Abstract
Membrane proteins conduct many important biological functions essential to the survival of organisms. However, due to their inherent hydrophobic nature, it is very difficult to obtain structural information on membrane-bound proteins using traditional biophysical techniques. We are developing a new approach to probe the secondary structure of membrane proteins using the pulsed EPR technique of Electron Spin Echo Envelope Modulation (ESEEM) Spectroscopy. This method has been successfully applied to model peptides made synthetically. However, in order for this ESEEM technique to be widely applicable to larger membrane protein systems with no size limitations, protein samples with deuterated residues need to be prepared via protein expression methods. For the first time, this study shows that the ESEEM approach can be used to probe the local secondary structure of a 2H-labeled d8-Val overexpressed membrane protein in a membrane mimetic environment. The membrane-bound human KCNE1 protein was used with a known solution NMR structure to demonstrate the applicability of this methodology. Three different α-helical regions of KCNE1 were probed: the extracellular domain (Val21), transmembrane domain (Val50), and cytoplasmic domain (Val95). These results indicated α-helical structures in all three segments, consistent with the micelle structure of KCNE1. Furthermore, KCNE1 was incorporated into a lipid bilayer and the secondary structure of the transmembrane domain (Val50) was shown to be α-helical in a more native-like environment. This study extends the application of this ESEEM approach to much larger membrane protein systems that are difficult to study with X-ray crystallography and/or NMR spectroscopy.
Keywords: membrane protein, ESEEM, SDSL, α-helix, EPR, KCNE1
Introduction
Membrane proteins play a variety of essential roles in living organisms, such as ion transportation and signal transduction within cell membranes.1,2 Roughly 30% of all proteins encoded in human and E. coli genomes are predicted to be membrane proteins and greater than 50% of the membrane proteins are potential drug targets.3–5 Despite the vast number of membrane proteins and their functional significance, structural information on membrane proteins still lags behind those of soluble proteins, even with improved purification and crystallization methods.6 Challenges in determining membrane protein structures lie in the inherent hydrophobic nature of membrane proteins, making overexpression, purification, and crystallization difficult.6,7 Moreover, the structures of membrane proteins can be influenced by their solubilizing membrane mimetic. So far, a majority of characterized membrane protein structures have been determined in detergent micelles, which are not ideal membrane mimetics.8–10 A lipid bilayer represents a much better environment to probe the structural and dynamic properties of an integral membrane protein.
EPR spectroscopy coupled with site-directed spin labeling (SDSL) has emerged as a powerful biophysical technique to provide robust solutions to these problems and gain pertinent structural and dynamic information for membrane proteins in native-like environments.11–26 Electron spin echo envelope modulation (ESEEM) is a powerful pulsed EPR spectroscopic technique that can detect weakly coupled NMR active nuclei to a nearby unpaired electron spin. In this study, the unpaired electron spin is introduced via a MTSL (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) spin label (SL) and the NMR-active nucleus is a 2H-labeled d8-Val residue. Since the interaction between the unpaired electron spin and the 2H nucleus falls off proportionally to ∼ 1/r,6 the detection limit for this system is ∼ 8 Å.16,17,27 Thus, if the SL and 2H-labeled d8-Val are positioned close enough (<8 Å), the weak dipolar-coupling interaction will produce 2H modulation in the time domain of the ESEEM experiment, and a Fourier transformation (FT) of the time domain spectrum will then yield a peak at the 2H Larmor frequency (∼2.2 MHz at X-band). Accordingly, if a peak at the 2H Larmor frequency is observed from an ESEEM experiment, it indicates that the two labels (SL and 2H-labeled d8-Val) are positioned within 8 Å of each other.
