fixed in 4% paraformaldehyde/PBS for 1 hour 20 minutes at 4°C prior to dehydration overnight in 20% sucrose/PBS at 4°C. The tissues were cut and placed into Tissue-Tek Crymold (Sakura Finetek, Torrence, CA, USA) containing one part Tissue-Tek OCT Compound (Sakura Finetek) and one part 20% sucrose/PBS prior to flash freezing with liquid nitrogen. Embedded molds were sectioned with a cryostat at 8 μm thickness onto glass slides and stained with primary antibodies for SRF (1:100, Cat No: sc-13029; Santa Cruz Biotechnology, Dallas, TX, USA), Ki67 (1:100, Cat No: RM-9106-S0; Thermo Scientific, Fremont, CA, USA) overnight at 4°C followed by staining with the secondary antibody Alexa Fluor 594-Conjugated AffiniPure Donkey Anti-Rabbit IgG (1:500, Cat No: 711-585-152; Jackson Immuno Research, West Grove, PA, USA) for 1 hour at room temperature prior to mounting onto slides with 4,6-diamidino-2-phenylindole (DAPI)-Prolong Gold (Cat No: P36931; Invitrogen, Carlsbad, CA, USA). Images were collected using the Fluoview FV10-ASW 3.1 Viewer software (Olympus, Tokyo, Japan) with an Olympus FV1000 confocal laser scanning microscope.
Western Blot Analysis
SRF and MYH11 protein expression in the jejunum tissues from the hypertrophy and sham mice were analyzed by Western blot. Hypotonic lysis buffer was directly applied to treated tissue, which were then grinded. Supernatants were boiled for 5 minutes, and the protein samples (30 μg) were then subjected to electrophoresis on 10% sodium dodecyl sulfate polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (0.45 μM, Cat No. 162-0115; Bio-Rad Laboratories) for overnight in cold-room. After blocking, blots were incubated overnight in a cold-room with rabbit anti-SRF (1:1000, Cat No: sc-13029; Santa Cruz Biotechnology, Dallas, TX, USA), goat anti-MYH11 (1:1500, Cat No: sc-79079; Santa Cruz Biotechnology), and monoclonal mouse anti-human smooth muscle actin (SM22α, Cat No. M0851; Dako, Glostrup, Denmark) diluted in 5% blocking buffer. The blots were then washed 3 times (10 minutes each) in TBS-T (Tris buffered saline containing 0.1% Tween 20) followed by incubation with peroxidase-conjugated AffiniPure bovine anti-goat IgG (1:5000, Cat No: 805-035-180; Jackson Immuno Research), peroxidase-conjugated AffiniPure donkey anti-mouse IgG (1:5000, Cat No: 715-035-150; Jackson Immuno Research) and peroxidase-conjugated AffiniPure donkey anti-rabbit IgG (1:5000, Cat No: 711-035-152; Jackson Immuno Research) for 1 hour. After washing, the blot was finally detected by exposure to ECL (Cat No: RPN 2135; Amersham Biosciences, Piscataway, NJ, USA) for between 10 seconds and 2 minutes.
Cell Culture
The jejunum was dissected from 3-week-old transgenic Myh11-Cre-EGFP mice, which expressed EGFP in SMCs, and the muscularis (n = 3) was gently peeled off from the mucosa and incubated for 30 minutes at 37°C in a collagenase-based dissociation solution containing 1.3 mg/mL collagenase (type II; Worthington Biochemical, Freehold, NJ, USA), 2 mg/mL bovine serum albumin (BSA; Sigma, St. Louis, MO, USA), 2 mg/mL trypsin inhibitor (Sigma), and 0.27 mg/mL ATP (Sigma), 10 mL of calcium-free Hank’s balanced salt solution (pH 7.0). The tissue was then bluntly triturated with a glass pipette until all of the tissue was broken down in the media. Single cells were obtained by using a 30 μm Pre-Separation Filter (Miltenyi Biotec, Bergisch Gladbach, Germany) and seeded in a 4-well glass slide Lab-Tek II CC2 glass chamber slide (Thermo Scientific) at a concentration of 1.9 ×106 cells/mL per well in Medium199 (Invitrogen) with 10% heat inactivated fetal bovine serum (Invitrogen), 1% antibiotic-antimycotic (Invitrogen), and incubated at 37°C with 5% CO2 for 1, 2, 4, and 8 days with daily change of fresh media. Cultured slides were used for epifluorescence imaging or harvested for quantitative polymerase chain reaction (QPCR) analysis.
Quantitative Polymerase Chain Reaction
Total RNAs were extracted from the cultured cells, cDNAs were reverse transcribed from the total RNAs, and QPCR was performed on the cDNAs as previously described.12 All primers used for RT-PCR are shown in Table.
Table.
Oligonucleotides Used for Quantitative Polymerase Chain Reaction in This Study
| Name | Sequence (5′ to 3′) | Tm | Gene | Size (c/gDNA, bp) |
|---|---|---|---|---|
| Srf-2 | CCACTGGCTTTGAAGAGCCAGATC | 59°C | Srf | 132/1997 |
| Srf-2r | CTGTCTGGATTGTGGAGGTGGTAC | 59°C | Srf | |
| Myh-1 | GATGACCTGGTCGTGGACTTGG | 59°C | Myh11 | 121/1537 |
| Myh-1r | CATCCGCATACTTGGAGGAGATG | 57°C | Myh11 | |
| Myo-5 | GTTCAGCAGTTTCAGATATCACCCAG | 58°C | Myocd | 155/4440 |
| Myo-5r | CTGAAGACATGAATGATCTTCCCTGG | 58°C | Myocd | |
| Elk-1 | GTTCAAGTTGGTGGATGCAGAGG | 57°C | Elk1 | 146/4136 |
| Elk-1r | CAAACTTCTGGCCGCTCACCTTG | 59°C | Elk1 | |
| Gapdh-1 | AATGGTGAAGGTCGGTGTGAACG | 57°C | Gapdh | 155/1989 |
| Gapdh-1r | CGTGAGTGGAGTCATACTGGAAC | 57°C | Gapdh |
bp, base pair.
Statistical Methods
Western blot and QPCR data were analyzed for statistical significance by using Student’s test. Measured variables were expressed as mean ± SEM. The differences in mean values between 2 animal groups (sham and hypertrophy) were evaluated and considered significantly different when P < 0.05 or P < 0.01.
