Abstract
Hfq proteins in Gram-negative bacteria play important roles in bacterial physiology and virulence, mediated by binding of the Hfq hexamer to small RNAs and/or mRNAs to post-transcriptionally regulate gene expression. However, the physiological role of Hfqs in Gram-positive bacteria is less clear. Bacillus anthracis, the causative agent of anthrax, uniquely expresses three distinct Hfq proteins, two from the chromosome (Hfq1, Hfq2) and one from its pXO1 virulence plasmid (Hfq3). The protein sequences of Hfq1 and 3 are evolutionarily distinct from those of Hfq2 and of Hfqs found in other Bacilli. Here, the quaternary structure of each B. anthracis Hfq protein, as produced heterologously in Escherichia coli, was characterized. While Hfq2 adopts the expected hexamer structure, Hfq1 does not form similarly stable hexamers in vitro. The impact on the monomer–hexamer equilibrium of varying Hfq C-terminal tail length and other sequence differences among the Hfqs was examined, and a sequence region of the Hfq proteins that was involved in hexamer formation was identified. It was found that, in addition to the distinct higher-order structures of the Hfq homologs, they give rise to different phenotypes. Hfq1 has a disruptive effect on the function of E. coli Hfq in vivo, while Hfq3 expression at high levels is toxic to E. coli but also partially complements Hfq function in E. coli. These results set the stage for future studies of the roles of these proteins in B. anthracis physiology and for the identification of sequence determinants of phenotypic complementation.
Keywords: Hfq, small RNA, sRNA, Bacillus anthracis, anthrax, pXO1
Introduction
Hfq is a small, RNA-binding protein involved in post-transcriptional gene regulation in eubacteria, with roles in binding and facilitating interactions between mRNAs and small non-coding RNAs (sRNAs),1,2 as well as in modulating the stability of certain mRNAs.3
Hfq belongs to the family of Sm and Sm-like RNA-binding proteins, identified in all domains of life.4 In Gram-negative eubacteria, RNA binding by Hfq is important in cell physiology; Δhfq Escherichia coli exhibit pleiotropic defects, including reduced growth rate and impaired oxidative stress response.5 Δhfq strains of many Gram-negative pathogens, including Salmonella, also exhibit decreased virulence.6 In contrast, the role of hfq in Gram-positive bacteria is less clear; for example, regulation by small RNAs in Staphylococcus aureus is unaffected in an hfq null mutant.7 However, small RNAs are abundant in Gram-positive bacteria, and a recent report identified the first evidence for an hfq-dependent growth phenotype in (Gram-positive) Clostridium difficile.8
Structurally, Hfq monomers consist of a single α-helix followed by five β-strands and a long, unstructured C-terminal extension, although this extension is not present in the Hfqs of most low-GC Gram-positive bacteria.9 Eubacterial members of this protein family form well-conserved homohexameric rings that can bind RNA on the distal and proximal faces; rings are stabilized by interactions between the β4 and β5 strands that create a continuous β-sheet around the ring.10,11 In contrast, eukaryotic LSm/Sm proteins form heteroheptamers.12
While the majority of bacteria, including Bacillus subtilis, carry only one copy of hfq, Bacillus anthracis (the causative agent of anthrax) carries three. The roles of these multiple Hfqs are not understood; however, recent work in B. anthracis has identified small RNAs upregulated under virulence-related conditions,13 providing additional impetus for exploring the role of Hfqs and sRNAs in B. anthracis physiology. One of the few bacteria expressing multiple Hfq homologs is Burkholderia cenocepacia,14 in which one of the two Hfq proteins is unusually large, forms trimers instead of hexamers, and plays a role in virulence and stress response.15
Here, we characterize the biochemical and biophysical properties of the three B. anthracis Hfqs, as produced recombinantly in E. coli. We observe differences in quaternary structure and probe the role of Hfq sequence in hexamer formation. We also explore the implications of Hfq sequence variation on complementation of the Δhfq phenotype in E. coli. In addition to providing the first step in examining roles of these proteins in B. anthracis physiology, our work contributes to the overall understanding of structure–function relationships for the Hfq family.
