Abstract
Cytoplasmic domains of transmembrane bacterial chemoreceptors are largely extended four-helix coiled coils. Previous observations suggested the domain was structurally dynamic. We probed directly backbone dynamics of this domain of the transmembrane chemoreceptor Tar from Escherichia coli using site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy. Spin labels were positioned on solvent-exposed helical faces because EPR spectra for such positions reflect primarily polypeptide backbone movements. We acquired spectra for spin-labeled, intact receptor homodimers solubilized in detergent or inserted into native E. coli lipid bilayers in Nanodiscs, characterizing 16 positions distributed throughout the cytoplasmic domain and on both helices of its helical hairpins, one amino terminal to the membrane-distal tight turn (N-helix), and the other carboxyl terminal (C-helix). Detergent solubilization increased backbone dynamics for much of the domain, suggesting that loss of receptor activities upon solubilization reflects wide-spread destabilization. For receptors in either condition, we observed an unanticipated difference between the N- and C-helices. For bilayer-inserted receptors, EPR spectra from sites in the membrane-distal protein-interaction region and throughout the C-helix were typical of well-structured helices. In contrast, for approximately two-thirds of the N-helix, from its origin as the AS-2 helix of the membrane-proximal HAMP domain to the beginning of the membrane-distal protein-interaction region, spectra had a significantly mobile component, estimated by spectral deconvolution to average approximately 15%. Differential helical dynamics suggests a four-helix bundle organization with a pair of core scaffold helices and two more dynamic partner helices. This newly observed feature of chemoreceptor structure could be involved in receptor function.
Keywords: bacterial chemotaxis, chemoreceptors, transmembrane receptors, helical coiled-coils, EPR spectroscopy, helical dynamics, protein dynamics
Introduction
Chemoreceptors are central components in the high-performance biological signaling system that mediates bacterial chemotaxis.1–3 Of the thousands of chemoreceptors identified by genome sequencing in a wide range of bacterial species, most are transmembrane proteins.4 Their amino-terminal, periplasmic signal-input modules have a variety of structural organizations.5–9 In contrast, their carboxyl-terminal, cytoplasmic output modules share a common structure of a four-helix, coiled-coil rod formed by two extended helical hairpins, one from each protomer of the chemoreceptor dimer [Fig. 1(A)].4,10–14 For many chemoreceptors,6 transmembrane domains are connected to the coiled-coil rods by a signal-conversion module in the form of a HAMP (histidine kinase, adenylyl cyclase, methyl-accepting chemoreceptors, phosphatases) domain.15 HAMP domains are parallel four-helix bundles formed from two pairs of helices (AS1/AS2 and AS1′/AS2′), each pair contributed by a receptor subunit [Fig. 1(A)].16–18 Biochemical and genetic studies have identified functional regions along the coiled-coil rod [Fig. 1(A)].1–3 Adjacent to the HAMP domain is an adaptational modification region containing the methyl-accepting glutamates that mediate sensory adaptation. Farther from the membrane is a “glycine hinge,” a cluster of functionally important glycines at which alpha-helical angles can be perturbed to create a slight bend in the coiled coil and which may be involved in receptor flexibility and signaling.19 At the membrane-distal tip is a protein-interaction region that includes ∼40 very highly conserved residues. The conserved residues are centered on each side of the hairpin turn and are involved in interaction of receptor homodimers to form trimers of those dimers as well as being involved in receptor interaction with the chemotaxis histidine kinase CheA and the coupling protein CheW to form core signaling complexes.4,20,21
Figure 1.
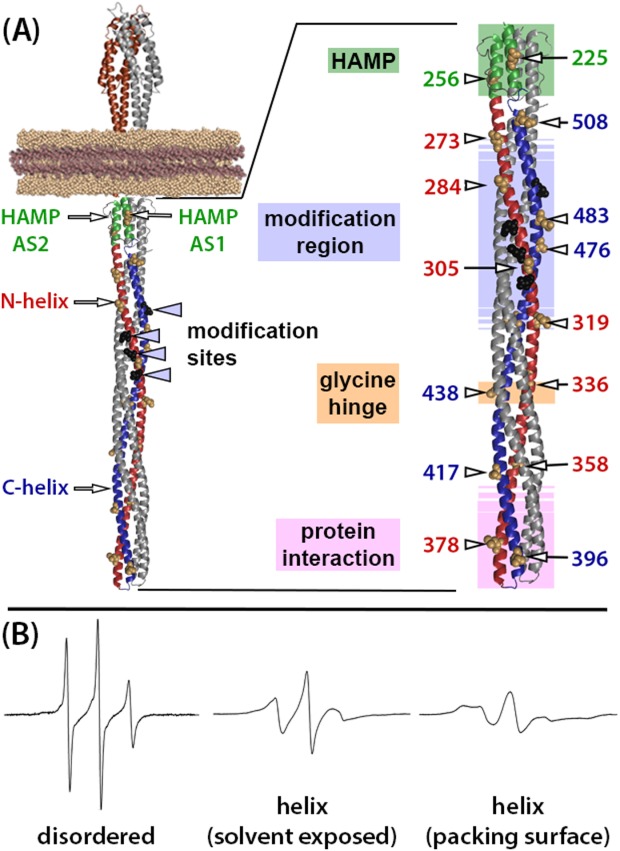
Chemoreceptor structural organization and example EPR spectra. (A) Ribbon diagrams of a molecular model of a chemoreceptor dimer based on X-ray and NMR structures of receptor fragments.1–3 The left-hand image is a model of an intact, single chemoreceptor dimer inserted into a Nanodisc with relevant structural features labeled (see text for explanations). The right-hand image is an enlargement of the receptor cytoplasmic domain with positions of the mutagenically introduced cysteines indicated by residue number and an arrow pointing to the CPK representation of the respective native side-chains on the surface of the Tar helices. Functional regions are indicated by labels to the left of the structure and by shaded colored boxes (HAMP pale green, modification region pale blue, glycine hinge pale orange, and protein interaction region pale pink). Shaded boxes for the modification and interaction regions are shown with borders that fade out to symbolize a lack of definition for their precise positions. In both images, one receptor protomer is color coded: brown for the periplasmic, ligand-binding domain, green for HAMP, red for the N-helix in the coiled coil, and blue for its C-helix. Sites of adaptational modification are indicated by black spheres, which are labeled in the left-hand diagram. (B) Example EPR spectra for a nitroxide spin label coupled by methanethiosulfonate chemistry to a cysteine introduced into chemoreceptor Tar (from left to right): in a disordered segment (position 549 in urea-denatured Tar51), on a solvent-exposed, noninteracting surface of a well-ordered alpha helix (Tar position 396, this study) or on a packing surface of a well-ordered alpha helix (Tar position 401, N.L. Bartelli, unpublished observation).
Experimental characterizations of the transmembrane chemoreceptors of Escherichia coli and Salmonella enterica have suggested that along a significant portion of their length the helices of the coiled-coil segment of cytoplasmic domain are not tightly packed but instead dynamic.10,22–27 In addition, several lines of evidence suggest that dynamics varies along the length of the cytoplasmic domain and may be involved in chemoreceptor signaling.3,26–31
To investigate directly helical dynamics in the cytoplasmic domain of an intact, membrane-inserted chemoreceptor, we used site-directed spin labeling and continuous-wave electron paramagnetic resonance (EPR) spectroscopy32–35 to characterize the E. coli transmembrane chemoreceptor Tar (Taxis to aspartate and repellents) as an intact, functional native dimer inserted into bilayers of native E. coli lipid contained in individual Nanodiscs. Nanodiscs are small, ∼10 nm plugs of lipid bilayer rendered water-soluble by an annulus of “membrane scaffold protein” into which transmembrane proteins can be incorporated36 and which provide a natural membrane environment in which chemoreceptors exhibit essentially native activities.37–42 EPR is well suited for investigation of helical dynamics because MTSL (1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate) spin labels placed on solvent-exposed faces of alpha helices, at positions devoid of side-chain tertiary contacts, report differences in fluctuations of the peptide backbone [Fig. 1(B)].33,35,43–45 This sensitivity to peptide backbone fluctuations has been utilized to map regional protein flexibility43,46 and probe changes in protein conformation induced by backbone-modulating osmolytes and denaturants.47–51 Our laboratory has utilized site-directed spin labeling and EPR spectroscopy to characterize the organization and packing of a chemoreceptor transmembrane segment52 as well as peptide backbone dynamics along the carboxyl-terminal, 40-residue segment of a chemoreceptor inserted into lipid bilayers.51 The latter study documented distinct differences in mobility between spin labels on the helical coiled-coil body of the cytoplasmic domain of a chemoreceptor and the disordered segment of ∼35 residues at the receptor carboxyl terminus.51
We have now expanded our characterization of chemoreceptor structure by EPR spectroscopy to probe helical dynamics along the length of the chemoreceptor cytoplasmic domain.