An α-helix has a unique helical structure of 3.6 residues per turn and has a rise of roughly 5.4 Å per turn.28 Taking these structural characteristics into account, two residues located 3 or 4 amino acids apart in a linear sequence are actually closer together than 2 residues located 2 amino acids apart when in an α-helix. Furthermore, when the two side chain labels (SL and 2H-labeled d8-Val) are positioned 3 or 4 residues (i + 3 or i + 4) apart on a typical α-helix, the distance between the two will be small enough (<8 Å) to pick up the weak dipolar-coupling interaction, and reveal a FT peak in the ESEEM frequency domain spectrum.27–32 Conversely, if the two labels are positioned 2 amino acids (i + 2) apart, they will be located too far away (>8 Å) and no FT peak would be observed. The pattern of 2H ESEEM peaks present in i + 3 and i + 4 samples, while absent or minimal in i + 2 is unique to α-helical secondary structure. In contrast, when the two labels are located on a β-strand, the distance of the two labels in i + 3 and i + 4 will be too far away (>8 Å) to show 2H Larmor peaks in the ESEEM frequency domain spectrum. In principle, by introducing a MTSL spin label and a 2H-labeled residue at different positions (i + 2, i + 3, or i + 4) within a small segment of a full-length membrane protein, the presence of 2H ESEEM peaks in i + 3 and i + 4, while not at the i + 2 positions can be used to probe the local α-helical secondary structure of membrane proteins.
The application of ESEEM spectroscopy to probe the secondary structure of small peptides has been successful. However, several challenges exist for this methodology to work on larger membrane proteins prepared via bacterial overexpression methods. Membrane proteins are difficult to express and sample conditions must be optimized for 2H-labeling on the side chain of amino acids such as Val, and minimizing isotope scrambling. To test the feasibility of this ESEEM approach for investigating the secondary structures of spin-labeled membrane proteins in their native-like membrane environments, the full-length human KCNE1 protein was used as a model membrane protein system. KCNE1 is a single-transmembrane protein essential for the function of the voltage-gated KCNQ1 potassium channel in the cardiac action potential.33–36 The solution NMR structure of KCNE1 was previously determined in LMPG (lyso-myristoylphosphatidylglycerol) micelles and the transmembrane domain of KCNE1 was validated using DEER EPR methods in lipid bilayers.25,36 The local secondary structures derived from the ESEEM data will be directly compared with the existing structure to validate the ESEEM technique.
Results and Discussions
The solution NMR structure of KCNE1 in LMPG micelles (PDB entry 2K21) has α-helical segments present in the extracellular, transmembrane, and cytoplasmic domains.36 To assess the feasibility of this ESEEM approach for probing the α-helical content of a membrane protein, all three KCNE1 helical domains were probed. Single Cys KCNE1 variants (Cys is the site of MTSL labeling) were overexpressed in E. coli grown in the presence of a minimal medium spiked with 2H-labeled d8-Val and purified (see Materials and Methods), giving rise to dual-labeled proteins suitable for ESEEM analysis. SDSL has been used extensively for KCNE1 studies.13,14,16,22,25 However, the specific 2H-labeling of Val for KCNE1 is challenging in the bacterial system due to the potential amino acid isotope scrambling. This problem was overcome by adding a large excess amount of non-labeled amino acids at both the culture scale-up and the protein induction stages as well as shortening the induction time to 1 hour.37
Figure 1 shows a structural representation of KCNE1 in a LMPG micelle with the probed region highlighted in green [Fig. 1(A)] and the corresponding ESEEM frequency domain spectra of i + 2, i + 3, and i + 4 samples with normalized FT intensity [Fig. 1(B)]. As seen from Figure 1(A), 2H-labeled d8-Val 50 (i) located in the transmembrane domain of KCNE1 is being probed at different spin-labeled positions. The frequency domain spectra of the i + 2 sample shows that there is no FT peak present at the 2H Larmor frequency (∼2.2 MHz at X-band), indicating that the distance between the 2H-nuclei on the Val50 side chain and the unpaired electron on the MTSL spin label is greater than 8 Å. Conversely, there are obvious ESEEM peaks at the 2H Larmor frequency in the i + 3 and i + 4 samples, indicating a distance smaller than 8 Å between the two labels. These observations suggest that the probed transmembrane segment of KCNE1 is α-helical and agrees with NMR structure of KCNE1 in LMPG micelles.36
Figure 1.