Results
Sequence analysis of Hfqs
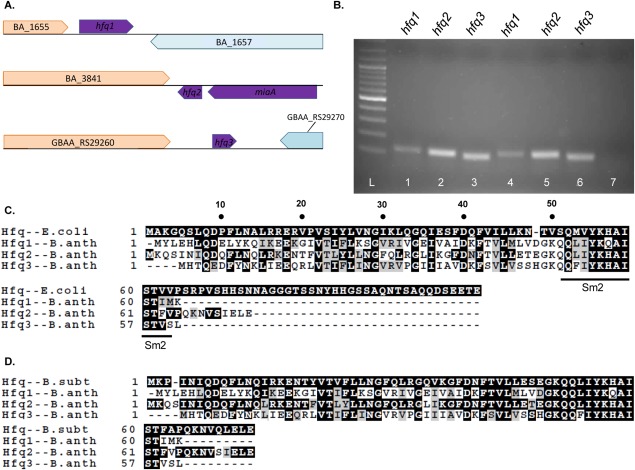
The genome of B. anthracis is unusual in that it carries three hfq homologs [Fig. 1(A)]. hfq2 (BA_3842) is syntenic with the single hfq locus in most bacteria. hfq1 (BA_1656) is located in a distinct section of the chromosome, while hfq3 (GBAA_RS29265) is encoded by the virulence plasmid pXO1. hfq1 and hfq2 are also conserved in the close relatives Bacillus cereus and Bacillus thuringiensis [Supporting Information Fig. 1(A,B)]. Certain strains of B. cereus, like 03BB108, carry a pXO1-like plasmid that encodes hfq3. All three mRNAs are expressed in mid- to late-log phase cultures of B. anthracis [Fig. 1(B)].
Figure 1.
Hfq genes and proteins. (A) Location of each Hfq homolog in B. anthracis genome. (B) 2% agarose gel of RT-PCR products amplified from B. anthracis RNA, using hfq primers as indicated. Lanes 1–3, RNA collected at OD600 = 0.5; Lanes 4–6, OD600 = 0.7; Lane 7, negative control including all three primers. (C–D) Alignment of protein sequences of B. anthracis Hfqs with sequences of (C) E. coli Hfq and (D) B. subtilis Hfq using ClustalW. Images were generated with BOXSHADE3.2, with the upper sequence used as the consensus, dark boxes designating identity to consensus, and light boxes designating biochemical similarity to consensus.
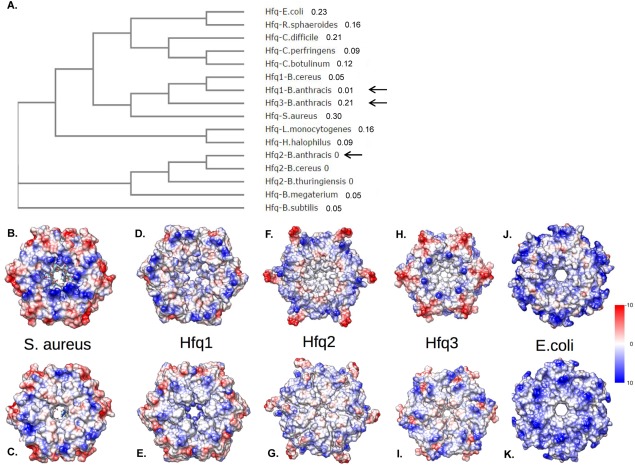
When B. anthracis Hfq protein sequences are aligned against E. coli and B. subtilis Hfqs [Fig. 1(C,D)], Hfq2 exhibits highest protein sequence conservation with B. subtilis Hfq, consistent with its conserved genomic location. All three B. anthracis Hfqs lack the long C-terminal tail of E. coli Hfq [Fig. 1(C)]. The C-terminus of Hfq2 is similar in length to those of S. aureus [Supporting Information Fig. 1(C)] and B. subtilis [Fig. 1(D)] Hfqs, but Hfq1 and Hfq3 exhibit additional truncations. In an unrooted cladogram, the evolutionary distances of Hfq1 and Hfq3 from both Hfq2 and Hfqs of related species are evident [Fig. 2(A); Supporting Information Fig. 2].