Results
Experimental approach
We used oligonucleotide-directed, site-specific mutagenesis to create 16 single-cysteine variants of the Escherichia coli aspartate receptor Tar. Each cysteine was placed on a solvent-exposed helical surface of the cytoplasmic domain. The positions were distributed along the length of this rod-shaped domain, among the functional regions and between the two antiparallel helices of the helical hairpins of the homodimer. One of these helices is on the amino terminal side of the membrane-distal hairpin turn (the N-helix) and the other on the carboxyl side (the C-helix) [Fig. 1(A)]. Cysteine-containing, detergent-solubilized receptors were purified utilizing a 6-histidine tag placed by mutagenesis at their extreme Tar carboxyl terminus.53 These purified receptors were spin labeled and X-band, continuous-wave EPR spectra collected. Each spin-labeled receptor was incorporated into Nanodiscs made with natural E. coli lipids in conditions that favored one receptor dimer per disc.37,40,54 Dimers are the minimal chemoreceptor structural unit10,37 and the minimal functional unit for adaptational covalent modification and transmembrane signaling, i.e., the coupling of ligand binding in the periplasmic domain to conformational changes in the cytoplasmic domain.38 Receptor-containing Nanodiscs were purified utilizing the 6-histidine tag on Tar and EPR spectra taken of the bilayer-inserted receptor. Isolation of dimers in separate Nanodiscs insured that measurements were not complicated by higher-order interactions among chemoreceptors, yet reflected the mobility of spin labels on helices in intact, functionally active molecules. In addition, since spin labels were placed on the helical surfaces, extending into the solvent at noninteracting positions, the two spin labels on each dimer (one on each protomer) were oriented 180° apart on opposite sides of a bundle of four helices and thus were sufficiently separated to avoid mutual influence in the form of spin–spin coupling. We tested for possible structural perturbations of the receptors generated by introduction of a cysteine and its subsequent spin labeling by determining the ability of the chemotaxis methylesterase/deamidase to recognize the respective cysteine-substituted, spin-labeled, Nanodisc-inserted Tar dimers and thus deamidate glutamines at their four sites of adapatational modification. Functionally active chemoreceptors in their natural environment of the cytoplasmic membrane or inserted in reconstituted bilayers of E. coli lipids are efficiently deamidated; receptors functionally perturbed because of insertion in non-native lipid bilayers are minimally deamidated; and receptors rendered functionally inactive by being solubilized in detergent micelles are not deamidated at all.37,40,41 By the deamidation test, all but two of the 16 cysteine-substituted, spin-labeled forms of Tar reconstituted into native-lipid Nanodiscs had over half the activity of the cysteine-less receptor without a spin label and those two had at least 29% of that activity (Table1).
Table 1.
Enzymatic Deamidation of Cysteine-Substituted, Spin-Labeled Tar in a Nanodisc
| Cysteine position | Deamidationa | Cysteine position | Deamidationa |
|---|---|---|---|
| None | 1.00 ± 0.03 | 378 | 0.86 ± 0.05 |
| 225 | 0.77 ± 0.02 | 396 | 0.64 ± 0.01 |
| 256 | 0.82 ± 0.01 | 417 | 0.64 ± 0.05 |
| 273 | 0.61 ± 0.02 | 438 | 0.75 ± 0.03 |
| 284 | 0.69 ± 0.06 | 456 | 0.80 ± 0.03 |
| 305 | 0.38 ± 0.04b | 476 | 0.91 ± 0.03 |
| 319 | 0.88 ± 0.03 | 483 | 0.87 ± 0.02 |
| 336 | 0.61 ± 0.02 | 508 | 0.52 ± 0.03 |
| 358 | 0.29 ± 0.03 |
Relative to Nanodisc-embedded Tar without a cysteine and thus without a spin label.
Value likely artificially low because spin-labeled C305 is centered one helical turn between deamidation sites Q302 and Q309 and thus likely to interfere with deamidase access to those sites and possibly the nearby residues Q309 and Q491 [Fig. 1(A)].
Mobility of spin labels on chemoreceptor Tar inserted in natural E. coli lipid bilayers
Figure 2 shows the continuous-wave EPR spectra taken at room temperature for spin labels at the 16 indicated positions on the solvent-exposed surface of the cytoplasmic domain of intact chemoreceptor Tar dimers. These dimers were each inserted into a Nanodisc-provided bilayer of native E. coli phospholipids. The spectra have features consistent with contributions from two label populations, one with relatively restricted mobility, and the other with greater mobility. Considering the overall spectral pattern for each position, 10 (positions 225, 358, 378, 396, 417, 438, 456, 476, 483, and 508) resembled spectra of spin labels on the surface of well-structured alpha helices43 and 6 (positions 256, 273, 284, 305, 319 and 336) exhibited greater mobility than typical, well-ordered alpha helices.43,51,55 To assess the pattern of spin-label mobility as a function of position along the rod-like cytoplasmic domain, we expressed mobility as a single numerical parameter. We used two such parameters, h(+1)/h(0), the ratio of the magnitudes of the low-field and central line in an EPR spectrum and Δ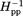 , the inverse of the magnitude of the central line width. These parameters are commonly used as a single numeric representation of relative spin-label mobility.32,34,43,44,46–48,51,52,56,57 In Figure 3, h(+1)/h(0) [Fig. 3(A)] and Δ
, the inverse of the magnitude of the central line width. These parameters are commonly used as a single numeric representation of relative spin-label mobility.32,34,43,44,46–48,51,52,56,57 In Figure 3, h(+1)/h(0) [Fig. 3(A)] and Δ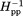 [Fig. 3(B)] values for the 16 spectra in Figure 2 are plotted as a function of the distance of the spin label from the membrane surface. Both parts of Figure 3 include selected values from our previous characterization of the 35-residue disordered flexible arm at the Tar carboxyl terminus.51 As can be seen in Figure 3(A,B), expressing label mobility in terms of h(+1)/h(0) spreads the range of values for higher mobilities and expressing mobility in terms of Δ
[Fig. 3(B)] values for the 16 spectra in Figure 2 are plotted as a function of the distance of the spin label from the membrane surface. Both parts of Figure 3 include selected values from our previous characterization of the 35-residue disordered flexible arm at the Tar carboxyl terminus.51 As can be seen in Figure 3(A,B), expressing label mobility in terms of h(+1)/h(0) spreads the range of values for higher mobilities and expressing mobility in terms of Δ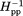 spreads the range for lower mobilities.
spreads the range for lower mobilities.
Figure 2.