Results
Serum Response Factor Is Required for Prenatal Gastrointestinal Tract and Cardiac Development
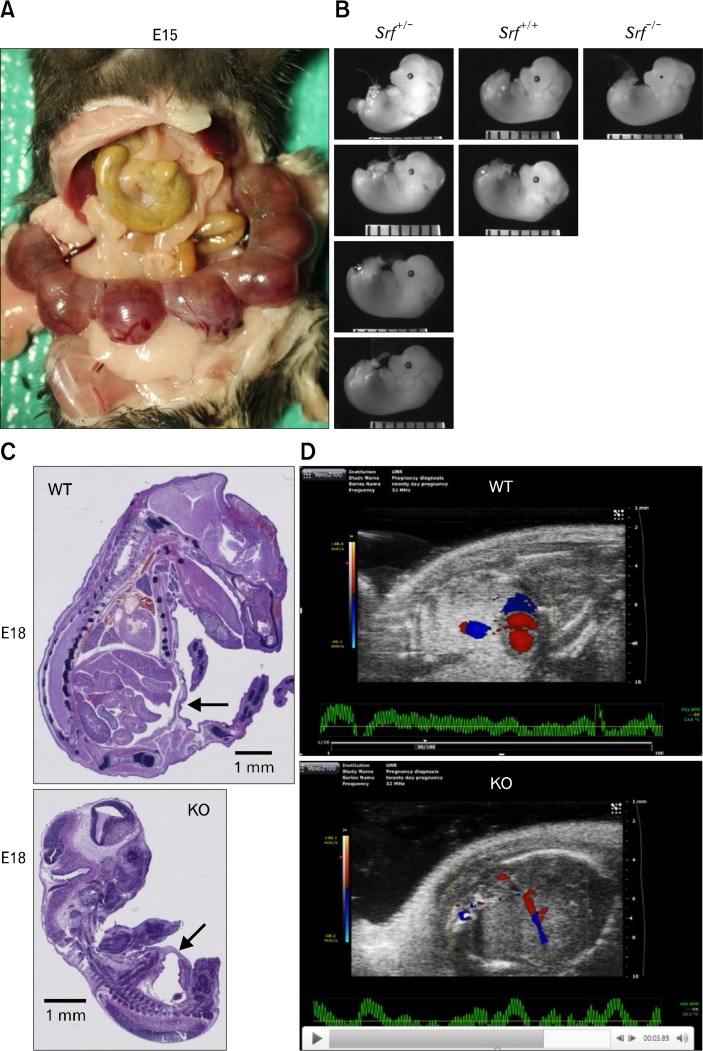
All congenital homozygous Srf KO (KO hereafter) mice died before birth while heterozygous Srf KO and wild type mice were viable. Multiple breeding combinations consisting of homozygous floxed (Srflox/lox) or heterozygous floxed (Srflox/+) mice with Cre males and females resulted in birth of ∼100 pups that were either wild type or Srf KO heterozygotes. We, therefore, examined the embryos of pregnant females. Figure 1A shows a representative mother that had 7 embryos at E15. Genotyping of the embryos identified 4 heterozygous KO (Srf+/−), 2 wild type (WT, Srf+/+), and 1 homozygous KO (Srf−/−) in agreement with Mendelian genetics. Grossly, there was no significant difference in size between homozygous KO, heterozygous KO, and WT embryos (Fig. 1B). However, sagittal cross sections of whole fetuses at E18 showed that internally, the KO fetus was grossly underdeveloped compared to the WT fetus, and the severe underdevelopment of the GI tract in the KO fetus was particularly striking (Fig. 1C).
Figure 1.
Fetal defects of congenital smooth muscle cell (SMC)-specific Srf knockout (KO). (A) A pregnant Srf KO mouse with the uterus containing 7 developing embryos at E15 days. (B) Genotypes and photomicrographs of fetuses showing wild type (WT), heterozygous (Srf+/−), and homozygous Srf KO (Srf−/−). (C) Sagittal cross sectional images of WT and KO fetuses with H&E staining at E18. Note significant global underdevelopment of the KO fetus with paucity of GI track development. (D) Doppler ultrasonography of viable WT and KO fetuses, respectively, showing red and blue areas indicating significantly lowered cardiovascular activity in KO fetus (see real-time images in Supplementary Video).
Interestingly, real-time doppler ultrasonography of the fetal heart (echocardiogram) also revealed that cardiac contractility, ejection fraction, and blood flow were significantly weaker in the KO fetus than WT fetus at E18 (Fig. 1D and Supplementary Video). The fetal defects of the congenital Srf KO mouse suggested that SRF is essential for development of the GI tract and heart as well as overall survival of the fetus in utero. In a parallel set of experiments, we also generated an inducible SMC-specific Srf KO mouse system, which showed that SRF is required for maintenance of a differentiated phenotype and prevention of degeneration and apoptosis in adult GI SMCs (unpublished data). Collectively, these animal studies also indicated that SRF is essential for development and maintenance of mature GI SMCs within the GI tract.