Figure 2.
Bacillus anthracis Hfq phylogeny and modeling. (A) Unrooted cladogram depicting relationship of Hfq1-3 protein sequences (black arrows) with those of other bacterial species. Numbers designate branch lengths. (B–K) Models of Hfq hexamers. Upper row depicts proximal face and lower row depicts distal face for the crystal structure of S. aureus Hfq (B,C); models of B. anthracis Hfq1, 2, and 3 (D–I); or the crystal structure of E. coli Hfq (J,K). Surfaces are colored by electrostatic surface potential, from blue (highly positive) to red (highly negative).
Predicted molar masses and pIs of the monomeric proteins are, respectively: Hfq1, 7.3 kDa, pI 8.2; Hfq2, 8.5 kDa, pI 8.3; and Hfq3, 6.8 kDa, pI 8.4. We used MODELLER to create structural and surface charge models of each B. anthracis Hfq as a hexamer [Fig. 2(B–K)]. The predicted surface charge patterns for Hfq1 and Hfq2 are distinct from those in S. aureus and E. coli. Hfq3 has the surface charge pattern most similar to that of S. aureus, with an internal ring of negative charges.
Characterization of Hfq1
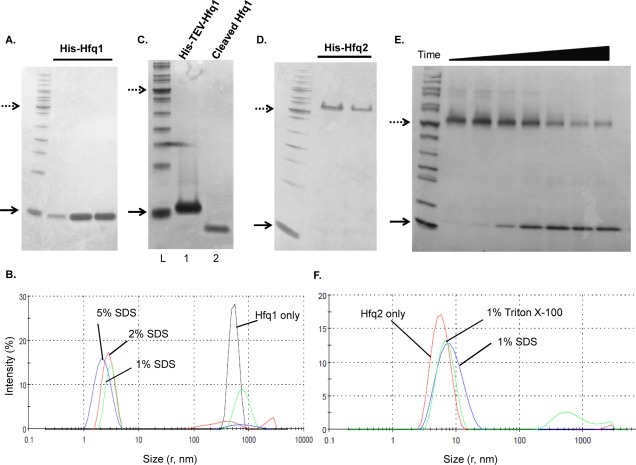
Bacillus anthracis Hfqs were expressed with N-terminal His-tags in E. coli from T5- or T7-based plasmids. His-Hfq1, as expressed from the T5 promoter, was recovered from the soluble fraction. In the absence of boiling but the presence of 4% SDS (Tricine semi-native PAGE), His-Hfq1 was monomeric [Fig. 3(A)]. This is in contrast to Hfqs from E. coli and most other bacteria, which migrate as hexamers in semi-native PAGE.16 Electrophoresis at lower SDS concentrations (sample 0.4%–4%, running buffer 0.1%) yielded the same results [Supporting Information Fig. 3(A)]. Analysis of His-Hfq1 by analytical ultracentrifugation (AUC) at multiple concentrations also demonstrated it to be predominantly monomeric [Fig. 4(A)]. Mass spectrometric analysis of His-Hfq1 matched the expected size, indicating that its monomeric nature is not due to degradation [Supporting Information Fig. 4(A)].