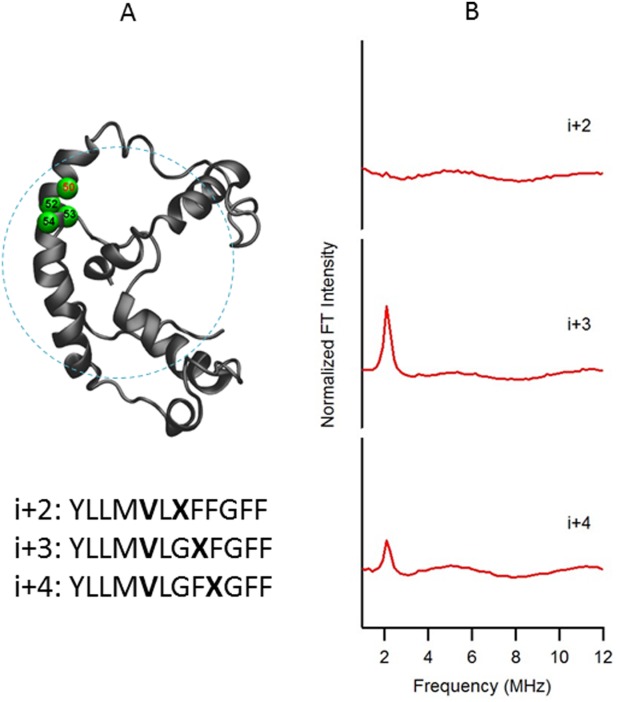
(A) Structural representation of KCNE1 in a LMPG micelle. The probed α-helical region is colored in green and located on the transmembrane domain of the full-length KCNE1. Residue 50 is side chain 2H-labeled Val (denoted i), Residues 52, 53, and 54 are independent Cys mutations (denoted i + 2, i + 3, and i + 4, respectively), which is subject to MTSL labeling. Sequences of fully engineered ESEEM mutants around probed regions were shown below the protein model. The targeted 2H-labeled Val is shown in boldface as V, and the MTSL spin-labeled cysteine is shown in boldface as X. (B) Frequency domain spectra of three-pulse ESEEM data of i + 2, i + 3, and i + 4 samples normalized in FT intensity.
The ESEEM data of the probed regions from the extracellular (Val21) and cytoplasmic domains (Val95) of KCNE1 in LMPG micelles are shown in Figures 2 and 3, respectively. In both figures when a SL is placed three or four residues away (i + 3 or i + 4) from the 2H-labeled d8-Val side chain, large peaks centered at the 2H Larmor frequency are observed in the ESEEM frequency domain spectra. The ESEEM data clearly indicate that the secondary structures of those regions are α-helical. However, for both data sets (Figs. 2 and 3) a small peak is observed at the 2H Larmor frequency for the i + 2 samples. The peak is much smaller than the i + 3 and i + 4 peaks and can be attributed to higher dynamics from the outside domain than the transmembrane domain. The pattern of FT peaks in the frequency domain spectra of Figures 2(B) and 3(B) indicate α-helical content in regions, and matches the solution NMR structure of KCNE1 in LMPG micelles.
Figure 2.

(A) Structural representation of KCNE1 in a LMPG micelle. The probed α-helical region is colored in green and located on the extracellular domain of the full-length KCNE1. Residue 21 is side chain 2H-labeled Val (denoted i), Residues 19, 18, and 17 are independent Cys mutations (denoted i−2, i−3, and i−4, respectively), which is subject to MTSL labeling. Sequences of fully engineered ESEEM mutants around probed regions were shown below the protein model. The targeted 2H-labeled Val is shown in boldface as V, and the MTSL spin-labeled cysteine is shown in boldface as X. (B) Frequency domain spectra of three-pulse ESEEM data of i−2, i−3, and i−4 samples normalized in FT intensity.
Figure 3.