Serum Response Factor and PDGFRα Are Linked to Smooth Muscle Cell Hypertrophy
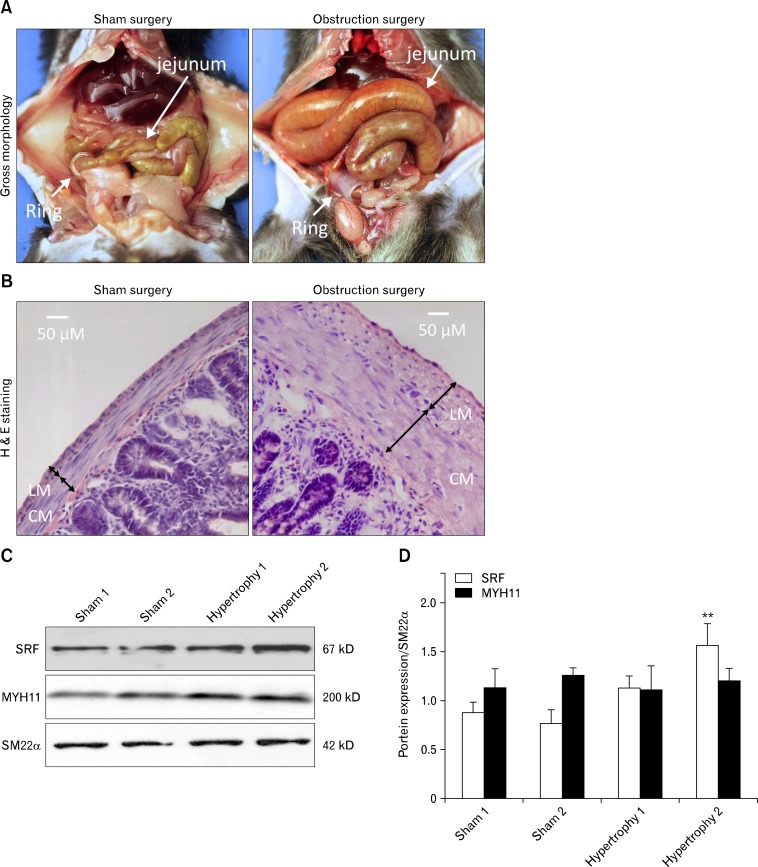
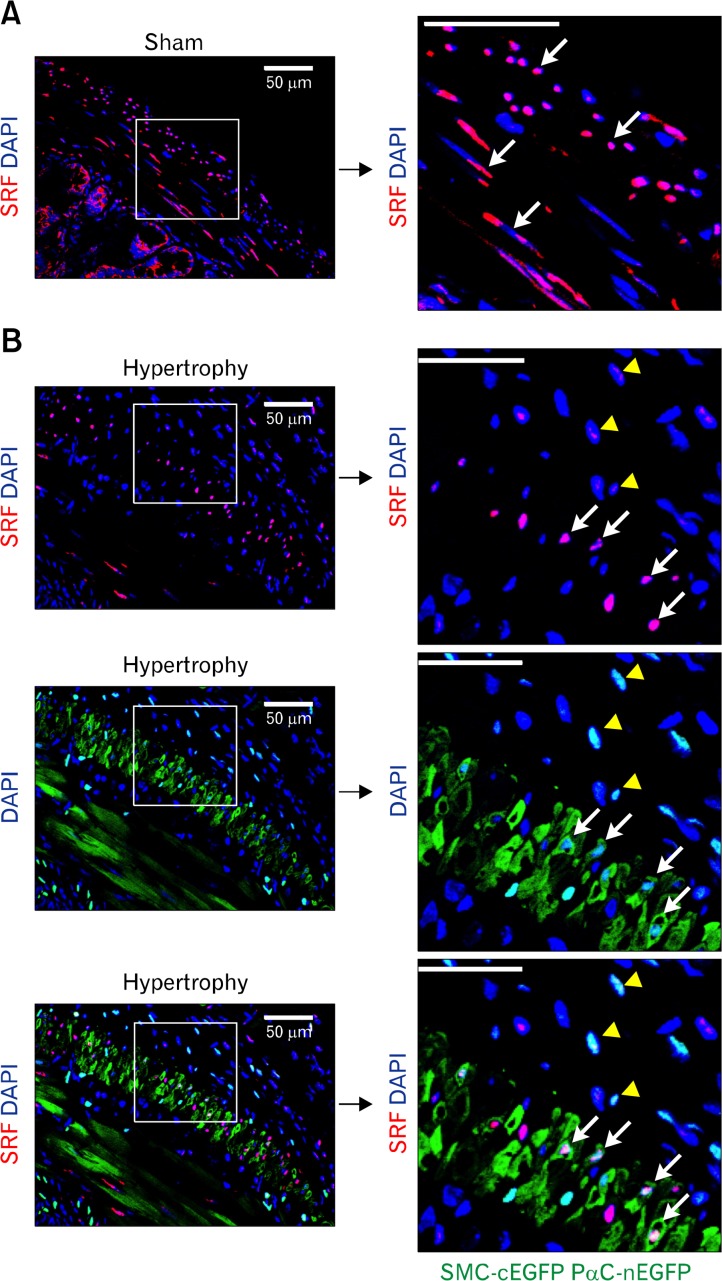
Since SMCs dedifferentiate into a proliferating phenotype and hypertrophy under pathological obstruction,13 we hypothesized that SRF expression in GI SMCs decreases during the resulting compensatory process. To test our hypothesis, we looked for changes in SRF expression in hypertrophic GI tissue using a surgically induced partial obstruction mouse model.10 Two weeks after placement of a small silicon ring on the distal ileum, the jejunum and ileum were severely distended compared to the large ring sham control (Fig. 2A). H&E staining confirmed the presence of significant hypertrophy in the longitudinal and circular muscle layers of partially obstructed jejunum (Fig. 2B), and western blot analysis showed that expression levels of SRF protein in the jejunum smooth muscle increased with partial obstruction, whereas expression levels of MYH11 and SM22α did not change (Fig. 2C and 2D). To examine the localization of SRF in the GI tract, we generated Myh11cytoplasmic EGFP; Pdgfranuclear EGFP mice and looked at SRF expression in the smooth muscle via immunofluorescence labeling. We discovered that there were at least 2 distinct populations of cells expressing SRF in the muscularis (Fig. 3A and 3B). The sham control jejunum had populations of SMCs expressing robust levels of SRF (SRFhigh cells; Fig. 3A, white arrows) within the circular and longitudinal muscle layers. However, a magnified view of the hypertrophied jejunum revealed the presence of cells expressing low levels of SRF (SRFlow cells; Fig. 3B, yellow arrowheads) adjacent to a population of SRFhigh cells (Fig. 3B, white arrows) located in the longitudinal muscle layer just outside the myenteric border. The layer of SRFhigh cells in hypertrophied jejunum was notably thinner than that of the sham control, and most of these SRFhigh cells expressed cytoplasmic EGFP (cEGFP) indicative of MYH11 expression and hence, SMC phenotype (Fig. 3B). However, these SMCs (cEGFP/SRFhigh) also expressed low levels of nuclear EGFP (nEGFP), indicating a PDGFRαlow SMC/myofibroblast-like phenotype (Fig. 3B, white arrows). In contrast to the cEGFP/SRFhigh SMCs of the longitudinal muscle, most cells in the subserosal hypertrophic region had little or no cEGFP expression along with low levels of SRF and nEGFP (PDGFRα) expression, indicating a non-SMC PDGFRαlow phenotype (Fig. 3B, yellow arrowheads). This transitional evidence suggests that SMCs dedifferentiate into PDGFRαlow cells during the hypertrophic process and is dependent on SRF expression levels.
Figure 2.
Gastrointestinal (GI) smooth muscle hypertrophy in mouse partial obstruction model. Partial obstruction related hypertrophy was surgically induced for ∼2 weeks by placing a small silicon ring on the distal ileum of mice. (A) Gross images of GI tract in sham (large ring) and obstruction (small ring) surgeries. (B) Representative H&E staining of jejunal cross sections from sham control and partially obstructed mice. Hypertrophied jejunum contained significantly thicker circular (CM) and longitudinal muscle (LM) layers compared to a sham control. (C) Western blot analysis of SRF, MYH11, and SM22α in hypertrophied and sham control jejunum. (D) Summary of Western blot analysis. Samples were run in triplicates for each animal (n = 2), and bars represent standard error of the mean. **P < 0.01.
Figure 3.
