Abstract
A 55-year-old man with well-controlled HIV had severe diarrhea for 3 weeks and developed multiorgan dysfunction and bacteremia due to Escherichia coli. The genome of the patient's isolate had features characteristic of extraintestinal pathogenic E. coli and genes distantly related to those defining enteropathogenic E. coli.
Keywords: case report, comparative genomics, virulence factors, type 3 secretion, hemolysin
A novel isolate of Escherichia coli with features of extraintestinal and enteropathogenic strains was isolated from the blood of a man with multiorgan dysfunction following 3 weeks of diarrhea.
Graphical Abstract Figure.
A novel isolate of Escherichia coli with features of extraintestinal and enteropathogenic strains was isolated from the blood of a man with multiorgan dysfunction following 3 weeks of diarrhea.
Escherichia coli colonizes most neonates within hours of birth, likely during delivery (Bettelheim et al. 1974). Escherichia coli is notoriously dual natured. Most strains coexist with their host without incident, while others are highly pathogenic. The difference in the ability of strains to cause disease resides in specific genes encoding virulence factors, the acquisition of which often depends on horizontal transfer. The core genome shared among all E. coli strains amounts to only ∼40% of its average 5000 gene total, while the total number of genes that exist in all E. coli strains is predicted to exceed 15 000 (Rasko et al. 2008; Touchon et al. 2009). While the nomenclature of E. coli remains in a state of flux as our understanding evolves, a variety of pathotypes have been described (Kaper, Nataro and Mobley 2004; Croxen et al. 2013).
Escherichia coli can cause disease at many sites outside the gastrointestinal tract, including the urinary tract, meninges, biliary tract, peritoneum, lungs, and skin and soft tissue (Russo and Johnson 2003). Strains that cause these infections often share several known and suspected virulence factors, and thus have been grouped into a single pathotype called extraintestinal pathogenic E. coli (ExPEC) (Russo and Johnson 2000). The known and suspected virulence factors common among ExPEC, whether from urinary tract infections (UTIs) or other sites, include capsule, iron-uptake systems and a variety of pili, most notably P fimbriae (Russo and Johnson 2000). Serotyping, randomly amplified polymorphic DNA genomic profiling, multilocus sequence typing and pulsed field gel electrophoresis reveal that closely related strains from the same lineage can cause UTIs and infections at other sites (Johnson and Russo 2002; Johnson et al. 2013), providing the rationale that ExPEC is an appropriate category.
Strains typically associated with diarrheal illness include enteropathogenic (EPEC), enterohemorrhagic (EHEC), enterotoxigenic (ETEC), enteroaggregative (EAEC) and enteroinvasive E. coli (EIEC) (Kaper, Nataro and Mobley 2004; Croxen et al. 2013). The clinical syndrome caused by infection varies among these organisms, but diarrhea is the predominant symptom. Very rarely, isolates from E. coli pathotypes typically associated with enteric infections have caused infections outside the gastrointestinal tract. For example, case reports of UTIs due to diffuse-adhering E. coli, Shiga-toxin-producing E. coli (STEC) and EAEC have been reported (Tarr et al. 1996; Germani et al. 1997; Olesen et al. 2012). Bacteremia secondary to such strains is even less common, with only a few such reports in the literature. Recently, Herzog et al. (2014) reported a case of an EAEC strain causing UTI and bacteremia in a renal transplant patient on immunosuppressive therapy. There has also been a report of EIEC bacteremia, in a patient with AIDS (Bessesen et al. 1991). Here we report the draft genome sequence of a strain that caused bacteremia in an adult with severe diarrhea.
A 55-year-old man with well-controlled HIV was admitted to the Veterans Affairs Maryland Health Care System in Baltimore, Maryland, with a 3-week history of progressive diarrhea and cramping abdominal pain. By the time of presentation, he was having 10–15 watery, non-bloody bowel movements per day. The pain was diffuse throughout the abdomen. He denied nausea, vomiting, subjective fever and chills.