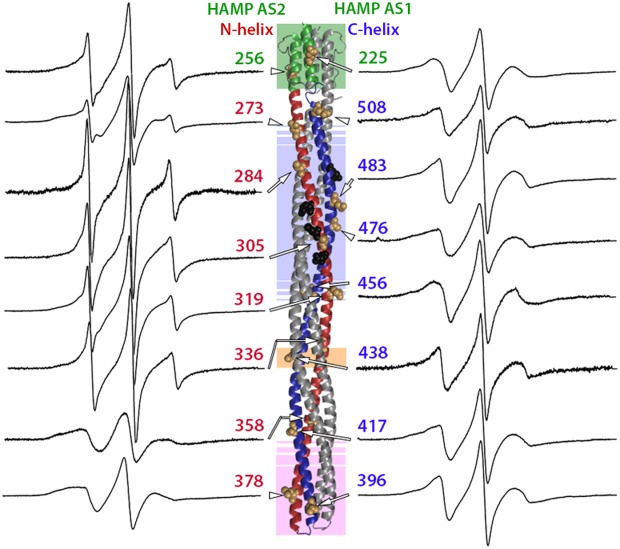
Spectra of purified, spin-labeled Tar dimers inserted in the native lipid bilayer of a Nanodisc. Normalized EPR spectra for 16 positions in the Tar cytoplasmic domain are displayed on each side of a ribbon diagram of that domain as shown in the right-hand image of Figure 1(A), with each spectrum near the spin-label position on the diagram and an arrow pointing to the position of a CPK representation for the native side-chain. Spectra for positions on the N-helix and HAMP AS2 helix are on the left, and spectra for positions on the C-helix and HAMP AS1 are on the right. As in Figure 1(A), shaded regions indicate functional regions: HAMP pale green, modification region pale blue, glycine hinge pale orange and protein interaction region pale pink. Sites of adaptational modification are shown as black CPK side-chains.
Figure 3.
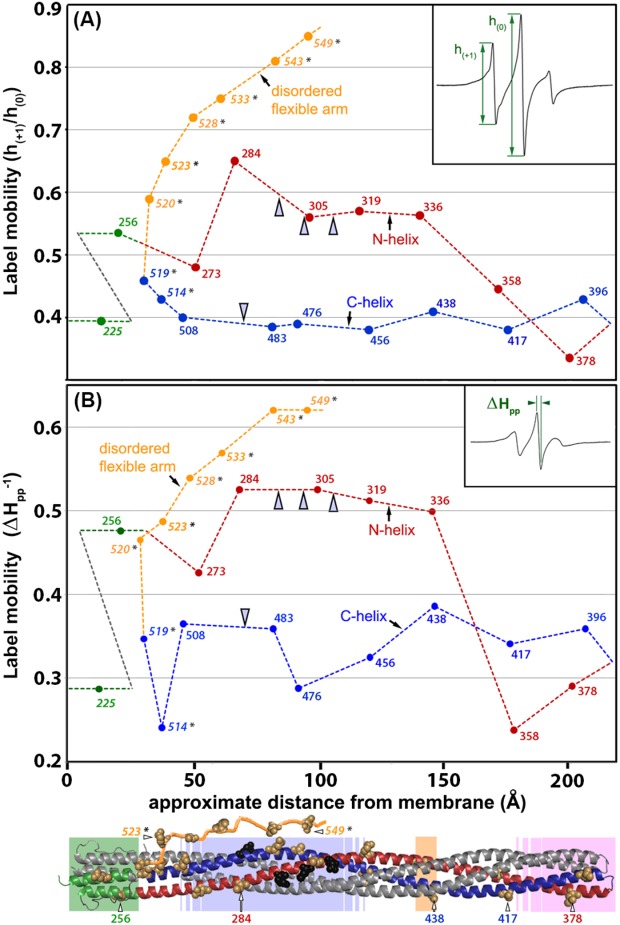
Spin-label mobility as a function of position along the length of the cytoplasmic domain for Tar dimers inserted in the native lipid bilayer of a Nanodisc. Mobility parameters h(+1)/h(0) [Fig. 3(A)] and Δ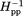 [Fig. 3(B)] (see insets and text) are plotted as a function of spin-label distance in Å from the membrane surface as determined from the three-dimensional model of the chemoreceptor cytoplasmic domain [Fig. 1(A)]. The model is shown as a ribbon diagram below the abscissa with spin-label positions marked by CPK models of the respective native side chains and functional regions indicated by shaded boxes as in the right-hand image of Figure 1(A). The model includes a representation of the 35-residue C-terminal, unstructured flexible arm. For reference, some positions are labeled with their residue number. Values for the mobility parameters for each of the 16 spectra in Figure 2 are plotted, as well as values for selected spectra of spin labels (marked by an asterisk on the residue number) at positions 514, 519, 520, 523, 528, 533, 543, and 549 that were obtained in a study of the Tar C-terminal disordered flexible arm (residues 520–554) and its boundary with the C-helix in the helical coiled coil structured body of the cytoplasmic domain.51 Since the disordered, flexible arm does not have a fixed position relative to the helical structure of the rest of the cytoplasmic domain, the segment was placed for clarity extending away from the membrane as a function of residue number in the ribbon diagram and the plot. A dotted line traces backbone connectivity between residue positions as well as the approximate spatial placement of the protomer backbone relative to distance from the membrane. Both the line and the data points are color coded by structural element: HAMP green, N-helix red, C-helix blue, and disordered, flexible arm orange; the two helices and the flexible arm are labeled on the plot. Sites of adaptational modification are indicated by open arrowheads. The data for position 225, and 514–549,51 were for Tar with two glutamates and two glutamines at its sites of adaptational modification and are distinguished by italics. All other data were for four-glutamine Tar. For 13 of the 16 spin-label positions for which spectra were acquired independently two to four times, h(+1)/h(0) and Δ
[Fig. 3(B)] (see insets and text) are plotted as a function of spin-label distance in Å from the membrane surface as determined from the three-dimensional model of the chemoreceptor cytoplasmic domain [Fig. 1(A)]. The model is shown as a ribbon diagram below the abscissa with spin-label positions marked by CPK models of the respective native side chains and functional regions indicated by shaded boxes as in the right-hand image of Figure 1(A). The model includes a representation of the 35-residue C-terminal, unstructured flexible arm. For reference, some positions are labeled with their residue number. Values for the mobility parameters for each of the 16 spectra in Figure 2 are plotted, as well as values for selected spectra of spin labels (marked by an asterisk on the residue number) at positions 514, 519, 520, 523, 528, 533, 543, and 549 that were obtained in a study of the Tar C-terminal disordered flexible arm (residues 520–554) and its boundary with the C-helix in the helical coiled coil structured body of the cytoplasmic domain.51 Since the disordered, flexible arm does not have a fixed position relative to the helical structure of the rest of the cytoplasmic domain, the segment was placed for clarity extending away from the membrane as a function of residue number in the ribbon diagram and the plot. A dotted line traces backbone connectivity between residue positions as well as the approximate spatial placement of the protomer backbone relative to distance from the membrane. Both the line and the data points are color coded by structural element: HAMP green, N-helix red, C-helix blue, and disordered, flexible arm orange; the two helices and the flexible arm are labeled on the plot. Sites of adaptational modification are indicated by open arrowheads. The data for position 225, and 514–549,51 were for Tar with two glutamates and two glutamines at its sites of adaptational modification and are distinguished by italics. All other data were for four-glutamine Tar. For 13 of the 16 spin-label positions for which spectra were acquired independently two to four times, h(+1)/h(0) and Δ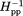 were determined for each spectrum and averaged to provide the values plotted here. Percent standard deviations for those average h(+1)/h(0) and Δ
were determined for each spectrum and averaged to provide the values plotted here. Percent standard deviations for those average h(+1)/h(0) and Δ values were, respectively, position 225, 1.8 and 9.5; 256, 1.3 and 1.6; 273, 2.9 and 7.2; 305, 12 and 0; 336, 0.9 and 1.2; 358, 1.6 and 6.1; 378, 1.8 and 3.1; 396, 5.2 and 4.7; 417, 7 and 0; 438, 0 and 0; 476, 0 and 9.5; 483, 5.5 and 9.6; 508, 3.4 and 5.4.
values were, respectively, position 225, 1.8 and 9.5; 256, 1.3 and 1.6; 273, 2.9 and 7.2; 305, 12 and 0; 336, 0.9 and 1.2; 358, 1.6 and 6.1; 378, 1.8 and 3.1; 396, 5.2 and 4.7; 417, 7 and 0; 438, 0 and 0; 476, 0 and 9.5; 483, 5.5 and 9.6; 508, 3.4 and 5.4.