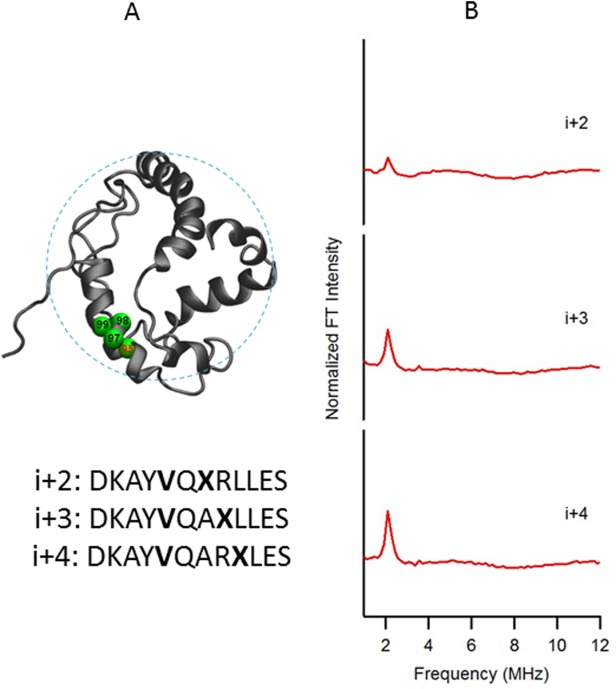
(A) Structural representation of KCNE1 in a LMPG micelle. The probed α-helical region is colored in green and located on the cytoplasmic domain of the full-length KCNE1. Residue 95 is side chain 2H-labeled Val (denoted i), Residues 97, 98, and 99 are independent Cys mutations (denoted i + 2, i + 3, and i + 4, respectively), which is subject to MTSL labeling. Sequences of fully engineered ESEEM mutants around probed regions were shown below the protein model. The targeted 2H-labeled Val is shown in boldface as V, and the MTSL spin-labeled cysteine is shown in boldface as X. (B) Frequency domain spectra of three-pulse ESEEM data of i + 2, i + 3, and i + 4 samples normalized in FT intensity.
The ESEEM data of Figures 1–3 are from KCNE1 solubilized in LMPG micelles, that is, the same environment in which the solution NMR structure was obtained. However, the structure of a membrane protein directly depends on its lipid environment.8,10 It is important to study the structure of a membrane protein in a lipid bilayer environment since it represents a better model of a cell membrane than a micelle. A native-like, membrane-mimetic bicelle formed from DMPC (1,2-dimyristoyl-sn-glycero-3-phosphorylcholine) and DPC (n-dodecylphosphocholine) lipids was used in this study. The same transmembrane domain (Val50) as in Figure 1 of the full-length KCNE1 was probed in a DMPC/DPC (q = 3.2) bicelle environment (Fig. 4). The ESEEM data obtained for KCNE1 in bicelles are shown in Figure 4(B). Strong 2H ESEEM peaks are clearly observed at the i + 3 and i + 4 positions, but not at the i + 2 position. The pattern of FT peaks in the frequency domain spectra clearly indicate that this transmembrane region of KCNE1 contains an α-helical secondary structure in a bicelle membrane environment, and matches the KCNE1 structure obtained in LMPG micelles.36
Figure 4.
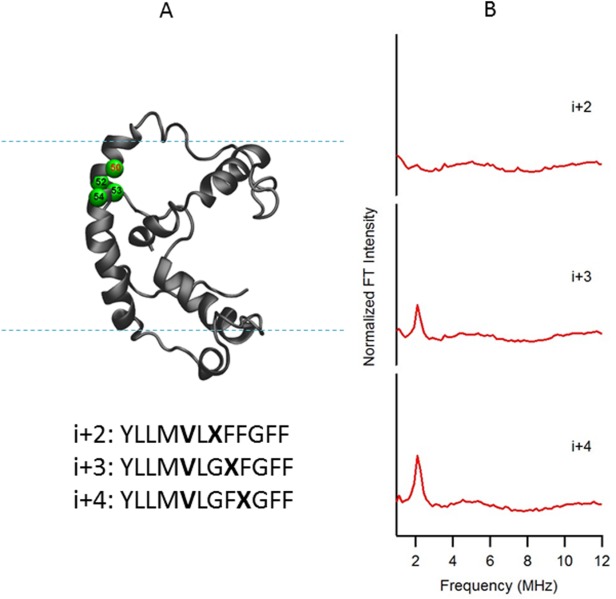
(A) Structural representation of KCNE1 in a DMPC/DPC bicelle (q=3.2). The probed α-helical region is colored in green and located on the transmembrane domain of the full-length KCNE1. Residue 50 is side chain 2H-labeled Val (denoted i), Residues 52, 53, and 54 are independent Cys mutations (denoted i + 2, i + 3, and i + 4, respectively), which is subject to MTSL labeling. Sequences of fully engineered ESEEM mutants around probed regions were shown below the protein model. The targeted 2H-labeled Val is shown in boldface as V, and the MTSL spin-labeled cysteine is shown in boldface as X. (B) Frequency domain spectra of three-pulse ESEEM data of i + 2, i + 3, and i + 4 samples normalized in FT intensity.