His HIV had been treated for the past six years with atazanavir and lopinavir/ritonavir, with an undetectable viral load and recent CD4 count of 271 mm−3. He had chronic hepatitis C infection. He was a current tobacco user and had a remote history of cocaine use.
His vital signs and oxygen saturation were normal. Orthostatic blood pressure and pulse were not recorded. He was cachectic and ill appearing with dry mucous membranes. His abdomen was soft, but tender in all quadrants. His peripheral white blood cell count was elevated (Table S1, Supporting Information) with 91% neutrophils and 2% band forms. He was anemic and had hyponatremia (120 mEq/L) and acute kidney injury. His liver function parameters were also abnormal with marked hyperbilirubinemia.
Computed tomography scan without intravenous contrast revealed non-specific gallbladder wall thickening and mild ascites. Magnetic resonance cholangiopancreatography showed no abnormality of the biliary tree. Later in his hospitalization, he also underwent liver biopsy, which revealed moderate to marked activity with early cirrhosis (modified Batts–Ludwig Classification: Grade 3–4, Stage 3–4) in addition to marked chronic cholestasis.
Fecal Clostridium difficile toxin gene PCR was negative. Fecal tests for leukocytes and ova and parasites were also negative. Stool culture revealed heavy growth of an oxidase-negative, non-lactose fermenting Gram-negative bacillus, but tests for Salmonella, Shigella, Yersinia, Campylobacter, Aeromonas, Plesiomonas and Vibrio species were negative and the isolate was discarded.
At presentation, he received multiple liters of intravenous fluid and his antiretroviral medication was held in consideration of his hepatic insult. Blood cultures drawn in the emergency department grew a non-lactose fermenting Gram-negative rod. A suspected diagnosis of Salmonella bacteremia was excluded when biochemical testing of the blood isolate indicated E. coli (API® 20E code 5144502 Biomerieux, Hazelwood, MO). Although the strain did not ferment lactose, it converted tryptophan to indole and displayed lysine decarboxylase activity.
Antibiotic therapy with piperacillin-tazobactam was initiated on admission. Based on sensitivities of the blood isolate, antibiotic coverage was narrowed to ciprofloxacin. The patient's diarrhea resolved, his metabolic parameters began to return to baseline and his antiretroviral medications were resumed. He was discharged 7 days after his presentation to finish a 2-week course of oral ciprofloxacin. On follow-up 1 month later, he felt well, his liver enzymes and total bilirubin were normal. His creatinine had improved, but remained elevated.
In view of the patient's bacteremia, we tested the ability of the strain isolated from blood (VACI-14) to invade epithelial cells using a standard gentamicin protection assay (Donnenberg, Donohue-Rolfe and Keusch 1989). As controls, we included a human commensal isolate and an EIEC isolate (Donnenberg, Donohue-Rolfe and Keusch 1989). We detected minimal invasive ability in the VACI-14 isolate compared to the EIEC strain (Fig. S1, Supporting Information).