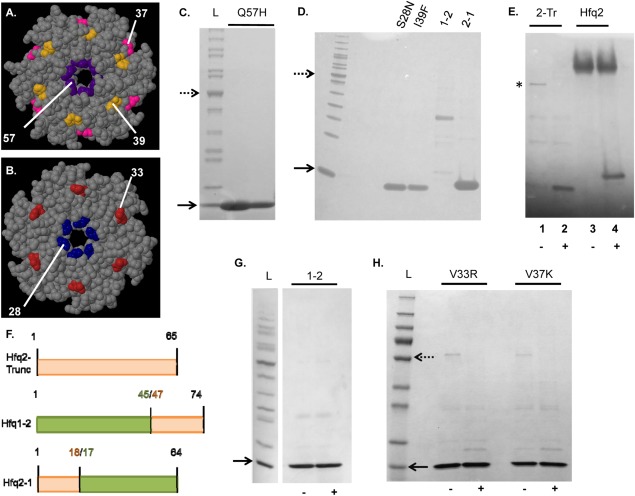
Figure 3.
Purification and characterization of Hfq1 and Hfq2. Dotted (50 kDa) and solid (10 kDa) arrows mark protein ladder reference bands; samples are run as semi-native unless indicated. (A) Monomeric appearance of His-Hfq1; fractions from nickel column on 16% Tricine. (B) DLS of His-Hfq1 samples, incubated overnight at room temperature. Black curve—no additives; Green curve—1% SDS; Red curve—2% SDS; Blue curve—5% SDS. (C) Monomeric appearance of His-TEV-Hfq1 and Hfq1; 16% Tricine. Lane 1, uncleaved protein; Lane 2, cleaved and purified protein. (D) Hexameric appearance of His-Hfq2; 4%–20% Tris–Glycine. (E) Appearance and dissociation of His-TEV-Hfq2 hexamers, with increasing boiling time before loading to 4%–20% Tris–Glycine. From left, boiling times were 0″, 30″, 1′, 2′, 4′, 8′, and 12′. (F) DLS of His-TEV-Hfq2, performed in Glycerol Buffer (Supporting Information) + 400 mM imidazole post-elution from nickel. Results are the average of two independent accumulations at 25°C and are displayed here under the standard viscosity setting; RH values in Results are corrected for the theoretical effect of the 5% glycerol on solution viscosity.
Figure 4.
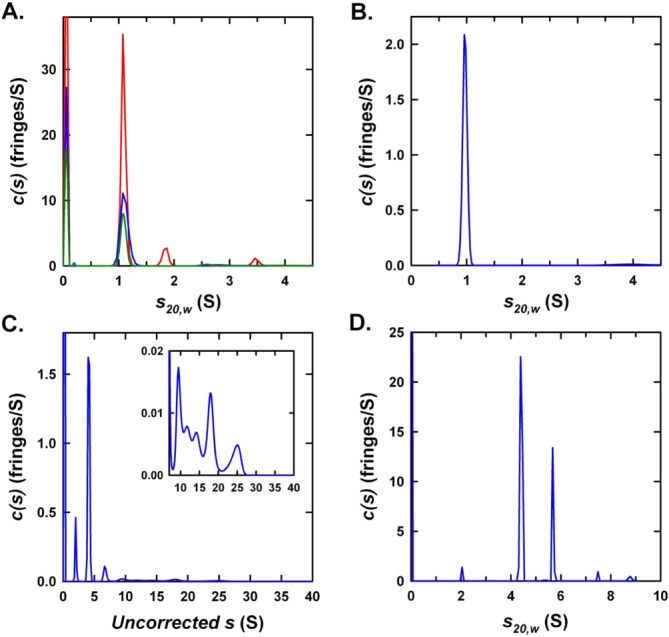
Analytical ultracentrifugation of Hfq1 and Hfq2. Rayleigh interference c(s) distributions for (A) His-Hfq1 at 120 (red), 60 (blue), and 30 µM (green) in 500 mM NaCl and 20 mM Tris–HCl (pH 7.4); (B) Hfq1 at 8.3 µM in 500 mM NaCl and 50 mM Tris–HCl (pH 7.4); (C) His-Hfq2 in 500 mM NaCl and 20 mM Tris–HCl (pH 7.4); and (D) His-TEV-Hfq2 at 125 µM in 1M NaCl, 50 mM Tris–HCl (pH 7.4) and 5% (v/v) glycerol. Inset in (C) expands intensity scale for the faster sedimenting species, which have disproportionate absorbance contributions. Data for (A) show a major species at 1.09S with estimated molar mass of 9.8 kDa, indicative of a His-Hfq1 monomer (expected monomer mass = 8797 Da). Data for (B) show a single species at 0.96S with estimated molar mass of 7.5 kDa (expected monomer mass = 7325 Da).