This ESEEM protein expression method for investigating the secondary structure of membrane proteins can be complicated by the existence of multiple 2H-labeled d8-Val residues in the protein. The location of the SL must be strategically placed so that it does not detect 2H nuclei from several different Val residues within the ∼8 Å detection limit. KCNE1 has 9 Val residues, and SLs in this study were placed near residues Val21, Val50, and Val95 to probe the secondary structure. Val108 and Val109 would be a poor region of the protein to study with this method, because the residues are right next to each other and a SL at a nearby position could potentially detect both 2H-labeled d8-Val residues.
In this study, we successfully demonstrated the feasibility of using ESEEM spectroscopy to directly probe the local α-helical secondary structure of a recombinant, overexpressed, 2H-labeled d8-Val, and reconstituted membrane protein in its native-like bilayer environment. This powerful technique has no protein size limitations and can be easily applied to investigate the secondary structure of specific segments of membrane proteins or globular proteins of unknown structure. This ESEEM secondary structure approach is very sensitive and can be studied at lower protein concentrations (µg) with shorter acquisition times (minutes) in a lipid bilayer. Also, this pulsed EPR ESEEM approach is one of the few biophysical techniques that can be used to compare the local secondary structure of a membrane protein in both a micelle and a lipid bilayer environment. The protocol for expressing 2H-labeled membrane protein KCNE1 can easily be adapted to other membrane proteins.
Materials and Methods
Engineering of KCNE1 ESEEM mutants
Site-directed mutagenesis was carried out to generate all designed SDSL Cys mutants using the QuickChange Lightning Site-Directed Mutagenesis Kit (Strategene) as previously described.8,22,25,36,38 For probing the KCNE1 transmembrane domain, all KCNE1 Val residues were 2H-labeled and Val at position 50 was chosen as the target site (denoted i) to probe with ESEEM. Cys mutations were made independently at positions 52, 53, and 54 (denoted i + 2, i + 3, and i + 4, respectively), which were then spin-labeled with MTSL. Special attention was paid to amino acids within 5 residues on each side of the probed region to ensure that only one Val was present in order to avoid false positives due to interfering 2H-labeled Val residues with the SL. Fully engineered ESEEM mutants around probed regions were as follows: i + 2 (YLLMVLXFFGFF), i + 3 (YLLMVLGXFGFF), and i + 4 (YLLMVLGFXGFF). The targeted 2H-labeled d8-Val is shown in boldface as V, and the MTSL spin-labeled cysteine is shown in boldface as X. The ESEEM mutants for the α-helical regions of the cytoplasmic domain and extracellular domains were engineered in a similar manner.
Expression and purification of KCNE1 with 2H-labeled d8-val
The overexpression of KCNE1 with 2H-labeled d8-Val was optimized from the previously described method by Tanaka's group.37,39 In brief, the plasmid containing the KCNE1 site-directed mutant as described above was transformed into E. coli BL21-CodonPlus(DE3)-RP competent cells (Stratagene). A single colony was inoculated into 5 mL of Luria broth (LB) medium containing 50 μg/mL of ampicillin. Pre-culture was grown at 37°C overnight. Cells from pre-culture were pelleted at 3,000 g and transferred into 500 mL of M9 minimal medium containing 50 mg of each non-labeled amino acid excluding Val. When the OD600 reached 0.8, 500 mg of each non-labeled amino acid and 50 mg of 2H-labeled d8-Val were added. To assist the solubilization of non-labeled amino acids, Tyr was dissolved in 1N NaOH instead of adding the solids directly. The culture was then allowed to grow for 15 min. IPTG (isopropyl β-D-thiogalactopyr-anoside) was then added to a final concentration of 1 mM and the culture was induced for 1 hour. Cells were harvested at 8,000 g for 10 minutes and stored at −80°C. Purification of KCNE1 was carried out according to a previous method38 with a final elution of the pure protein into 250 mM imidazole (pH 7.0) containing 1.43 mM β-mercaptoethanol and 0.2% LMPG or 0.2% SDS detergent. The protein concentration was determined by measuring the OD280 on a Nano Drop 200c (Thermo Scientific). The purity of the KCNE1 protein was confirmed by SDS-PAGE analysis. Also, MALDI-TOF (Bruker Autoflex III Smartbeam) was used to verify the 2H isotope labeling of the KCNE1 Val residues.