Differentiated serum response factor (SRF)high smooth muscle cells (SMCs) become SRFlow platelet-derived growth factor receptor alpha positive (PDGFRα+) cells during hypertrophy. Representative confocal laser scanning images of jejunal cross sections from sham control (A) and partial obstruction (B) mice following immunohistochemical staining with anti-SRF antibody (red) and counterstaining with DAPI (blue). Note robust levels of SRF expression in SMC nuclei of sham control (A; SRFhigh cells, white arrows) compared to significantly decreased SRF expression in hypertrophied muscle layers (B; SRFlow cells, yellow arrowheads). Panels on right are high-resolution images magnified from the square boxes. Cytoplasmic enhanced green fluorescent protein (cEGFP; MYH11) is expressed in differentiated SMCs while nuclear EGFP (nEGFP; PDGFRα) is expressed in PDGFRα+ cells (PαC) from double transgenic Myh11-Cre-EGFP; Pdgfra-EGFP mice. Note that longitudinal SMCs expressing cEGFP with high levels of SRF also express low levels of nEGFP (SRFhigh/cEGFP [MYH11]high/nEGFP [PDGFRα]low, white arrows]. These cells are located in the vicinity of SRFlow/cEGFP (MYH11)−/nEGFP (PDGFRα)low cells in the subserosal area of hypertrophied smooth muscle (yellow arrowheads). The latter may represent a transitional phenotype between mature SMCs and PDGFRα+ cells. All scale bars are 50 μm.
Smooth Muscle Cells May Dedifferentiate and Gain PDGFRα Expression to Proliferate During Intestinal Smooth Muscle Hypertrophy and Hyperplasia
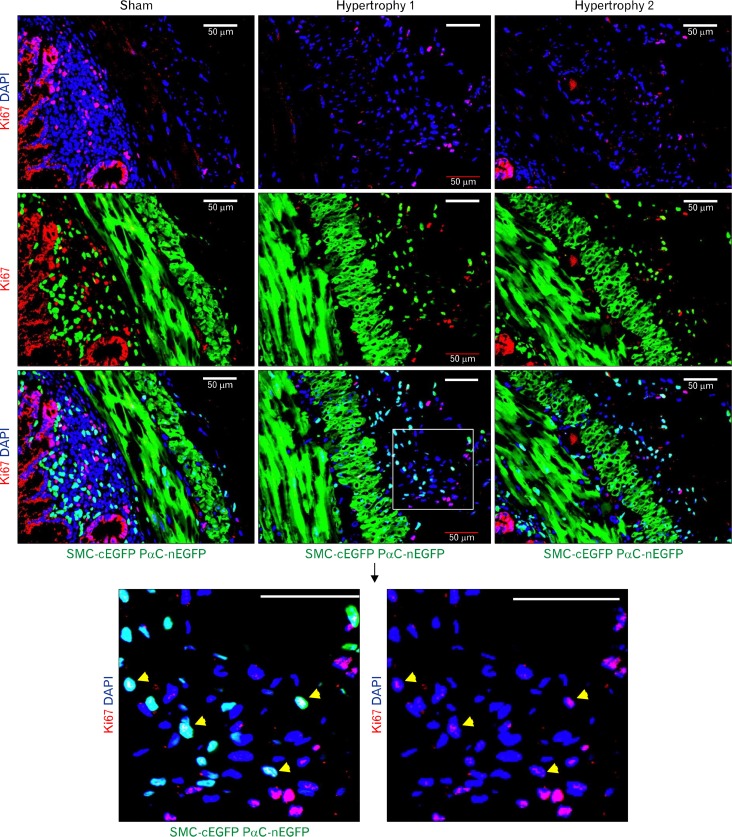
In both surgically obstructed and sham control mice, there was little proliferative activity, as detected by anti-Ki67 staining, in differentiated SMCs expressing cEGFP (Fig. 4). However, cells expressing low levels of nEGFP (PDGFRα) in hypertrophied jejunum, which were located just outside of the longitudinal muscle layer (subserosal area), displayed higher levels of cell cycling although the Ki67 signal was relatively weaker in this population of cells (Fig. 4, bottom panels). Other areas of the cross section that showed high levels of Ki67 staining included the mucosal epithelia and submucosa in both obstructed and sham control jejunum (Fig. 4).
Figure 4.
Dedifferentiated platelet-derived growth factor receptor alpha (PDGFRα+) cells proliferate during gastrointestinal smooth muscle hypertrophy and hyperplasia. Proliferative activity as detected by anti-Ki67 antibody staining in a sham control and partially obstructed jejunal tissues from double transgenic Myh11-Cre-EGFP; Pdgfra-EGFP mice. In sham controls, cycling cells are abundant in the mucosal epithelia and submucosal regions. However, smooth muscle layers show little proliferative activity. Two hypertrophic jejunal tissues samples show high proliferative activity in the hypertrophic region just outside of the longitudinal muscle layers. Bottom panels are high-resolution images magnified from the square boxes that show nuclear colocalization of Ki67, nEGFP, and DAPI signals. Note Ki67 positive cells also expressing low levels of PDGFRα (nEGFP; yellow arrow heads). All scale bars are 50 μm.