As illustrated in Figures 2 and 3, spin labels on or near the protein-interaction region (positions 358, 378, 396, and 417), at all C-helix positions (396 and 417 in the protein-interaction region plus positions 438, 456, 476, 483, 508, 514, and 519) and at the one position characterized in the AS1 helix of the HAMP domain (position 225) had spectra and corresponding mobility parameters characteristic of well-structured alpha helices. Strikingly, spin-label mobility was distinctly different along the N-helix. Mobility was consistently greater for spin labels on the N-helix than on its companion C-helix over approximately two-thirds of the N-helix, from position 256 on HAMP AS2 to position 336 near glycine residues 340 and 341 (Figs. 2 and 3). For positions in or near the protein-interaction region (358, 417, 378, and 396), there were no consistent difference between the mobility of spin labels of the two companion helices; both exhibited values characteristic of stable helices. Taken together, the data revealed an unexpected asymmetry in backbone dynamics between the N- and C-helices of the dimeric four-helix coiled coil over approximately two-thirds of the length of the cytoplasmic domain.
Analyzing the difference between N-helix and C-helix backbone dynamics
The higher mobilities for spin-labels on the membrane-proximal two-thirds of the N-helix in comparison to its companion C-helix could reflect differences in distribution between the more mobile and less mobile components of the respective spectra. We investigated this possibility by deconvoluting each spectrum into a more mobile and less mobile component. Since many of the relevant spectra had a significant contribution from what appeared to be a highly mobile component, we deconvoluted by subtracting a spectrum with mobility characteristics of a spin label on a disordered protein segment. We used three separate reference spectra as examples of spin labels on the disordered protein backbone of Tar. One was for a spin label very near the end of the disordered, carboxyl-terminal flexible arm of Tar at position 543,51 and two were for spin labels on urea-denatured Tar at positions 478 or 483. For each initial spectrum, subtracting any one of these reference spectra yielded a residual, less-mobile spectrum typical of spin labels on a noninteracting surface of a stable alpha-helix and a value for the percentage of highly mobile component subtracted (Fig. 4 and Table2). There was no more than a 4% contribution from a highly mobile component in the spectra of spin labels on AS1 (225), in or near the protein-interaction region (358, 378, 396, and 417) or on the entire C-helix (396 and 417 in the protein-interaction region plus positions 438, 456, 476, 483, and 508) (Fig. 4 and Table2). In contrast, spectra for N-helix positions from 256 to 336 had a larger contribution from a highly mobile state, averaging approximately 15% with the exception of position 284 which exhibited a significantly higher contribution of ∼60%, perhaps reflecting perturbation of the protein by introduction of a spin-labeled cysteine at this position. Taken together, the overall pattern of contributions by a highly mobile component suggest that the mobility differences we observed between the N-helix and the C-helix reflect a greater probability for approximately two-thirds of the N-helix to make excursions into a less ordered state, thus, in essence, to be more dynamic.
Figure 4.
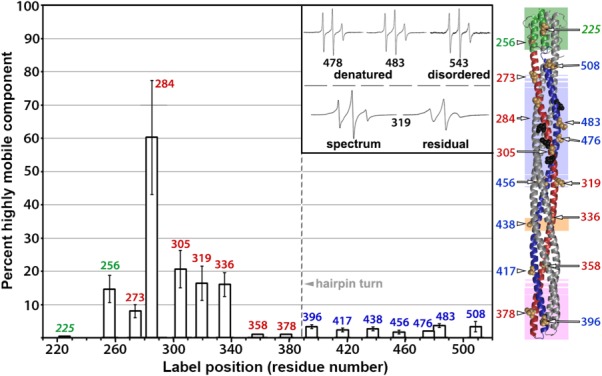
Results of spectra deconvolution. The bars show average contributions with standard deviations of a highly mobile component to the respective spectra of spin labels at each of 16 positions in the Tar cytoplasmic domain (Fig. 2) as determined by spectral deconvolutions using three different reference spectra of spin labels on a disordered Tar protein backbone (Table2). Inset (upper): Reference spectra. From left to right, urea-denatured, detergent-solubilized Tar labeled at position 478 or 483, and Tar labeled in the disordered carboxyl-terminal flexible arm at position 543.51 Inset (lower): An example deconvolution. On the left is the spectrum for Tar spin-labeled at position 319, and on the right is the residual spectrum after subtracting the highly mobile component. To the right of the bar graph is a cartoon of the Tar cytoplasmic domain with spin-label positions indicated by a CPK representation of the respective native side-chains and residue number. Labels and color coding are as in the right-hand image of Figure 1(A).
Table 2.
Deconvolution of EPR Spectra
| Spectrum | Percent contribution by a highly mobile component | ||||
|---|---|---|---|---|---|
| for spin label at position | 478 Urea | 483 Urea | 543 Intrinsically Disordered | Avg. | SD |
| 225 | 0a | 0a | 0a | 0a | 0 |
| 256 | 11 | 14 | 19 | 15 | 4 |
| 273 | 6 | 8 | 10 | 8 | 2 |
| 284 | 42b | 63b | 76b | 60b | 17 |
| 305 | 16 | 19 | 27 | 21 | 6 |
| 319 | 12 | 15 | 22 | 16 | 5 |
| 336 | 13 | 15 | 20 | 16 | 4 |
| 358 | 1 | 1 | 1 | 1 | 0 |
| 378 | 1 | 1 | 1 | 1 | 0 |
| 396 | 3 | 3 | 4 | 3 | 0.6 |
| 417 | 2 | 2 | 3 | 2 | 0.6 |
| 438 | 3 | 2 | 3 | 3 | 0.6 |
| 456 | 1 | 2 | 2 | 2 | 0.6 |
| 476 | 2 | 2 | 2 | 2 | 0 |
| 483 | 3 | 4 | 4 | 4 | 0.6 |
| 508 | 3 | 2 | 5 | 3 | 1.5 |
Results of spectral deconvolution are shown for spectra of spin labels at the indicated positions of Tar inserted in a lipid bilayer provided by a Nanodisc (Fig. 2). Spectra were deconvoluted by subtracting a reference spectrum representative of a spin label on a disordered Tar polypeptide backbone (see text). The reference spectra were for Tar spin labeled at position 543 in the disordered carboxyl-terminal 35-residue Tar flexible arm51 and two examples of cholate-solubilized Tar denatured in 4M urea, labeled at position 478 and 483, respectively. The two right-most columns show average percent contributions of a highly mobile component determined for each example spectra (Avg.) and the respective standard deviations (SD) of that average.
Position 225 had no apparent mobile component.
A dominant mobile component made analysis difficult. Thus, the values are rough estimates of the high proportion of mobile component.
Mobility of spin labels in chemoreceptor Tar solubilized in detergent
Less than 7.5% of the mass of Tar and related chemoreceptors is embedded in the hydrophobic environment of the membrane lipid bilayer,4,58 yet detergent-solubilized receptors cannot perform most of their characteristic functions.37,59 Could inactivation by detergent solubilization be correlated with changes in backbone dynamics? We investigated by comparing spin-label mobility at our 16 positions on the surface of the cytoplasmic domain of intact Tar in two states: solubilized in the detergent cholate and inserted into a native lipid bilayer provided by a Nanodisc. EPR spectra (Fig. 5) and the corresponding pattern of h(+1)/h(0) and Δ as a function of position (Fig. 6) revealed that spin-label mobility for many but not all positions was increased in the detergent-solubilized state. For spin labels on the N-helix, increases in mobility were substantial when expressed in terms of h(+1)/h(0) but appeared smaller when expressed in terms of Δ
as a function of position (Fig. 6) revealed that spin-label mobility for many but not all positions was increased in the detergent-solubilized state. For spin labels on the N-helix, increases in mobility were substantial when expressed in terms of h(+1)/h(0) but appeared smaller when expressed in terms of Δ , probably reflecting the limited sensitivity of the latter parameter to further increases for already high label mobilities. For most positions on the C-helix, spin-labels exhibited mobilities as high as those for spin labels on the N-helix of the bilayer-inserted receptor. For the already dynamic positions in the HAMP AS2 helix and the N-helix, spin-label mobilities for detergent-solubilized receptor were even higher. One or both mobility parameters were not significantly increased at three of the four positions in the protein interaction region (358, 378, and 396) or at the one position tested in AS1 (225) (Figs. 5 and 6). EPR line shapes for these positions were typical for spectra of spin-labels at relatively stable sites,43–45 although with a slightly increased mobile component relative to Tar in a lipid bilayer for the protein-interaction positions and slightly decreased for the AS1 position. The lack of significant mobility changes at several positions upon detergent solubilization indicates that the increased mobility observed at other positions cannot be the result of a difference in rotational motion of entire protein in Nanodiscs versus detergent. Interestingly, detergent-solubilized and thus functionally inactivated Tar maintained the differential dynamics of higher mobility of surface spin labels over approximately two-thirds of the cytoplasmic domain for the N-helix versus the C-helix. This implies that important features of the receptor structural organization were maintained in the detergent-solubilized state, even though function had been lost.