MTSL spin labeling and reconstitution of KCNE1 into bicelles
The MTSL spin labels were dissolved in methanol to a concentration of 250 mM, added directly to the concentrated KCNE1 in elution buffer at a 10:1 MTSL:protein molar ratio, and reacted for 24 hours with rigorous shaking at room temperature in the dark. Excess/unreacted MTSL spin labels were removed through rebinding with Ni-NTA resin following the previously described method.25,40 The reconstitution of spin-labeled KCNE1 into DMPC/DPC bicelles (q = 3.2:1) was optimized from a method described previously.41 In brief, DMPC lipid powder was added directly into KCNE1 solubilized in elution buffer containing 0.5% DPC. The bicelles were formed by incubating on ice and 42°C alternatively with gentle vortexing until the sample became clear. KCNE1 incorporated bicelles were concentrated to obtain the desired spin concentration for ESEEM measurements (∼150 µM). The final MTSL spin label concentration was determined using a CW-EPR X-Band (∼9 GHz) spectrometer.
Three-pulse ESEEM spectroscopy measurements
All three-pulse ESEEM data were collected using a Bruker ELEXSYS E580 spectrometer and a standard Bruker X-Band MS3 split-ring resonator.31,32,42 A τ of 200 ns was selected in order to suppress 1H modulation. All data were collected under identical parameters at a microwave frequency of ∼9.269 GHz and at a temperature of 80 K. Before Fourier Transformation, the ESEEM time domain data were fit to a normalized exponential decay curve, which was subtracted from the experimental spectra as described in the literature.31,32,42 A cross-term averaged FFT was used to obtain the frequency domain spectra.
References
- Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. 2006. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Fruh V, Zhou Y, Chen D, Loch C, Ab E, Grinkova YN, Verheij H, Sligar SG, Bushweller JH, Siegal G. Application of fragment-based drug discovery to membrane proteins: identification of ligands of the integral membrane enzyme DsbB. Chem Biol. 2010;17:881–891. doi: 10.1016/j.chembiol.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Han L, Yap CW, Xie B, Chen Y. Progress and problems in the exploration of therapeutic targets. Drug Discov Today. 2006;11:412–420. doi: 10.1016/j.drudis.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Doerr A. Membrane protein structures. Nat Methods. 2008;6:35–35. [Google Scholar]
- Baker M. Making membrane proteins for structures: a trillion tiny tweaks. Nat Methods. 2010;7:429–434. doi: 10.1038/nmeth0610-429. [DOI] [PubMed] [Google Scholar]
- Coey AT, Sahu ID, Gunasekera TS, Troxel KR, Hawn JM, Swartz MS, Wickenheiser MR, Reid RJ, Welch RC, Vanoye CG, Kang C, Sanders CR, Lorigan GA. Reconstitution of KCNE1 into lipid bilayers: comparing the structural, dynamic, and activity differences in micelle and vesicle environments. Biochemistry. 2011;50:10851–10859. doi: 10.1021/bi2009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. Eur Biophys J. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, McHaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- Berliner LJ. From spin-labeled proteins to in vivo EPR applications. Eur Biophys J. 2010;39:579–588. doi: 10.1007/s00249-009-0534-x. [DOI] [PubMed] [Google Scholar]
- Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzi NL, Fleissner MR, Anthis NJ, Kalai T, Hideg K, Hubbell WL, Clore GM. A rigid disulfide-linked nitroxide side chain simplifies the quantitative analysis of PRE data. J Biomol NMR. 2011;51:105–114. doi: 10.1007/s10858-011-9545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kalai T, Hideg K, Hubbell WL. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci USA. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DJ, Hubbell WL, Klug CS. Probing protein secondary structure using EPR: investigating a dynamic region of visual arrestin. Appl Magn Reson. 2012;43:405–419. doi: 10.1007/s00723-012-0369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Liu Y, Borbat P, Zweier JL, Freed JH, Hubbell WL. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J Am Chem Soc. 2012;134:9950–9952. doi: 10.1021/ja303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith HJ, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA. DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry. 2013;52:6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch MT, Yang Z, Brooks EK. Hubbell WL. Mapping protein conformational heterogeneity under pressure with site-directed spin labeling and double electron-electron resonance. Proc Natl Acad Sci USA. 2014;111:E1201–E1210. doi: 10.1073/pnas.1403179111. , and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CJ, Fleissner MR, Brooks EK, Hubbell WL. Stationary-phase EPR for exploring protein structure, conformation, and dynamics in spin-labeled proteins. Biochemistry. 2014;53:7067–7075. doi: 10.1021/bi5011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu ID, Kroncke BM, Zhang R, Dunagan MM, Smith HJ, Craig A, McCarrick RM, Sanders CR, Lorigan GA. Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry. 2014;53:6392–6401. doi: 10.1021/bi500943p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jimenez-Oses G, Lopez CJ, Bridges MD, Houk KN, Hubbell WL. Long-range distance measurements in proteins at physiological temperatures using saturation recovery EPR spectroscopy. J Am Chem Soc. 2014;136:15356–15365. doi: 10.1021/ja5083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force DA, Randall DW, Lorigan GA, Clemens KL, Britt RD. ESEEM studies of alcohol binding to the manganese cluster of the oxygen evolving complex of Photosystem II. J Am Chem Soc. 1998;120:13321–13333. [Google Scholar]
- Heming M, Narayana M, Kevan L. Analysis of nuclear-quadrupole interaction effects in electron spin-echo modulation spectra by 2nd-order perturbation-methods. J Chem Phys. 1985;83:1478–1484. [Google Scholar]
- Mims W. Envelope modulation in spin-echo experiments. Phys Rev B. 1972;5:2409. [Google Scholar]
- Voet D, Voet JG. Biochemistry. 4th ed. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- Liu L, Sahu ID, Mayo DJ, McCarrick RM, Troxel K, Zhou A, Shockley E, Lorigan GA. Enhancement of electron spin echo envelope modulation spectroscopic methods to investigate the secondary structure of membrane proteins. J Phys Chem B. 2012;116:11041–11045. doi: 10.1021/jp304669b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo D, Zhou A, Sahu I, McCarrick R, Walton P, Ring A, Troxel K, Coey A, Hawn J, Emwas AH, Lorigan GA. Probing the structure of membrane proteins with electron spin echo envelope modulation spectroscopy. Protein Sci. 2011;20:1100–1104. doi: 10.1002/pro.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- McCrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, Vanoye CG, Kim HJ, Sanders CR. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Yamamoto M, Tanaka T. A simple method for amino acid selective isotope labeling of recombinant proteins in E. coli. J Biomol NMR. 2008;42:59–67. doi: 10.1007/s10858-008-9264-0. [DOI] [PubMed] [Google Scholar]
- Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr, Sanders CR. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Yamamoto M, Tanaka T. Selective isotope labeling of recombinant proteins in Escherichia coli. Methods Mol Biol. 2012;896:439–448. doi: 10.1007/978-1-4614-3704-8_30. [DOI] [PubMed] [Google Scholar]
- Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolandt OV, Walther TH, Grage SL, Ulrich AS. Magnetically oriented dodecylphosphocholine bicelles for solid-state NMR structure analysis. Biochim Biophys Acta. 2012;1818:1142–1147. doi: 10.1016/j.bbamem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Zhou A, Abu-Baker S, Sahu ID, Liu L, McCarrick RM, Dabney-Smith C, Lorigan GA. Determining alpha-helical and beta-sheet secondary structures via pulsed electron spin resonance spectroscopy. Biochemistry. 2012;51:7417–7419. doi: 10.1021/bi3010736. [DOI] [PMC free article] [PubMed] [Google Scholar]