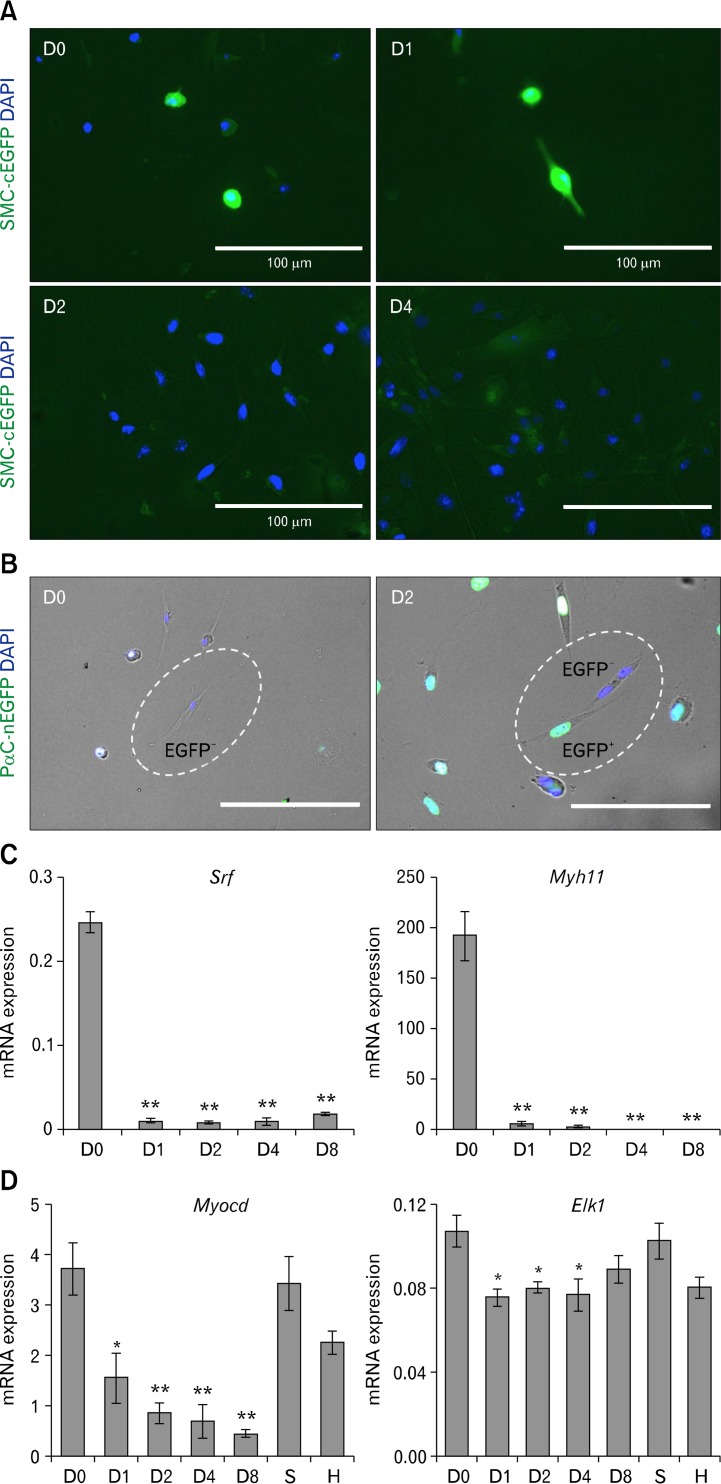
To recapture the possible phenotypic switch from SMCs to PDGFRα+ cells in vitro, we cultured primary jejunal SMCs from Myh11-Cre-EGFP or Pdgfra-EGFP mice for up to 8 days (Fig. 5). cEGFP expression in differentiated SMCs from Myh11-Cre-EGFP mice was detectable by epifluorescence microscopy at day 0 and day 1 of cell culture but was abruptly lost by day 2 (Fig. 5A). In contrast, dispersed SMCs from Pdgfra-EGFP mice began to express nEGFP at day 2 indicating a phenotypic transition to PDGFRα+ cells (Fig. 5B). QPCR analysis of the dispersed cells in culture at differing time points revealed that transcription of Srf and its target gene Myh11 essentially shut down within 24 hours of incubation (Fig. 5C). Srf expression began to resume at low levels on day 8, whereas Myh11 expression remained silent. The data indicated that both Srf and Myh11 genes turned off quickly in cell culture conditions as SMCs began to proliferate and that the residual Myh11 promoter driven cEGFP remained in SMCs up to 1 day following silencing of the genes. The cell culture and QPCR data also showed that SMCs grown in culture may lose contractile proteins, such as MYH11, with the gain of PDGFRα expression as early as day 2 of culture and that the SRFlow cells expressing PDGFRα seen in the partial obstruction model (Figs. 3 and 4) may represent a population of cells that may have dedifferentiated from SMCs to proliferate as a hypertrophic and hyperplastic response.
Figure 5.
Loss of Srf, Myocd, and Myh11 with gain of platelet-derived growth factor receptor alpha (PDGFRα) expression in dedifferentiated smooth muscle cells (SMCs) during cell culture. Jejunal smooth muscle was dissected from 2-week-old Myh11-Cre-EGFP or Pdgfra-EGFP mice, and the cells were dispersed and cultured for 8 days. At designated time points, cultured cells were examined by epifluorescence imaging and/or harvested for quantitative polymerase chain reaction (QPCR) analysis. (A) Epifluorescence imaging of SMCs from a Myh11-Cre-EGFP mouse showing loss of cEGFP (green) after day 2 in culture. DAPI (blue) marks nuclei in cells. (B) Epifluorescence imaging showing gain of nEGFP (green) expression in SMCs from a Pdgfra-EGFP mouse after 2 days in culture indicating activation of PDGFRα expression in SMCs. (C) QPCR analysis showing immediate and dramatic loss of Srf and Myh11 mRNA transcripts in primary SMCs after day 1 of culture. (D) QPCR analysis showing gradual loss of Myocd mRNA transcripts and sustained expression of Elk1 in primary SMCs after day 1 of growth in culture, in hypertrophic jejunum (H), and in sham controls (S). Expression of each gene was normalized by housekeeping gene, Gapdh (n = 3). *P < 0.05, **P < 0.01.
The vascular SMC phenotype is regulated by SRF and its cofactors myocardin (MYOCD) and ELK1, an ETS domain-containing protein.14 Therefore, we also examined the Myocd and Elk1 mRNA transcript levels of cultured SMCs and hypertrophied jejunum smooth muscle. Myocd expression declined gradually during cell culture and was also significantly decreased in hypertrophic tissue (Fig. 5D). However, expression levels of Elk1 mRNA did not change significantly in cell culture although there was a decrease in hypertrophic tissue. Collectively, the QPCR data was consistent with the dedifferentiated proliferative phenotype of SRFlow/nEGFP(PDGFRα)low cells in Figures 5A, 5B, and 3B and indicated that SRF by itself may directly modulate the SMC phenotype.