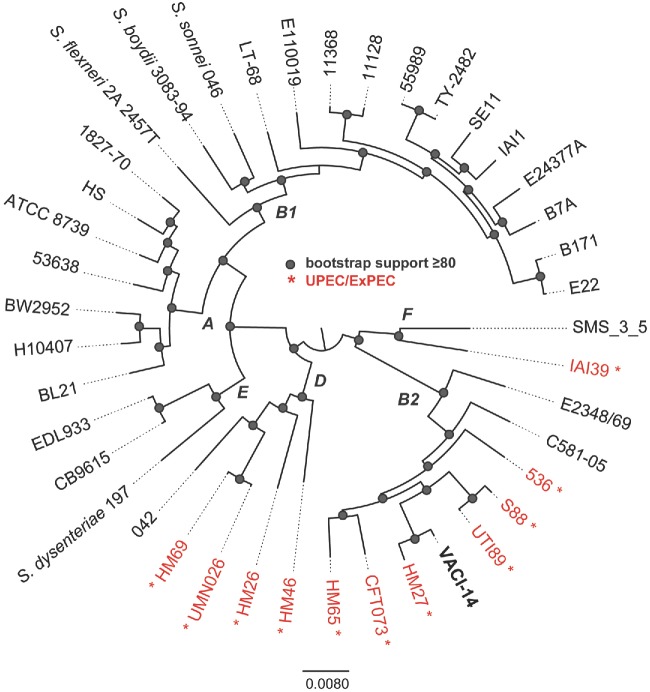
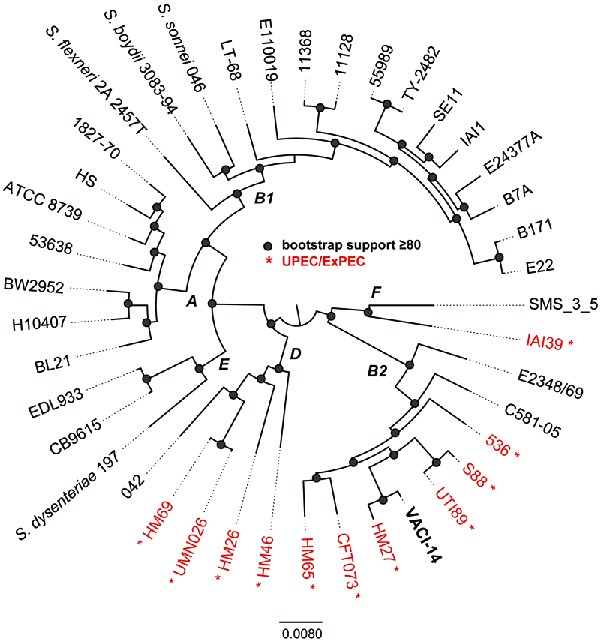
To gain further insight into the apparent ability of the blood isolate to cause both severe diarrhea and bacteremia, we determined its draft genome sequence. Genomic DNA was extracted from the isolate using the Sigma GenElute genomic kit (Sigma-Aldrich) and sequenced using Illumina MiSeq sequencing of a paired-end library at the University of Maryland School of Medicine, Institute for Genome Sciences, Genome Resource Center with standard operating procedures (http://www.igs.umaryland.edu/resources/grc/). The paired-end Illumina reads were subsampled to an approximate genome coverage of 120-fold, and the subsampled reads were assembled using MaSuRCA v.1.9.2 (Zimin et al. 2013). The assembly statistics are as follows: total number of contigs, 109; number of bases in assembly, 5241 165; average contig length, 48 084; N50, 232 331 bp; 97.07% of the bases in assembly are included in contigs >10 kb. Overall, the draft assembly quality is excellent for these types of comparative analyses. The resulting assembly was compared to representative genomes from each of the pathotypes and phylogenetic lineages of E. coli and Shigella species. A total of 41 genomes were aligned with Mugsy (Angiuoli and Salzberg 2011). Homologous blocks present in each genome were concatenated with the bx-python toolkit (https://bitbucket.org/james_taylor/bx-python); the total amount of conserved sequence in the core genome was ∼2.5 Mb. Columns that contained one or more gaps in the alignment were removed with Mothur (Schloss et al. 2009), and a maximum-likelihood tree was inferred with RAxML v.7.2.8 (Stamatakis 2006); bootstrap support values were calculated with 100 replicates. The resulting phylogeny was visualized with FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) (Fig. 1). The genomic sequence of this isolate has been deposited in GenBank under accession number LASE00000000.
Figure 1.
Phylogenomic analysis of 41 E. coli and Shigella genomes that represent members of each of the sequenced pathotypes and phylogenetic groups. All genomes were aligned using Mugsy (Angiuoli and Salzberg 2011) and homologous blocks were concatenated as previously described (Hazen et al. 2013). A phylogenetic tree was inferred with RAxML v7.2.8 (Stamatakis 2006) on this alignment, with 100 bootstrap replicates and nodes that have bootstrap support ≥80 are indicated by a shaded circle. Letters at nodes indicate the E. coli phylogroup as previously defined by MLST or phylogenomic analyses. Isolates indicated in red are from urinary tract sources of isolation.