Analysis of His-Hfq1 by analytical size-exclusion chromatography (data not shown) as well as dynamic light scattering (DLS) detected large aggregates; these could be disrupted by incubation with increasing SDS concentrations [Fig. 3(B), black curve]. At 1% or 2% SDS [Fig. 3(B); green, red curves], particle radius is ≈3 nm, the size expected for hexamers. These complexes may represent true but unstable hexamers, or disordered aggregates derived by detergent disruption of larger particles. Further disruption of complexes occurred at 5% SDS [Fig. 3(B), blue curve]. After overnight incubation with 0.5% CHAPS or 1M urea, a subpopulation with radius ≈1.6 nm was observed [Supporting Information Fig. 4(B)], consistent with the calculated 1.7-nm size of a 76-amino acid globular protein, but smaller than the expected 2.6-nm size of an unfolded polypeptide chain of this length.17 Therefore, we propose that His-Hfq1 folds, then forms soluble aggregates, which may sediment too quickly to be observed in AUC or are less stable under the buffer conditions of AUC.
To demonstrate that the monomeric behavior of Hfq1 is not due to effects of the His-tag, we prepared a T7-driven, TEV-cleavable construct; Hfq1 cleaved to remove the His-tag still separated as a monomer in semi-native PAGE [Fig. 3(C)]. This was confirmed by AUC of Hfq1, which gave a sedimentation coefficient consistent with a monomeric species with a mass of 7.5 kDa, corresponding to the calculated molar mass of 7.3 kDa [Fig. 4(B)].
Characterization of Hfq2
T5-expressed His-Hfq2 separated on semi-native Tris–glycine gels at the expected hexameric size, ≈60 kDa [Fig. 3(D)]. High A260/A280 ratios (>2) and higher molecular weight complexes with absorbance at 260 nm in AUC [Fig. 4(C); Supporting Information Fig. 3(B)] indicated binding of nucleic acids, as expected for Hfq hexamers. Larger yields of His-TEV-Hfq2 were generated by T7 expression. Purified His-TEV-Hfq2 protein migrated as a hexamer; with increasing boiling time in SDS, hexameric and higher-order complexes dissociated to monomers [Fig. 3(E)]. The absence of complete liberation of monomer may be due to interhexameric aggregation after purification, as observed here above the hexamer [left side, Fig. 3(E)], just as in the case of E. coli Hfq.16 An on-column 2M urea wash of His-TEV-Hfq2 was sufficient to remove nucleic acids, yielding an A260/A280 ratio of 0.9 (vs. predicted = 0.8 for pure His-TEV-Hfq2). Preliminary results indicated binding of urea-washed Hfq2 to purine-rich oligoribonucleotides [Supporting Information Fig. 3(C)]. Primary and secondary structures were as predicted [Supporting Information Figs. 3(D) and 4(C)]. Further studies utilized the uncleaved protein because of difficulty in cleavage of tags off the hexamers, presumably due to interhexameric aggregation.
AUC was used to further characterize quaternary structure of His-TEV-Hfq2. Interference [Fig. 4(D)] and absorbance [280 nm; Supporting Information Fig. 3(E)] profiles identified two major species, one at 4.40S (estimated mass of 48 kDa) and one at 5.59S (estimated mass of 70 kDa). Neither species matches the calculated hexameric molar mass of 64 kDa. However, the addition to the buffers of both glycerol and 1M NaCl (to reduce aggregation) is expected to influence the effective partial specific volume such that corrections to standard conditions and estimated molar masses become unreliable in the absence of further information.18,19 These effects may be exacerbated by the presence of a central cavity within the homohexamer that excludes glycerol and NaCl cosolutes.