Loss of Serum Response Factor in Smooth Muscle Cells Is Implicated in the Pathophysiology of Rectal Prolapse in Humans
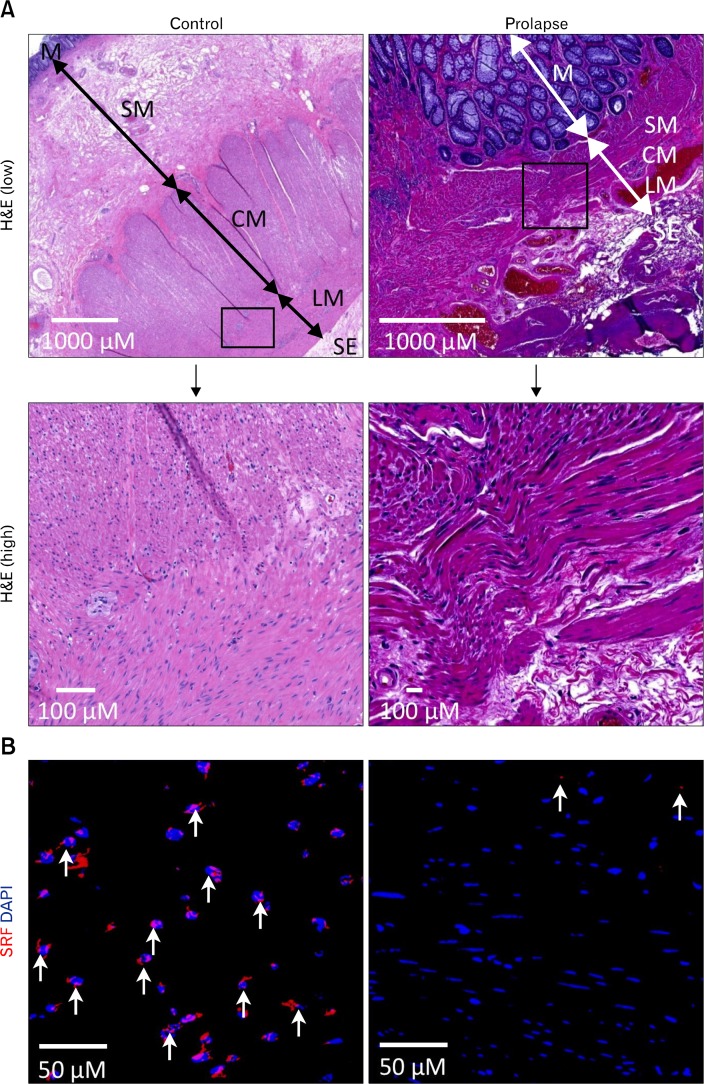
To assess the relevance of our studies to human GI pathologies, we examined expression of SRF in human colorectal prolapse tissue. The histological morphology of healthy colon was well organized with distinct circular and longitudinal muscle layers present in between the submucosa and subserosa (Fig. 6A). In contrast, prolapse tissue contained disorganized and dysplastic muscle layers that were in abnormal contact with the mucosa juxtaposed to a highly disorganized and dysplastic serosa layer. Robust levels of SRF expression were detected by immunohistochemistry in the nuclei of SMCs of control colon but barely detectable in rectal prolapse tissue (Fig. 6B). These findings mirrored those of the mouse partial obstruction model (Fig. 3B). Taken together, our human rectal prolapse tissue studies support the hypothesis that SRF is required for SMC differentiation and maintenance and that pathological loss of SRF in SMCs may trigger SMC dedifferentiation and growth leading to hypertrophy and hyperplasia in the GI smooth muscle.
Figure 6.
Loss of serum response factor (SRF) protein expression in dysplastic smooth muscle cells (SMCs) of human rectal prolapse tissue. (A) H&E staining of control colon and rectal prolapse tissue. Control colon sections show a well-organized muscularis with distinctive circular (CM) and longitudinal muscle (LM) layers, whereas prolapse tissue contains a disorganized and disrupted muscularis. (B) Immunohistochemical staining of SRF in control colon and rectal prolapse tissues. SMCs in control colon express abundant SRF (red) in nuclei counterstained with DAPI (blue). Rectal prolapse SMCs display minimal SRF expression (n = 3). M, mucosa; SM, submucosa; SE, serosa.
Discussion
To our knowledge, this is the first animal study that has demonstrated a developmental defect of the GI smooth muscle caused by a congenital Srf deficiency. SRF is an essential regulator for muscle cell proliferation and differentiation in skeletal,15 cardiac,16 and smooth muscle,5 and a global Srf gene mutagenesis results in a severe gastrulation defect that is embryonic lethal.1 Cardiac-specific (Myh6 promoter driven) and vascular-specific (SM22α promoter driven) knockouts of Srf are also embryonic lethal due to a severe cardiac defect.8,17 The expression of SM22α (Tagln) is relatively restricted to vascular SMCs in adults18 in addition to cardiac muscle cells in embryos.19 However, SM22α is also expressed in proliferating and differentiated SMCs20 as well as PDGFRα+ cells in GI smooth muscle (transcriptome analysis, unpublished data). To target SMCs specifically, we generated a Myh11 promoter driven Srf KO mouse model. MYH11, which is expressed in SMCs of the GI tract by E13.5,9 is widely considered the most selective marker for differentiated SMCs.21 In this KO system, the Srf gene is deleted by Cre under the control of the Myh11 promoter at E13.5. However, SRF protein synthesized prior to E13.5 may continue to reside within cells for several days. In our congenital studies, the cardiac and GI defects from Myh11 promoter driven Srf KO manifested at E18, which is later than the time of manifestation for Myh6 (E12.5) and SM22α (E11.5) promoter driven KOs.8,17 This phenotypic discrepancy may be partially explained by the differential timing of promoter activation in different cell populations during development. Since MYH11 is the most distinctive marker for differentiated SMCs, the prenatal GI and cardiac defects revealed in this study most accurately represent the phenotypes that result from congenital deletion of Srf in SMCs. The manifestation of cardiac dysfunction was not anticipated since MYH11 is not expressed in cardiomyocytes. However, it is conceivable that the Srf KO may have disrupted the normal vascularization of the developing heart in utero since MYH11 is expressed in vascular SMCs. Our congenital Srf KO studies along with the 2 previously described phenotypes emphasize the critical nature of SRF in the development of prenatal GI smooth muscle and cardiac muscle.
The phenomena of SMC hypertrophy and hyperplasia, which occurs during partial bowel obstruction, has been previously well described.13,22,23 Inflammation may also cause SMC hypertrophy within the GI tract.24,25 However, the molecular mechanism remains less understood. Our in vivo and in vitro data indicated that under the physiologic stress of partial obstruction, intestinal SMCs may lose expression of SRF and characteristic contractile proteins in favor of a dedifferentiated phenotype capable of proliferation. In vascular SMCs, SRF regulates expression of contractile proteins mainly through its cofactors MYOCD (differentiation) and ELK1 (dedifferentiation).14 In contrast, however, our data in Figures 3 and 5 suggest that the SMC phenotype in intestinal obstruction may be regulated directly by SRF rather than via ELK1. Elk1 expression was minimal at the beginning and did not increase in cell culture and hypertrophied jejunal smooth muscle (Fig. 5) although previous studies have reported ELK1 induction in vascular injury.26 In this vascular injury model, phosphorylated ELK1 repressed the SM22α gene. However, in our partial intestinal obstruction model, SM22α protein levels were not affected by hypertrophy of the jejunal smooth muscle (Fig. 2C). In addition, our transcriptome data showed that little or no Elk1 was expressed in both jejunal SMCs and colonic SMCs, whereas Srf was abundantly expressed in a SMC-specific manner.27 These observations support a model for direct SRF regulation of the GI SMC phenotype during intestinal obstruction, which differs from that of vascular SMCs.