Analysis of seven standard genes indicates that this strain belongs to multilocus sequence type 12 (https://cge.cbs.dtu.dk/services/MLST/). Phylogenetic analysis based on conservation of core E. coli genome revealed that the strain belongs to a lineage that includes other members of the ExPEC pathotype (highlighted in red and indicated by an asterisk in Fig. 1). Indeed, features characteristic of ExPEC, including complete sets of genes for P fimbriae and hemolysin and the kpsM gene used to identify group II capsule loci (Johnson and O'Bryan 2004), were present (Table 1). The predicted serotype of the strain, as inferred in silico from the sequence (https://cge.cbs.dtu.dk/services/SerotypeFinder/), is O4:H1. Sur-prisingly, the results of this analysis also revealed that the isolate has features defining EPEC strains, including the presence of locus of enterocyte effacement (LEE) genes encoding the ability to produce the attaching and effacing (A/E) phenotype and the absence of stx1 and stx2 genes encoding Shiga toxins, which are found in addition to the LEE in EHEC strains (Kaper 1996). However, some essential LEE genes were not identified and the others displayed low degrees of sequence similarity, raising doubt as to whether the T3SS is fully functional in this isolate. Interestingly, genes distantly related to some bfp genes encoding the bundle-forming pilus (BFP) were also present. Additional common ExPEC and tEPEC virulence factors such as other iron acquisition systems and non-LEE-encoded effectors were not found in the VACI-14 strain.
Table 1.
Potential virulence features identified in VACI-14.
| Gene1 | Associated pathotype | Protein | peptide Identity (%)2 | BLAST e-value3 | Reference Protein ID |
|---|---|---|---|---|---|
| hlyC | ExPEC | hemolysin-activating lysine-acyltransferase | 100 | 2E-99 | AAS93636.1 |
| hlyA | ExPEC | Hemolysin, chromosomal | 100 | 0 | WP_001620144.1 |
| hlyB | ExPEC | Alpha-hemolysin translocation ATP-binding protein | 100 | 0 | KGM64139.1 |
| hlyD | ExPEC | Hemolysin secretion protein D, chromosomal | 69 | 0 | WP_000860472.1 |
| kpsM | ExPEC | KpsM | 97 | 3E-169 | AAN82146.1 |
| kpsT | ExPEC | KpsT | 90 | 2E-124 | AAN82145.1 |
| papH | ExPEC | PAP fimbrial minor pilin protein | 100 | 2E-108 | KEN78663.1 |
| papC | ExPEC | Outer membrane usher protein papC | 99 | 0 | CAM84417.1 |
| papD | ExPEC | Chaperone protein papD | 99 | 1E-176 | WP_001758925.1 |
| papK | ExPEC | Fimbrial adapter papK | 87 | 1E-13 | ENB84384.1 |
| papK | ExPEC | Fimbrial adapter papK | 100 | 9E-45 | WP_029404503.1 |
| papE | ExPEC | Fimbrial protein papE | 100 | 5E-124 | WP_001621802.1 |
| papF | ExPEC | Minor pilin subunit PapF | 100 | 5E-124 | WP_001621802.1 |
| papK | ExPEC | Fimbrial adapter papK | 100 | 2E-41 | WP_029404503.1 |
| papK | ExPEC | Fimbrial adapter papK | 87 | 1E-13 | ENB84384.1 |
| papD | ExPEC | Chaperone protein papD | 100 | 5E-178 | WP_000265729.1 |