, probably reflecting the limited sensitivity of the latter parameter to further increases for already high label mobilities. For most positions on the C-helix, spin-labels exhibited mobilities as high as those for spin labels on the N-helix of the bilayer-inserted receptor. For the already dynamic positions in the HAMP AS2 helix and the N-helix, spin-label mobilities for detergent-solubilized receptor were even higher. One or both mobility parameters were not significantly increased at three of the four positions in the protein interaction region (358, 378, and 396) or at the one position tested in AS1 (225) (Figs. 5 and 6). EPR line shapes for these positions were typical for spectra of spin-labels at relatively stable sites,43–45 although with a slightly increased mobile component relative to Tar in a lipid bilayer for the protein-interaction positions and slightly decreased for the AS1 position. The lack of significant mobility changes at several positions upon detergent solubilization indicates that the increased mobility observed at other positions cannot be the result of a difference in rotational motion of entire protein in Nanodiscs versus detergent. Interestingly, detergent-solubilized and thus functionally inactivated Tar maintained the differential dynamics of higher mobility of surface spin labels over approximately two-thirds of the cytoplasmic domain for the N-helix versus the C-helix. This implies that important features of the receptor structural organization were maintained in the detergent-solubilized state, even though function had been lost.
Figure 5.
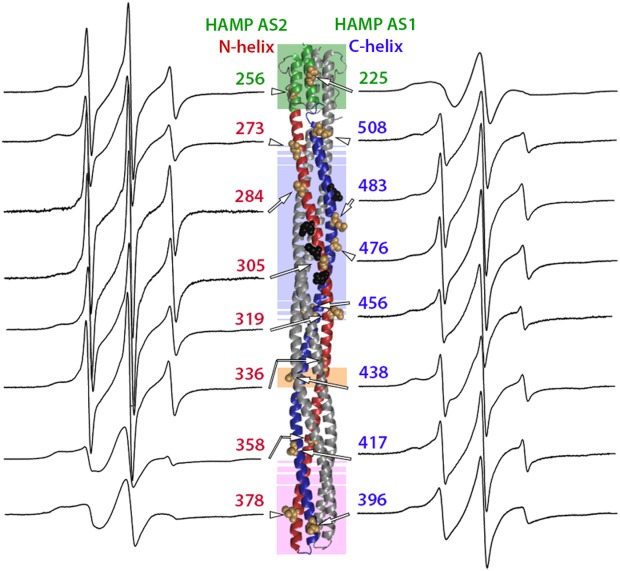
Spectra of purified, spin-labeled Tar solubilized in the detergent cholate. Normalized EPR spectra for 16 positions in the Tar cytoplasmic domain are displayed as in Figure 2 on each side of a ribbon-diagram of that domain, with each spectrum near the spin label position on the diagram and an arrow pointing to the position of a CPK representation for the native side-chain. Spectra for positions on the N-helix and HAMP AS2 helix are on the left, and spectra for positions on the C-helix and HAMP AS1 are on the right. As in Figures 1(A) and 2, shaded regions indicate functional regions: HAMP pale green, modification region pale blue, glycine hinge pale orange and protein interaction region pale pink. Sites of adaptational modification are shown as black CPK side-chains.
Figure 6.
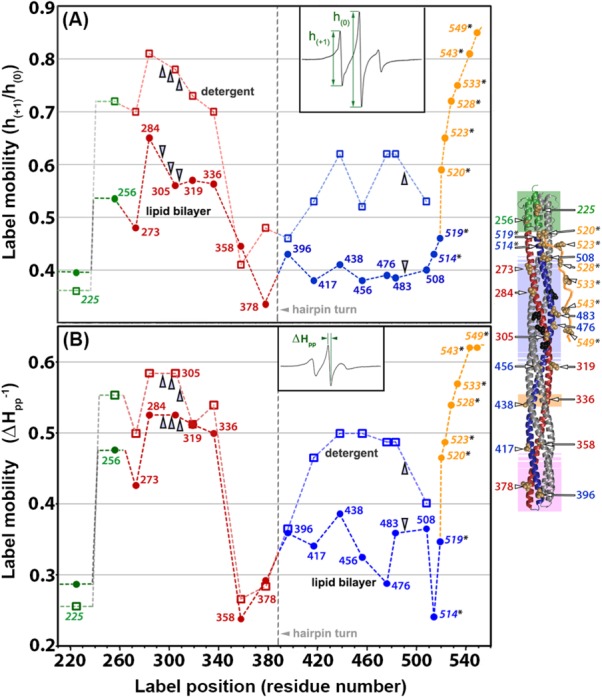
Spin-label mobility for Tar in a native lipid bilayer versus solubilized in detergent. Mobility parameters h(+1)/h(0) [Fig. 6(A)] and Δ [Fig. 6(B)] (see insets and text) for intact Tar inserted in a native lipid bilayer are plotted as a function of residue number (circles and dashed line labeled “lipid bilayer”) and compared to the respective mobility parameters for the same protein solubilized in the detergent cholate (squares and lighter dashed line labeled “detergent”). The spin-label positions are shown in the ribbon diagram of the receptor cytoplasmic domain to the right of the plot, in which each position is indicated by CPK models of the side chains and a residue number. Sites of adaptational modification are indicated by open arrowheads. A vertical grey line marks the position of the hairpin turn, which is at the membrane-distal tip of the receptor cytoplasmic domain and appears at the bottom of the ribbon diagram to the right. Dashed lines illustrate backbone connectivity between residue positions. Both lines and data points are color coded by structural element: HAMP green, N-helix red, C-helix blue, and disordered, flexible arm orange; the two helices and the flexible arm are labeled on the plot. The data for position 225 and 514–54951 are as shown in Figure 3.
[Fig. 6(B)] (see insets and text) for intact Tar inserted in a native lipid bilayer are plotted as a function of residue number (circles and dashed line labeled “lipid bilayer”) and compared to the respective mobility parameters for the same protein solubilized in the detergent cholate (squares and lighter dashed line labeled “detergent”). The spin-label positions are shown in the ribbon diagram of the receptor cytoplasmic domain to the right of the plot, in which each position is indicated by CPK models of the side chains and a residue number. Sites of adaptational modification are indicated by open arrowheads. A vertical grey line marks the position of the hairpin turn, which is at the membrane-distal tip of the receptor cytoplasmic domain and appears at the bottom of the ribbon diagram to the right. Dashed lines illustrate backbone connectivity between residue positions. Both lines and data points are color coded by structural element: HAMP green, N-helix red, C-helix blue, and disordered, flexible arm orange; the two helices and the flexible arm are labeled on the plot. The data for position 225 and 514–54951 are as shown in Figure 3.