The quaternary structure of Hfq2 was also analyzed by DLS [Fig. 3(F)]. A primarily monodisperse peak with a hydrodynamic radius (RH) of ≈4.8 nm was observed, with a small increase in radius after incubation with denaturant (1% SDS; 6.4 nm). This RH is larger than values from crystal structure measurements and DLS measurements (Rg and Rs ≈ 3.5 nm) for E. coli Hfq hexamers,10,20 but smaller than the Rg ≈ 7 nm predicted from small-angle X-ray scattering on E. coli Hfq, as well as the large RH estimated by DLS in the same study.16 Our measurement reflects the presence of multimers of the hexameric rings, consistent with their predominance at Hfq concentrations greater than 1 µM for E. coli Hfq.16 Taken together, our results suggest that, unlike Hfq1, His-TEV-Hfq2 forms discrete, stable higher molecular weight complexes consistent with the typical Hfq hexameric structure and higher-order multimers.
Characterization of Hfq3
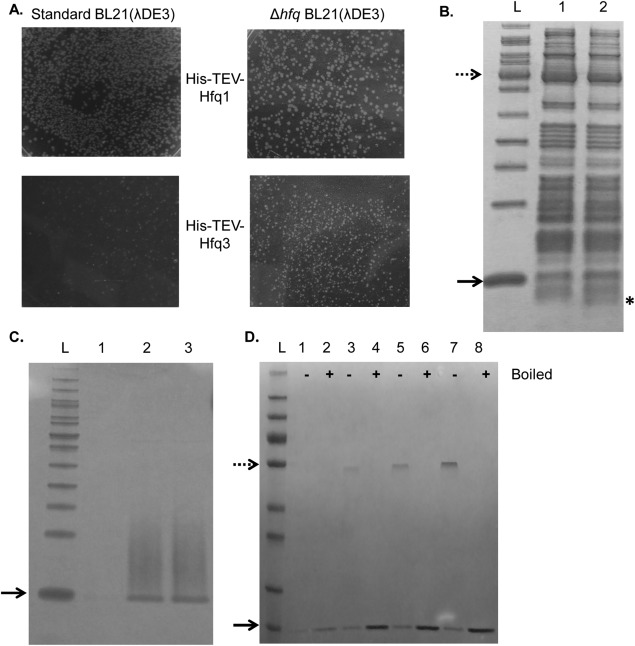
Upon transformation of a T7-His-Hfq3 expression vector into E. coli BL21(λDE3), only sparse, sickly colonies were obtained [Fig. 5(A), lower left], as compared with the larger colonies of bacteria carrying the corresponding T7-His-Hfq1 vector [Fig. 5(A), upper left], suggesting that leaky production of His-Hfq3 is toxic to E. coli. Δhfq E. coli transformed with the T7-His-Hfq3 vector exhibited larger colonies [Fig. 5(A), lower right], suggesting that the toxic effect of Hfq3 involves interaction with E. coli Hfq.
Figure 5.
Characterization of Hfq3. Dotted (50 kDa) and solid (10 kDa) arrows mark protein ladder reference bands; samples are run as semi-native unless indicated. (A) Expression toxicity of His-TEV-Hfq3. Images from a section at the middle of each transformation plate are depicted. (B) Cell-free expression of Hfq3; 16% Tricine. Lane 1, negative control; Lane 2, reaction with Hfq3 template; location of the expressed protein band in Lane 2 indicated with asterisk. (C) On-column refolding trial of His-Hfq3; 16% Tricine. Lanes 1–3: Fractions eluted from nickel after refolding. (D) Additional on-column refolding trial of His-Hfq3; 4%–20% Tris–Glycine. Sample were run as semi-native (−) or boiled (+). Note that the dotted arrow represents 55 kDa, and not 50 kDa, in size in (D).