| papC | ExPEC | Outer membrane usher protein papC | 100 | 0 | EOV01938.1 |
| papH | ExPEC | PAP fimbrial minor pilin protein | 98 | 1E-138 | KEN81350.1 |
| prsA | ExPEC | PRS fimbrial major pilin protein | 100 | 4E-121 | EFJ72838.1 |
| – | ExPEC | TonB-dependent siderophore receptor | 99 | 0 | WP_021542660.1 |
| flu | ExPEC | Antigen-43 | 100 | 0 | WP_021517784.1 |
| flu | ExPEC | Antigen-43 | 100 | 0 | EQN15149.1 |
| – | ExPEC | Yersiniabactin biosynthetic protein | 99 | 0 | WP_021521384.1 |
| tia | ETEC | Tia adhesin | 99 | 8E-179 | EFJ81905.1 |
| sen | EIEC | shET2 enterotoxin, N-terminal region family protein | 99 | 2E-112 | CDL47274.1 |
| espF | EPEC, EHEC | LEE-encoded effector EspF | 46 | >1 | YP_002331392.1 |
| – | EPEC, EHEC | Component of T3SS | 27 | >1 | YP_002331393.1 |
| escF | EPEC, EHEC | T3SS structure protein EscF | 34 | 0.7 | YP_002331394.1 |
| EPEC, EHEC | 35 | 0.21 | |||
| espB | EPEC, EHEC | Translocon EspB | NSM | NSM | YP_002331396.1 |
| espD | EPEC, EHEC | Translocon EspD | 35 | 0.15 | YP_002331397.1 |
| espA | EPEC, EHEC | Translocon EspA | NSM | NSM | YP_002331398.1 |
| speL | EPEC, EHEC | Secretion switching protein SpeL | NSM | NSM | YP_002331399.1 |
| escD | EPEC, EHEC | T3SS structure protein EscD | 31 | >1 | YP_002331400.1 |
| eae | EPEC, EHEC | Intimin | 41 | 7E-96 | YP_002331401.1 |
| cesT | EPEC, EHEC | Chaperone CesT | 29 | 0.25 | YP_002331402.1 |
| tir | EPEC, EHEC | Translocated intimin receptor Tir | 36 | >1 | YP_002331403.1 |
| map | EPEC, EHEC | LEE-encoded effector Map | 26 | 0.16 | YP_002331404.1 |
| cesF | EPEC, EHEC | Chaperone CesF | 29 | >1 | YP_002331405.1 |
| espH | EPEC, EHEC | LEE-encoded effector EspH | 29 | 0.51 | YP_002331406.1 |
| sepQ | EPEC, EHEC | T3SS structure protein SepQ | 35 | 2.2 | YP_002331407.1 |
| – | EPEC, EHEC | Hypothetical protein | 28 | 0.14 | YP_002331408.1 |
| – | EPEC, EHEC | Hypothetical protein | 28 | >1 | YP_002331409.1 |
| escN | EPEC, EHEC | Translocator EscN | 40 | 1E-79 | YP_002331410.1 |
| escV | EPEC, EHEC | Translocator EscV | 33 | 5E-84 | YP_002331411.1 |
| mpc | EPEC, EHEC | Regulator Mpc | 32 | >1 | YP_002331412.1 |
| espZ | EPEC, EHEC | LEE-encoded effector EspZ | 32 | 0.68 | YP_002331413.1 |
| rorf8 | EPEC, EHEC | Chaperone of T3SS Rorf8 | 24 | >1 | YP_002331414.1 |
| escJ | EPEC, EHEC | T3SS structure protein EscJ | 26 | 0.38 | YP_002331415.1 |
| sepD | EPEC, EHEC | Secretion switching protein SepD | 29 | >1 | YP_002331416.1 |
| escC | EPEC, EHEC | T3SS structure protein EscC | 29 | 4E-20 | YP_002331417.1 |
| cesD | EPEC, EHEC | Chaperone CesD | 44 | 0.007 | YP_002331418.1 |
| grlA | EPEC, EHEC | Positive regulator GrlA | 29 | 0.008 | YP_002331419.1 |
| grlR | EPEC, EHEC | Negative regulator GrlR | 56 | >1 | YP_002331420.1 |
| – | EPEC, EHEC | Hypothetical protein | 39 | 6E-28 | YP_002331421.1 |
| escU | EPEC, EHEC | Secretion system apparatus protein SsaU | 26 | 3E-30 | YP_002331422.1 |
| escT | EPEC, EHEC | T3SS structure protein EscT | 31 | 0.69 | YP_002331423.1 |