Discussion
We used site-directed spin labeling and continuous-wave EPR spectroscopy to probe helical dynamics in the cytoplasmic domain of a full-length bacterial chemoreceptor inserted in a bilayer of its native lipid. Analysis of 16 spin-labeled sites distributed along the solvent-exposed, noninteracting surface of the extended helical structure revealed segments of well-structured helices and of helices with significant dynamics. The considerable helical dynamics we detected is consistent with previous observations (see following section). In addition, we observed a striking and unanticipated difference in dynamics between the two companion helices of the helical hairpins that form the domain's four-helix, coiled-coil structure (Figs. 4). Differential dynamics over approximately two-thirds of the length of the two helices in the helical hairpins suggests that the extended four-helix bundle is organized over that length as a scaffold core of two C-helices plus two dynamic partner N-helices. This concept provides a new way to view the structure of the chemoreceptor cytoplasmic domain and may have implications for the understanding of receptor function. More generally, organization of this four-helix bundle as a scaffold core with dynamic partners highlights the possibility of structural asymmetry in helical bundles. These observations and implications are considered in more detail in the following sections.
Correlations with previous observations
The significant helical dynamics we observed in the chemoreceptor cytoplasmic domain is consistent with previous biochemical and structural observations. These include characterization of intact, functional chemoreceptors contained in native membrane vesicles.22,25,60–62 This series of studies observed that the cytoplasmic domain was sufficiently dynamic that disulfides between engineered cysteines placed at homologous positions in the two protomers of the chemoreceptor dimer formed rapidly and essentially quantitatively in conditions of mild oxidation, independent of sequence position and thus of distance in the average three-dimensional structure. Indications of a dynamic nature of the chemoreceptor cytoplasmic domain have also been provided by structural and biophysical characterization of fragments of the extended coiled-coil portion of that domain and of similar fragments fused to HAMP domains from heterologous species. These studies include characterizations of a fragment containing most of the coiled-coil portion of an E. coli chemoreceptor cytoplasmic domain by NMR23 and by hydrogen/tritium exchange.24 An X-ray crystallographic structure of a similar fragment from a closely related E. coli chemoreceptor (1QU710) or of that fragment fused to a HAMP domain from a distantly related bacterium (3ZX614) had high crystallographic temperature factors for residues in the modification region, implying that in these constructs the region was less firmly structured than other receptor regions. Mass spectrometry was used recently to characterize exchange of amide hydrogens in the hydrogen bonds of the peptide backbone of a cytoplasmic domain fragment of E. coli chemoreceptor Tar incorporated into functional signaling complexes created by vesicle-template assembly.26 The data showed that the adaptation region of the cytoplasmic domain fragment had the highest exchange relative to other operational regions, indicating that the region had the highest backbone dynamics in the domain, a pattern consistent with our observations. A recent study using pulsed dipolar electron spin resonance spectroscopy to characterize a cytoplasmic domain fragment of an E. coli chemoreceptor fused to one or two tandem HAMP domains from a different organism observed broader distributions of distances for spin labels at the fusion joint and at two positions in the modification region than for labels at four positions in the remainder of the domain and inferred that the broad distributions reflected greater structural dynamics.27
Coupling of the HAMP domain to the helical coiled coil
The HAMP domain is thought to convert the piston motion of the TM2 transmembrane domain helix that becomes HAMP helix AS1 into a different conformational change in the cytoplasmic coiled coil.1–3 Using a diagnostic site on the solvent-exposed surface of each of the two helices of the HAMP domain, AS1 and AS2, we found that spin-label mobility at the AS2 site was significantly greater than at the AS1 site or any site on the C-helix. Its higher mobility approached that of sites on the dynamic segment of the N-helix. This suggests that AS2 and its extension as the N-helix could be dynamically coupled. Such coupling could play a role in conformational signaling.
Detergent-solubilized chemoreceptor Tar has a destabilized cytoplasmic domain
We found that loss of chemoreceptor activity in the detergent-solubilized state is correlated with a significant and general increase in helical backbone dynamics for many but not all of the 16 positions we examined in the cytoplasmic domain (Figs. 5 and 6). Whether the increased dynamics reflects reduced interaction of neighboring helices, reduced stability of the backbone hydrogen bonds that create helical secondary structure, or a combination of the two, it seems likely that such structural perturbations would be a major if not the principle contributor to the loss of receptor activity in this domain upon detergent solubilization. The structural observations reported here and previous functional observations41 document a structural and functional coupling between the transmembrane domain and the cytoplasmic domain. This coupling emphasizes the importance of characterizing chemoreceptors in their natural condition of insertion in a native lipid bilayer.
Differential mobility of companion helices in helical coiled coils
Our data document that the spin labels on the C-helix of the chemoreceptor cytoplasmic domain have mobility characteristic of surface-facing positions on a well-structured alpha-helix, whereas spin labels on approximately two-thirds of the companion N-helix have significantly greater mobility, from AS2 of the HAMP domain to the membrane-proximal beginning of the protein-interaction region (Figs. 4). Deconvolution of the relevant spectra suggests that the increased mobility is the result of a higher probability of excursions of the helical backbone into a nonhelical state. These results are consistent with and complimented by patterns of hydrogen/deuterium exchange for amide hydrogens in the hydrogen bonds of the peptide backbone of a cytoplasmic domain fragment of E. coli chemoreceptor Tar incorporated into functional signaling complexes created by vesicle-template assembly.26 For both low- and high-density assemblies of that fragment, N-helix amide hydrogens had higher extents of exchange at 3 min than the companion C-helix amide hydrogens even though the magnitude of those extents and the magnitude of the differences changed from high in the adaptation region to lower in the more membrane-distal parts of the domain.26 These data imply that there is differential stability of the hydrogen bonds of the helical backbone in the two companion helices of the helical hairpin of each receptor protomer, with the C-helix more stable and the N-helix more dynamic. This parallels the conclusion we reach from our characterization by EPR spectroscopy of intact, bilayer-inserted receptor dimers. The authors of the hydrogen exchange study did not comment on the differential exchange rates they observed for companion helices, perhaps because differences could have been an artifactual situation for a receptor fragment missing the HAMP, transmembrane, and periplasmic domains.
Yet their data and ours are complementary. Taken together, the two independent assessments of helical dynamics argue that there are structurally relevant differences in helical dynamics between the companion helices in the helical hairpins of the chemoreceptor cytoplasmic domain. The hydrogen exchange experiments have the strength of probing chemoreceptors in active chemotaxis signaling complexes but the weaknesses that the receptors were fragments, missing the HAMP, transmembrane, and periplasmic domains and their potential structural influences. Our EPR studies have the strength of characterizing intact, functional chemoreceptor dimers inserted into bilayers of native lipids but the potential weakness that these dimers were not in chemotaxis signaling complexes and thus might not be in the same structural state as receptors in signaling complexes. Thus the weakness of one experimental approach is addressed by the strength of the complementary approach. Considering both sets of data, we conclude that differential dynamics of companion helices along approximately two-thirds of the length of the cytoplasmic domain is a structural property of the E. coli chemoreceptor Tar and likely a common feature of bacterial chemoreceptors in general.
Conventionally, helical bundles are considered symmetrical units of equivalently stable helices, rather than helical partners with differential dynamics. However, detailed analysis of a three-helix bundle revealed differential dynamics and thus asymmetry among the three constituent helices.63,64 The chemoreceptor cytoplasmic domain provides an example of a four-helix bundle with differential dynamics and asymmetry among its helices.
Notably, in the periplasmic, ligand-binding domain, there is also a movement-related asymmetry among the helices of a four-helix helical bundle. The bundle begins in the ligand-binding site at the membrane-distal tip and crosses the membrane as the transmembrane domain. It contains two helices from each protomer, TM1/α-1 and α-4/TM2. A large body of data has identified the interdimer interface between TM1/α-1 and TM1'/α-1' as static65–68 and the intradimer interface between TM1/α-1 and α-4/TM2 (as well as the symmetric intradimer interface TM1'/α-1' and α-4'/TM2') as mobile.65–71 Specifically there is a relative piston sliding between those two helices. Thus, the four-helix bundles of the chemoreceptor periplasmic/transmembrane and cytoplasmic domains each contain one pair of helices that move and one more static. The mobile helices, α-4/TM2 in the periplasmic/transmembrane domain and the N-helix from AS-2 to the membrane-distal protein-interaction region in the cytoplasmic domain, span much of the ∼300 Å length of the chemoreceptor. It is tantalizing to consider that the two sets of dynamic helices could be conformationally coupled via the HAMP domain.