Whether expression of SRF and contractile proteins (myosin heavy chains) increases or decreases with intestinal smooth muscle hypertrophy has been the subject of controversy.13,28 The discrepancy between contradictory findings by different investigators may be explained by the dynamic stages of the hypertrophic response at which the tissues were examined. For instance, Chen et al13 found that SRF and contractile protein in whole muscle tissue were down-regulated at the beginning of hypertrophy but later restored to the control levels. Our Western blot data indicated that SRF levels increased after 2 weeks of partial obstruction but that MYH11 levels remained unchanged. The immunohistochemistry actually showed a thinner band of SMCs expressing high levels of SRF in hypertrophied tissue relative to sham controls. Furthermore, we identified 2 distinctive SMC populations that appeared to be responsible for these changes: 1) mature hypertrophic SMCs that are SRFhigh/cEGFP(MYH11)high located near the myenteric border and 2) less differentiated SMCs that are SRFlow or ‒/cEGFP(MYH11)low or ‒ juxtaposed to the longitudinal smooth muscle layer. Collectively, our data indicated that after 2 weeks of intestinal partial obstruction, there were overall more cells (SMCs and hyperplastic PDGFRαlow cells) expressing SRF, albeit at lower levels. The increased number of cells expressing SRF resulted in a higher total amount of SRF expression compared to non-obstructed intestinal tissue.
A hybrid phenotype between fibroblasts (similar to PDGFRα+ cells) and SMCs (myofibroblast) has been reported in fibrosis of Crohn’s disease,29 and vascular SMCs have been shown to display plasticity from a quiescent contractile phenotype to a myofibroblast-like proliferative synthetic state during vascular repair.6 However, it has been technically difficult to validate and study dedifferentiated SMCs since they lose their distinctive cell markers. We partially overcame this limitation with a double transgenic mouse line that labeled SMCs with cEGFP and PDGFRα+ cells with nEGFP. Using this murine intestinal injury model, we report the first in vivo evidence of mature SMCs converting into proliferative PDGFRα+ cells. This finding offers a new opportunity to isolate and study dedifferentiated SMCs after loss of their characteristic markers.
Interestingly, we discovered that the dedifferentiated SRFlow or ‒/ cEGFP(MYH11)low or ‒ cells in hypertrophic jejunum robustly expressed the proliferative marker Ki67 and low levels of PDGFRα (nEGFPlow) implicating their role in smooth muscle hyperplasia. Although it is possible that these SRFlow or ‒/cEGFP(MYH11)low or ‒/nEGFP(PDGFRα)low cells could represent fibroblast-like cells in situ that may be differentiating into mature SMCs to compensate for the increased physiologic stress of partial obstruction, our cell culture and immunofluorescence data (thinner band of SRF+ SMCs) indicate that this cell population likely represents SMCs that have dedifferentiated to accommodate for cellular proliferation and creation of more mature SMCs. The discovery of nEGFP(PDGFRα)low cells was particularly intriguing because these cells were a separate and distinct population from that of nEGFP(PDGFRα)high cells and had higher levels of Ki67 expression. The latter finding suggested that nEGFP(PDGFRα)low cells may be more primitive than nEGFP (PDGFRα)high cells. Moreover, previous studies have implicated PDGF-AA, PDGF-BB, and their receptors in the proliferation of intestinal subepithelial myofibroblasts (ISEMFs) although a similar interplay in the subserosa has been less characterized.29,30
Although we have demonstrated that nEGFP(PDGFRα)low cells may derive from dedifferentiated SMCs, other potential sources for these cells include subserosal PDGFRα+ cells,31,32 myofibroblasts,29,33 or bone marrow-derived mesenchymal stem cells.34,35 In the GI tract, PDGFRα+ cells have a similar anatomical distribution as interstitial cells of Cajal (ICC), both of which have close contact with SMCs.36 Within the small intestine, PDGFRα+ cells are grouped into the subtypes PDGFRα+-IM (muscle bundles), PDGFRα+-MY (myenteric plexus), PDGFRα+-DMP (deep muscular plexus), and PDGFRα+-SS (subserosal surface).31,37 We found that each subtype was differentially remodeled during partial intestinal obstruction. However, further studies are needed to determine whether any of these sub-types can display plasticity in the remodeling response.
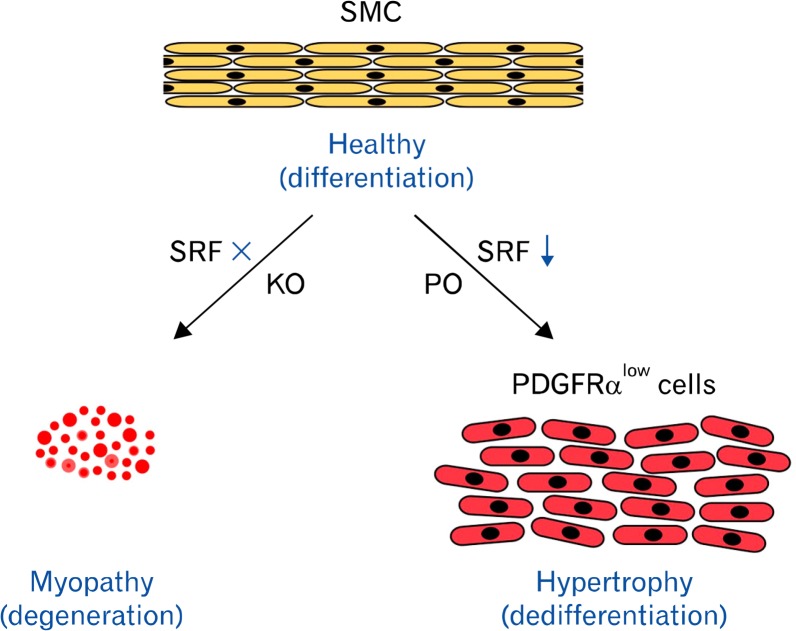
Finally, we have also demonstrated the pathologic loss of SRF expression in human rectal prolapse tissue. The question of whether loss of SRF in human SMCs is associated with a gain of PDGFRα expression and proliferative activity as is in mice should be addressed by further studies. In summary, we have shown that SRF is required for the development of prenatal SMCs and the maintenance of its differentiated phenotype in adult SMCs. The loss or down-regulation of SRF is directly linked to the pathological transitions of SMCs, which includes degeneration during myopathy or proliferation of PDGFRlow cells during obstruction induced hypertrophy (Fig. 7). This study offers new insight into the direct role of SRF in the plasticity of pathologic SMCs.
Figure 7.
Model of smooth muscle cell (SMC) phenotype regulated by SRF. In absence (knockout [KO]) of serum response factor (SRF), SMCs cannot grow and differentiate, but they degenerate and undergo apoptosis. With down-regulation of SRF during intestinal injury (partial obstruction [PO]), SMCs dedifferentiate into proliferative SRFlow PDGFRαlow cells, which result in hypertrophy of smooth muscle. PDGFRα, platelet-derived growth factor receptor alpha.