| escS | EPEC, EHEC | T3SS structure protein EscS | 38 | 0.006 | YP_002331424.1 |
| escR | EPEC, EHEC | Type III secretion system protein | 40 | 3E-35 | YP_002331425.1 |
| – | EPEC, EHEC | Component of T3SS | 39 | >1 | YP_002331426.1 |
| – | EPEC, EHEC | Component of T3SS | 45 | >1 | YP_002331427.1 |
| – | EPEC, EHEC | Component of T3SS | 50 | >1 | YP_002331428.1 |
| – | EPEC, EHEC | Component of T3SS | 35 | 0.19 | YP_002331429.1 |
| ler | EPEC, EHEC | Transcriptional regulator Ler | 44 | 0.004 | YP_002331430.1 |
| – | EPEC, EHEC | Hypothetical protein | 39 | >1 | YP_002331431.1 |
| espG | EPEC, EHEC | LEE-encoded effector EspG | NSM | NSM | YP_002331432.1 |
| bfpA | EPEC | Bundlin | NSM | NSM | NSM |
| bfpG | EPEC | BfpG | NSM | NSM | NSM |
| bfpB | EPEC | BfpB | 26 | 2E-34 | NP_053067.1 |
| bfpC | EPEC | BfpC | NSM | NSM | NSM |
| bfpU | EPEC | BfpU | NSM | NSM | NSM |
| bfpD | EPEC | BfpD | 31 | 3E-49 | NP_053070.1 |
| bfpE | EPEC | BfpE | 30 | 3E-31 | NP_053071.1 |
| bfpF | EPEC | BfpF | 32 | 7E-21 | NP_053072.1 |
| bfpP | EPEC | Prepilin peptidase | 28 | 1E-19 | NP_053073.1 |
| bfpH | EPEC | BfpH | 28 | 6E-07 | NP_053074.1 |
| bfpI | EPEC | BfpI | NSM | NSM | NSM |
| bfpJ | EPEC | BfpJ | NSM | NSM | NSM |
| bfpK | EPEC | BfpK | NSM | NSM | NSM |
| bfpL | EPEC | BfpL | NSM | NSM | NSM |
| perA | EPEC | PerA | 37 | 3E-18 | NP_053086.1 |
| perB | EPEC | PerB | NSM | NSM | NSM |
| perC | EPEC | PerC | NSM | NSM | NSM |
1“–” indicates that there is no meaningful gene name or that the gene has more than one name.
2NSM – no significant match.
3The BLAST analysis is based on the TBLASTN algorithm.
A/E activity and localized adherence are defining phenotypes of typical EPEC strains (Kaper 1996). However, as the patient's isolate rapidly destroyed the tissue culture cells used in these assays (not shown), we were unable to test for either of these phenotypes. Strains that produce BFP also often exhibit autoaggregation when grown under conditions that induce BFP expression (Bieber et al. 1998). We observed such aggregates after incubation for three hours at 37°C in Dulbecco's Modified Eagle Medium, although they were smaller than those produced by the prototype EPEC strain, E2348/69 (Fig. S2, Supporting Information).
Here, we report the unique case of a patient with severe, protracted diarrhea and multiorgan dysfunction with bacteremia due to an ExPEC strain that also featured distant orthologs of genes characteristic of EPEC. We presume that the non-lactose-fermenting Gram-negative rod that dominated his stool culture was the same strain, although this cannot be confirmed. While chronic diarrhea is a well-described complication of EPEC infection (Rothbaum et al. 1982), the disease occurs almost exclusively in infants and bacteremia is exceptional. We attribute the unusual presentation of chronic diarrhea and bacteremia due to E. coli to a combination of host and bacterial factors.