Materials and Methods
Strains, plasmids, and proteins
Tar proteins characterized in this study were produced in E. coli K-12 strain RP3098,72 which contains a deletion from flhA to flhD that eliminates the presence or expression of all chemoreceptor and che genes, and which harbored a derivative of the Tar-encoding plasmid pAL67. This plasmid, which carries tar under control of a modified lac promoter as well as lacIq and codes for Tar with six histidines (Tar-6H) added to its carboxyl terminus,53 was altered by site-specific mutagenesis to produce pAL533, which codes for Tar-6H 4Q with glutamines (Q in the single letter code) at the four sites of adaptational modification. Site-specific mutagenesis of pAL533 created plasmids coding for Tar-6H 4Q with cysteines substituted at the indicated residues (native Tar contains no cysteines): pAL754, H256C; pAL726, D273C; pNB3, A284C; pAL728, A305C; pAL757, D319C; pAL759, Q336C; pAL751, K358C; pNB4, L378C; pAL760, V396C; pNB6, A417C; pAL752, E438C; pAL777, E456C; pAL756, S476C; pAL758, Q483C; and pNB8, Q508C. pAL568, which codes for Tar-6H A225C, was created by site-specific mutagenesis of pAL67 and thus it has two glutamines and two glutamates (QEQE) at the sites of adaptational modification. pAL plasmids were constructed by Angela Lilly in our laboratory and pNB plasmids by Mutagenex, Inc. (Piscataway, NJ).
Purification and spin labeling of cysteine-substituted Tar
Cytoplasmic membrane vesicles enriched for each form of cysteine-substituted Tar-6H were isolated from RP3098 harboring the appropriate plasmid using procedures and conditions described51,54 with the exception that all steps had 2 mM DTT present. Frozen membrane vesicles containing 1–2 mg of cysteine-substituted Tar-6H were thawed, added to 1 mL 50 mM Tris–HCl pH 7.5, 10% w/v glycerol, 5.5% octyl β-D-glucopyranoside, 2 µM pepstatin, 2 µM leupeptin, 5 µM TLCK, 100 µM PMSF, incubated 0.5 h on ice and centrifuged 5 min at 10°C in an Eppendorf 5415 D table-top centrifuge at 13,200 RPM to remove insoluble material. The supernatant was applied to a gravity-packed, 1.5 mL bed volume, gravity flow Ni-NTA (Qiagen, Valencia, CA) column pre-equilibrated with 50 mM Tris–HCl pH 7.5, 30 mM imidazole, 100 mM NaCl, 25 mM sodium cholate; the column washed with five bed volumes of the same buffer and His-tagged receptor eluted with three bed volumes of the same buffer with 300 mM imidazole. Spin label reagent (1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate, Toronto Research Chemicals, North York, Canada) was added to 25 µM to the ∼4.5 mL, receptor-containing elutant, providing a ∼fivefold molar excess over anticipated yield of Tar. The solution was incubated for 2 h in the dark on ice, concentrated to <1 mL with an Amicon Ultra-4 (Millipore, Darmstadt, Germany) centrifugal filter device, its buffer exchanged to 50 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 100 mM NaCl, 25 mM sodium cholate using a Nap10 column (GE Healthcare, Little Chalfont, UK), and concentrated to ∼200 µL with an Amicon Ultra-4 (Millipore, Darmstadt, Germany) centrifugal filter device. This procedure typically yielded 100–250 µL of 50–150 µM spin-labeled Tar-6H.
Reconstitution of spin-labeled Tar into single dimer per disc Nanodiscs
Tar-containing Nanodiscs were prepared essentially as described54 with differences noted in the following brief description. The MSP1D1(-)/lipid molar ratio was 1:60 and the Tar-6H/MSP1D1(-) ratio 1:5. MSP was in sufficient excess that most discs formed without containing a receptor and those that had incorporated a six-histidine-tagged receptor dimer, and thus were purified using the nickel column, had an average content of ∼1 receptor dimer per disc.37,38,54 Analysis by electron microscopy and negative staining has revealed that only 5% of ∼1 dimer per discs material contains more than one receptor dimer.40 The (-) in the notation for the form of MSP used indicates that the histidine tag was removed by TEV protease digestion. In a 500 µL volume, 15–25 µM cholate-solubilized, spin-labeled Tar-6H was mixed with 7.5 mM E. coli polar lipids (Avanti Polar Lipids, Alabaster, AL) that had been solubilized in 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 150 mM sodium cholate; 125 µM MSP1D1(-); 2 µM leupeptin; 2 µM pepstatin; and 100 µM PMSF, buffered at pH 7.5 by 50 mM Tris–HCl and incubated at room temperature for 0.5 h. Seven hundred and fifty microliters of SM-2 Biobeads (Bio-Rad, Hercules, CA) were added to remove the ∼40 mM cholate and induce Nanodisc formation, and the mixture incubated with rotation for 1 h at room temperature. The resulting Nanodiscs were separated from Biobeads by making holes smaller than the beads in a tube containing the solution with the tip of an 18 G syringe needle and centrifuging the perforated tube inside a larger tube. The resulting Nanodisc solution was applied to a gravity-packed, gravity-eluted 1 mL bed volume Ni-NTA (Qiagen, Valencia, CA) column equilibrated with 50 mM Tris–HCl pH 7.5, 30 mM imidazole, 100 mM NaCl; the column washed with 5 mL of the same buffer to elute empty Nanodiscs and Tar-6H Nanodiscs eluted with 4 mL of the same buffer containing 300 mM imidazole. Eluted Tar-6H Nanodiscs were concentrated to ∼250 µL in an Amicon Ultra-4 (Millipore, Darmstadt, Germany) centrifugal filter device and buffer exchanged by adding 3 mL 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2 (TNKM 7.5) and concentrating to ∼250 µL using the same filter device. The resulting solution was further concentrated in a Nanosep 30 K Omega centrifugal concentrator (Pall Life Sciences, Port Washington, NY) to 30–45 µL, yielding spin-labeled Tar in Nanodiscs at 30–100 µM.
Electron paramagnetic resonance (EPR) spectroscopy
Continuous wave X-band spectra were collected at 20 mW incident microwave power at room temperature using a Brüker EMX spectrometer (Billerica, MA) equipped with a Brüker ER4123D resonator. Detergent-solubilized receptors were in 50 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 100 mM NaCl, and 25 mM sodium cholate, and receptors in Nanodiscs were in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2. Spectra were taken on 5 µL samples in silicon capillaries. Scans were performed with a 100-kHz field modulation that never exceeded the central peak-to-peak linewidth (ΔHpp) minus 0.5 Gauss. Scan width was a 100 G window centered at the central line-shape feature. Depending on signal strength, 8–64 scans were averaged. Independent acquisition of the spectrum for a spin-label at a given position was performed one to four times, yielding essentially the same spectrum for those acquired more than once, which was the case for 13 of 16 positions characterized for Nanodisc-inserted Tar. Data were processed using Labview-based software developed and distributed by Christian Altenbach (University of California, Los Angeles). Each spectrum was assessed for spectral distortions characteristic of free spin in the sample. For spectra lacking a significant mobile component, such distortions were removed empirically using a computational tool in the Altenbach software. This was possible for positions 225, 358, 378, 396, 417, 438, 456, 476, 483, and 508 of Tar in Nanodiscs and for positions 225, 358, 378, 396 of the receptor solubilized in detergent. For Nanodisc-embedded Tar-free spin was 0–0.9% total free spins and averaged 0.78%. For detergent-solubilized Tar-free spin was 0.7–1.73% total free spins and averaged 1.3%. For the other positions, the spectra exhibited a sufficiently significant mobile component that distortions contributed by free spin would have been difficult to recognize. Instead, we subtracted the average free spin for Nanodisc-embedded or detergent-solubilized receptors from the respective spectra but observed essentially no discernible change in the features of the spectra. For all positions, the free-spin-subtracted spectra and parameters derived from them were used for analysis and presented in Figures 6. For final processing and presentation, spectra were normalized to the same total spins.