Supplementary Materials
Note: To access the supplementary Video mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at http://dx.doi.org/10.5056/jnm15063.
Acknowledgments
The authors would like to thank Michael I. Kotlikoff for his kind donation of congenital Cre mouse lines, Lab Animal Medicine for trangenic mouse lines, and Nevada INBRE for the undergraduate research opportunities fund.
Footnotes
Financial support: This study was supported by the NIH grants R01DK103055 to Seungil Ro, P20 GM10351 and P01 DK41315 to Seungil Ro and Kenton M Sanders; Nevada INBRE funds, 5P20RR0146464-11 and 8 P20GM103440-11.
Conflicts of interest: None.
Author contributions: Chanjae Park, Moon Young Lee, Paul J Park, Se Eun Ha, Robyn M Berent, and Robert Fuchs, obtaining and/or analyzing data; Laren S Becker, interpreting data and providing human tissue samples; Joseph M Miano and Kenton M Sanders, interpreting data and reviewing the manuscript; and Seungil Ro, designing, directing, and writing the manuscript.
ORCID: Seungil Ro, http://orcid.org/0000-0003-0861-8334.
References
- 1.Arsenian S, Weinhold B, Oelgeschläger M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Liu Z, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 3.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 4.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 5.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/S0022-2828(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 6.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 7.Davis J, Molkentin JD. Myofibroblasts: trust your heart and let fate decide. J Mol Cell Cardiol. 2014;70:9–18. doi: 10.1016/j.yjmcc.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miano JM, Ramanan N, Georger MA, et al. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci USA. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics. 2002;10:211–215. doi: 10.1152/physiolgenomics.00054.2002. [DOI] [PubMed] [Google Scholar]
- 10.Chang IY, Glasgow NJ, Takayama I, Horiguchi K, Sanders KM, Ward SM. Loss of interstitial cells of Cajal and development of electrical dysfunction in murine small bowel obstruction. J Physiol. 2001;536(Pt 2):555–568. doi: 10.1111/j.1469-7793.2001.0555c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ro S, Park C, Jin J, et al. A model to study the phenotypic changes of interstitial cells of Cajal in gastrointestinal diseases. Gastroenterology. 2010;138:1068–1078. e1–e2. doi: 10.1053/j.gastro.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park C, Hennig GW, Sanders KM, et al. Serum response factor-dependent MicroRNAs regulate gastrointestinal smooth muscle cell phenotypes. Gastroenterology. 2011;141:164–175. doi: 10.1053/j.gastro.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Chen H, Sanders KM, Perrino BA. Regulation of SRF/CArG-dependent gene transcription during chronic partial obstruction of murine small intestine. Neurogastroenterol Motil. 2008;20:829–842. doi: 10.1111/j.1365-2982.2008.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 15.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 16.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micro-managing cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu Z, Yu W, Zhang SX, et al. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J Biol Chem. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Miano JM, Cserjesi P, Olson EN. SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ Res. 1996;78:188–195. doi: 10.1161/01.RES.78.2.188. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Miano JM, Mercer B, Olson EN. Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solway J, Seltzer J, Samaha FF, et al. Structure and expression of a smooth muscle cell-specific gene, SM22 alpha. J Biol Chem. 1995;270:13460–13469. doi: 10.1074/jbc.270.22.13460. [DOI] [PubMed] [Google Scholar]
- 21.Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–812. doi: 10.1161/01.RES.75.5.803. [DOI] [PubMed] [Google Scholar]
- 22.Gabella G. Hypertrophy of visceral smooth muscle. Anat Embryol. 1990;182:409–424. doi: 10.1007/BF00178906. [DOI] [PubMed] [Google Scholar]
- 23.Bertoni S, Gabella G. Hypertrophy of mucosa and serosa in the obstructed intestine of rats. J Anat. 2001;199(Pt 6):725–734. doi: 10.1046/j.1469-7580.2001.19960725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blennerhassett MG, Vignjevic P, Vermillion DL, Collins SM. Inflammation causes hyperplasia and hypertrophy in smooth muscle of rat small intestine. Am J Physiol. 1992;262(6 Pt 1):G1041–G1046. doi: 10.1152/ajpgi.1992.262.6.G1041. [DOI] [PubMed] [Google Scholar]
- 25.Masaoka T, Vanuytsel T, Vanormelingen C, et al. A spontaneous animal model of intestinal dysmotility evoked by inflammatory nitrergic dysfunction. PLoS One. 2014;9:e95879. doi: 10.1371/journal.pone.0095879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ Res. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MY, Park C, Berent RM, et al. Smooth muscle cell genome browser: enabling the identification of novel serum response factor target genes. PLoS One. 2015;10:e0133751. doi: 10.1371/journal.pone.0133751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lofgren M, Fagher K, Wede OK, Arner A. Decreased shortening velocity and altered myosin isoforms in guinea-pig hypertrophic intestinal smooth muscle. J Physiol. 2002;544(Pt 3):707–714. doi: 10.1113/jphysiol.2002.027060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol. 1999;277(2 Pt 1):C183–C201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson L, Lindahl P, Heath JK, Betsholtz C. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- 31.Chan F, Liu Y, Sun H, et al. Distribution and possible role of PDGF-AA and PDGFR-α in the gastrointestinal tract of adult guinea pigs. Virchows Arch. 2010;457:381–388. doi: 10.1007/s00428-010-0946-0. [DOI] [PubMed] [Google Scholar]
- 32.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 33.Brittan M, Hunt T, Jeffery R, et al. Bone marrow derivation of pericryptal myofibroblasts in the mouse and human small intestine and colon. Gut. 2002;50:752–757. doi: 10.1136/gut.50.6.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deans RJ, Moseley AB. Mesenchymal stem cells: biology and potential clinical uses. Exp Hematol. 2000;28:875–884. doi: 10.1016/S0301-472X(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 35.Ng F, Boucher S, Koh S, et al. PDGF, TGF-β, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 36.Sanders KM, Koh SD, Ro S, Ward SM. Regulation of gastrointestinal motility--insights from smooth muscle biology. Nat Rev Gastroenterol Hepatol. 2012;9:633–645. doi: 10.1038/nrgastro.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blair PJ, Rhee PL, Sanders KM, Ward SM. The significance of interstitial cells in neurogastroenterology. J Neurogastroenterol Motil. 2014;20:294–317. doi: 10.5056/jnm14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: To access the supplementary Video mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at http://dx.doi.org/10.5056/jnm15063.