The patient's immunocompromised status, as evidenced by his low CD4 count, cachexia and cirrhosis, likely played a role in his severe illness. CD4−/− mice infected with Citrobacter rodentium, which shares the A/E genes and phenotype with EPEC, suffer rapid mortality and splenic and hepatic abscesses, indicating systemic spread of the bacteria (Bry and Brenner 2004). In animal models, protein malnutrition can facilitate translocation of bacteria across the intestinal mucosa (Deitch et al. 1987). Cirrhosis, a well-recognized risk factor for numerous serious infections, contributes to poor pathogen clearance by a variety of mechanisms (Christou, Pappas and Falagas 2007).
We believe that the patient's acute kidney injury and hyponatremia were due to volume loss secondary to his severe and prolonged diarrhea. Similarly, it is possible that his liver derangements were also secondary to prolonged hypovolemia, exacerbated by chronic hepatitis C-associated liver disease and cirrhosis. However, cholestasis associated with bacteremia may have contributed to the clinical presentation.
The genomic sequence of this patient's isolate reveals features characteristic of both ExPEC and EPEC. The core genome of this isolate is most similar to other ExPEC isolates associated with urinary tract and other systemic infections. Indeed, this isolate had many features associated with ExPEC including capsule and iron-acquisition systems likely to have facilitated its survival in the blood. How it crossed from the intestinal lumen into the bloodstream is unclear. The fact that we did not observe evidence of cellular invasion is difficult to interpret, as we later observed that it caused extensive damage to tissue culture cells. This rapid cell destruction may be due to E. coli hemolysin, which could facilitate breach of the epithelial barrier (Trifillis et al. 1994). In contrast to the core genome sequence, the accessory genome of the patient's isolate includes highly divergent versions of genes found in EPEC and additional genes found in other diarrheagenic E. coli. These findings highlight the dynamic nature of these pathogens.
As noted above, hybrid strains of E. coli have been reported previously. Most notably, a 2011 outbreak in Germany due to a hybrid STEC-EAEC sickened 3816 people with 54 deaths (Frank et al. 2011). Genomic comparison with previously cultured isolates indicated that the outbreak was caused by a hybrid EAEC strain that developed increased potential for systemic illness following acquisition of the bacteriophage encoding Shiga toxin 2 (Rasko et al. 2011).
Previously, a patient with severe diarrhea and bacteremia due to what would now be called an atypical (bfp-) EPEC strain was described (Bratoeva et al. 1994). Although it produced uncharacterized pili, the presence of pap genes encoding P-fimbriae or other features characteristic of ExPEC was not reported. Genes characteristic of intestinal pathogenic E. coli have previously been noted among strains isolated from patients with UTIs (Toval et al. 2014). As revealed by its phylogenetic history, the strain described in the current report may have evolved from an ExPEC strain already capable of causing systemic infection, by the acquisition of mobile genetic elements distantly related to those encoding BFP and A/E ability, as has been described more generally in EPEC (Hazen et al. 2013). Indeed, EPEC strains closely related to ExPEC have been described previously (Hazen et al. 2013). However, characteristic ExPEC virulence genes are not present in those strains (data not shown). While bacteremia due to a hybrid ExPEC-EPEC strain appears to be rare, the total burden of such complex E. coli strains on human health remains to be determined.
Supplementary Material
Acknowledgments
The authors are grateful to Ms. Marianne Norfolk, MT (ASCP) for expert assistance in diagnostic microbiology.
SUPPLEMENTARY DATA
FUNDING
This work was supported by Public Health Service grants U19 AI090873 and U19 AI110820 from the National Institute of Allergy and Infectious Diseases.
Conflict of interest. None declared.