Spectral deconvolution
We assessed the contributions of the highly mobile and less mobile components of the spectra of spin-labeled, Nanodisc-inserted Tar using EPR130_NoDAQ, a tool in the Altenbach software. For each spectrum, we performed individual deconvolutions using spectra derived from spin labels on a disordered protein backbone of Tar. The examples were a spin label at position 543 which is very near the end of the disordered, carboxyl-terminal flexible arm of Tar51 and at positions 478 and 483 of Tar denatured with 4M urea. In three parallel analyses, each reference spectrum was subtracted from each experimental spectrum until no more fast component could be removed without producing an aberrant residual EPR spectrum. The contribution to the original spectrum by a highly mobile component was determined by double integral quantification and expressed as percent of total spin labels.
Receptor deamidation catalyzed by methylesterase CheB
Plateau extents of deamidation of the glutamines at the four sites of adaptational modification for each cysteine-substituted Tar-4Q variant were determined essentially as described,41 with the reaction performed in TNKM 7.5 and the extent of modification determined by densitometry of an immunoblot performed with anti-Tar serum.
Acknowledgments
We thank Angela Lilly for preparing numerous plasmids and Wing-Cheung Lai for preparation of the membrane scaffold protein used in Nanodisc preparations.
References
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Lai W-C. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422 doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal J, García-Fontana C, Muñoz-Martínez F, Ramos J-L, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol. 2010;12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Reyes-Darias J-A, Lacal J, Ramos JL, García-Ruiz JM, Gavira JA, Krell T. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci USA. 2012;109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks Kevin G, Foster Jeneva K, Parthasarathy R, Remington SJ, Guillemin K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Kim SH. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr Opin Struct Biol. 2000;10:462–469. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Zeth K, Hulko M, Dunin-Horkawicz S, Lupas AN. Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J Struct Biol. 2014;186:349–356. doi: 10.1016/j.jsb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Airola MV, Watts KJ, Bilwes AM, Crane BR. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol. 2013;425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J Biol Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley SK, Weis RM, Thompson LK. The cytoplasmic fragment of the aspartate receptor displays globally dynamic behavior. Biochemistry. 1996;35:5199–5206. doi: 10.1021/bi9524979. [DOI] [PubMed] [Google Scholar]
- Murphy OJ, Yi X, Weis RM, Thompson LK. Hydrogen exchange reveals a stable and expandable core within the aspartate receptor cytoplasmic domain. J Biol Chem. 2001;276:43262–43269. doi: 10.1074/jbc.M105585200. [DOI] [PubMed] [Google Scholar]
- Winston SE, Mehan R, Falke JJ. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshy SS, Li X, Eyles SJ, Weis RM, Thompson LK. Hydrogen exchange differences between chemoreceptor signaling complexes localize to functionally important subdomains. Biochemistry. 2014;53:7755–7764. doi: 10.1021/bi500657v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Borbat PP, Dzikovski B, Freed JH, Crane BR. Bacterial chemoreceptor dynamics correlate with activity state and are coupled over long distances. Proc Natl Acad Sci USA. 2015;112:2455–2460. doi: 10.1073/pnas.1414155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input–output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Piasta KN. Architecture and signal transduction mechanism of the bacterial chemosensory array: progress, controversies, and challenges. Curr Opin Struct Biol. 2014;29:85–94. doi: 10.1016/j.sbi.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Mol Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. In: Correia JJ, Detrich WH, editors. Methods Cell Biol. Vol. 84. New York: Academic Press; 2008. pp. 617–658. . In: III, editors. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 2013;23:725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin DN, Hazelbauer GL. The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J Bacteriol. 2010;192:1193–1200. doi: 10.1128/JB.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Khursigara CM, Subramaniam S, Hazelbauer GL. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol Microbiol. 2011;79:677–685. doi: 10.1111/j.1365-2958.2010.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin DN, Hazelbauer GL. Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J Biol Chem. 2012;287:41697–41705. doi: 10.1074/jbc.M112.415588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2014;111:15940–15945. doi: 10.1073/pnas.1415184111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4 − 58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: a case study of myoglobin. Biochemistry. 2012;51:6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirman NL, Milshteyn E, Galiano L, Hewlett JC, Fanucci GE. Characterization of the disordered-to-α-helical transition of IA3 by SDSL-EPR spectroscopy. Protein Sci. 2011;20:150–159. doi: 10.1002/pro.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores Jiménez RH, Cafiso DS. The N-terminal domain of a TonB-dependent transporter undergoes a reversible stepwise denaturation. Biochemistry. 2012;51:3642–3650. doi: 10.1021/bi300118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores Jiménez RH, Do Cao M-A, Kim M, Cafiso DS. Osmolytes modulate conformational exchange in solvent-exposed regions of membrane proteins. Protein Sci. 2010;19:269–278. doi: 10.1002/pro.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelli NL, Hazelbauer GL. Direct evidence that the carboxyl-terminal sequence of a bacterial chemoreceptor is an unstructured linker and enzyme tether. Protein Sci. 2011;20:1856–1866. doi: 10.1002/pro.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov A, Altenbach C, Barnakova L, Hubbell WL, Hazelbauer GL. Site-directed spin labeling of a bacterial chemoreceptor reveals a dynamic, loosely packed transmembrane domain. Protein Sci. 2002;11:1472–1481. doi: 10.1110/ps.0202502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W-C, Hazelbauer GL. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J Bacteriol. 2005;187:5115–5121. doi: 10.1128/JB.187.15.5115-5121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Structural features of the C-terminal domain of bovine rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- Morin B, Bourhis J-M, Belle V, Woudstra M, Carrière F, Guigliarelli B, Fournel A, Longhi S. Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling electron paramagnetic resonance spectroscopy. J Phys Chem B. 2006;110:20596–20608. doi: 10.1021/jp063708u. [DOI] [PubMed] [Google Scholar]
- Belle V, Rouger S, Costanzo S, Liquière E, Strancar J, Guigliarelli B, Fournel A, Longhi S. Mapping α-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins. 2008;73:973–988. doi: 10.1002/prot.22125. [DOI] [PubMed] [Google Scholar]
- Boldog T, Hazelbauer GL. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 2004;13:1466–1475. doi: 10.1110/ps.04648604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogonez E, Koshland DE., Jr Solubilization of a vectorial transmembrane receptor in functional form: aspartate receptor of chemotaxis. Proc Natl Acad Sci USA. 1985;82:4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass RB, Falke JJ. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J Biol Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass RB, Coleman MD, Falke JJ. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh STR, Cheng H, Bryson JW, Roder H, DeGrado WF. Solution structure and dynamics of a de novo designed three-helix bundle protein. Proc Natl Acad Sci USA. 1999;96:5486–5491. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh STR, Cheng RP, Wright WW, Alonso DOV, Daggett V, Vanderkooi JM, DeGrado WF. The hydration of amides in helices; a comprehensive picture from molecular dynamics, IR, and NMR. Protein Sci. 2003;12:520–531. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GF, Lebert MR, Lilly AA, Hazelbauer GL. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl cross-linking in vivo. Proc Natl Acad Sci USA. 1995;92:3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Lin CM, Falke JJ. Transmembrane signaling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughson AG, Hazelbauer GL. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc Natl Acad Sci USA. 1996;93:11546–11551. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- Peach ML, Hazelbauer GL, Lybrand TP. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 2002;11:912–923. doi: 10.1110/ps.4640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W-C, Beel BD, Hazelbauer GL. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol Microbiol. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