REFERENCES
- Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–42. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessesen MT, Wang E, Echeverria P, et al. Enteroinvasive Escherichia coli: a cause of bacteremia in patients with AIDS. J Clin Microbiol. 1991;29:2675–7. doi: 10.1128/jcm.29.11.2675-2677.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelheim KA, Breadon A, Faiers MC, et al. The origin of O serotypes of Escherichia coli in babies after normal delivery. J Hyg. 1974;72:67–70. doi: 10.1017/s0022172400023226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber D, Ramer SW, Wu CY, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–8. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- Bratoeva MP, Wolf MK, Marks JK, et al. A case of diarrhea, bacteremia, and fever caused by a novel strain of Escherichia coli. J Clin Microbiol. 1994;32:1383–6. doi: 10.1128/jcm.32.5.1383-1386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J Immunol. 2004;172:433–41. doi: 10.4049/jimmunol.172.1.433. [DOI] [PubMed] [Google Scholar]
- Christou L, Pappas G, Falagas ME. Bacterial infection-related morbidity and mortality in cirrhosis. Am J Gastroenterol. 2007;102:1510–7. doi: 10.1111/j.1572-0241.2007.01286.x. [DOI] [PubMed] [Google Scholar]
- Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–80. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch EA, Winterton J, Li M, et al. The gut as a portal of entry for bacteremia. Role of protein malnutrition. Ann Surg. 1987;205:681–92. doi: 10.1097/00000658-198706000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Donohue-Rolfe A, Keusch GT. Epithelial cell invasion: an overlooked property of enteropathogenic Escherichia coli (EPEC) associated with the EPEC adherence factor. J Infect Dis. 1989;160:452–9. doi: 10.1093/infdis/160.3.452. [DOI] [PubMed] [Google Scholar]
- Frank C, Werber D, Cramer JP, et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–80. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- Germani Y, Bégaud E, Duval P, et al. An Escherichia coli clone carrying the adhesin-encoding afa operon is involved in both diarrhoea and cystitis in twins. T Roy Soc Trop Med H. 1997;91:573. doi: 10.1016/s0035-9203(97)90031-6. [DOI] [PubMed] [Google Scholar]
- Hazen TH, Sahl JW, Fraser CM, et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. P Natl Acad Sci USA. 2013;110:12810–5. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog K, Engeler Dusel J, Hugentobler M, et al. Diarrheagenic enteroaggregative Escherichia coli causing urinary tract infection and bacteremia leading to sepsis. Infection. 2014;42:441–4. doi: 10.1007/s15010-013-0569-x. [DOI] [PubMed] [Google Scholar]
- Johnson JR, O'Bryan TT. Detection of the Escherichia coli group 2 polysaccharide capsule synthesis Gene kpsM by a rapid and specific PCR-based assay. J Clin Microbiol. 2004;42:1773–6. doi: 10.1128/JCM.42.4.1773-1776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Russo TA. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J Infect Dis. 2002;186:859–64. doi: 10.1086/342490. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207:919–28. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB. Defining EPEC. Rev Microbiol. 1996;27(Suppl. 1):130–3. [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Olesen B, Scheutz F, Andersen RL, et al. Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. J Clin Microbiol. 2012;50:3703–11. doi: 10.1128/JCM.01909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GS, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–93. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbaum R, McAdams AJ, Giannella R, et al. A clinicopathological study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology. 1982;83:441–54. [PubMed] [Google Scholar]
- Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–4. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect / Inst Pasteur. 2003;5:449–56. doi: 10.1016/s1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tarr PI, Fouser LS, Stapleton AE, et al. Hemolytic-uremic syndrome in a six-year-old girl after a urinary tract infection with shiga-toxin-producing Escherichia coli O103:H2. N Engl J Med. 1996;335:635–8. doi: 10.1056/NEJM199608293350905. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toval F, Kohler CD, Vogel U, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. 2014;52:407–18. doi: 10.1128/JCM.02069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifillis AL, Donnenberg MS, Cui X, et al. Binding to and killing of human renal epithelial cells by hemolytic P-fimbriated E. coli. Kidney Int. 1994;46:1083–91. doi: 10.1038/ki.1994.370. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Marcais G, Puiu D, et al. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–77. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.