First, we employed a cell-free expression system to produce untagged Hfq3. When synthesis reactions were separated by semi-native SDS-PAGE, a product having the expected 7-kDa size was observed from reactions done with [Fig. 5(B), Lane 2, asterisk], but not without [Fig. 5(B), Lane 1], the Hfq3 template, suggesting that Hfq3 exists as a monomer under these conditions. Next, while His-Hfq3 was successfully expressed from the T5 system, appreciable protein was recovered only from the 8M urea-soluble fraction at 30°C or 37°C. In two replicates of refolding on the nickel-chelate column and subsequent elution, the His-Hfq3 appeared monomeric on semi-native SDS-PAGE, but had a high propensity to aggregate, as seen by the streaky appearance [Fig. 5(C)]. Additionally, evidence of a faint band in the hexamer range is present [Fig. 5(C), Lanes 2–3]. In a third refolding experiment, the level of visible aggregation was lower, and a more abundant band that appears to be hexameric His-Hfq3 was observed in addition to monomeric His-Hfq3 [Fig. 5(D); monomeric size is consistent with His-Hfq3, but not E. coli Hfq]. In the refolding process, formation of nonspecific (monomeric) aggregates may compete with productive formation of Hfq3 hexamers.
Analysis of determinants of hexameric stability
Our results indicate that some feature(s) of the B. anthracis Hfq1 (and possibly Hfq3) sequence reduces hexamer stability. To identify residues that influence quaternary structure, we made a series of substitutions in His-Hfq1, selecting positions at which the Hfq1 residue differs from one shared by B. anthracis Hfq2 and E. coli Hfq. (Note that we are using E. coli residue numbering here, for easier comparison with the mutagenesis literature).
Hfq1 carries a Gln substitution at His57 in the Sm2-like motif [Fig. 1(C)], which is located in close proximity to hydrogen bond-forming residues at Hfq monomer interfaces in E. coli [Fig. 6(A)]. Mutation of H57 decreases hexamer stability in Pseudomonas aeruginosa Hfq.21 However, as observed by semi-native PAGE, Hfq1-Q57H was still monomeric [Fig. 6(C)]. We additionally examined the effect of S28N and I39F mutations [Fig. 6(A,B)]. In E. coli Hfq, F39 is on the RNA-binding surface around the ring pore. N28 is in the loop between the β1 and β2 strands, and is predicted to make intersubunit hydrogen bonds; however, P. aeruginosa N28A Hfq still forms hexamers.22 For His-Hfq1, both the S28N and I39F variants retained monomeric behavior [Fig. 6(D)].
Figure 6.
Analysis of sequence determinants for hexamer formation. Panels A and B depict Jmol-generated images of E. coli Hfq (PDB 3QHS). (A, B) Proximal and distal faces, respectively, of Hfq ring, with mutated residues highlighted; numbers provided are E. coli residue numbers. (C–H) depict semi-native SDS-PAGE, except as indicated. (C) His-Hfq1-Q57H; 4%–20% Tris–glycine. (D) His-Hfq1 mutants (S28N and I39F) and Hfq1-Hfq2 chimeras; 16% Tricine. (E) His-Hfq2-truncated protein (indicated with asterisk) in absence (−) versus presence (+) of boiling, next to His-TEV-Hfq2; 16% Tricine. (F) Construction of chimeric and truncated constructs. Hfq2 segments are depicted in orange and Hfq1 segments are depicted in green. “Split” residue numbers indicate numbering at each side of the junction. (G) Hfq1-2 chimera; 4%–20% Tris–Glycine; in absence (−) versus presence (+) of boiling. (H) His-Hfq1 mutants (V33R and V37K) in absence (−) and presence (+) of boiling; 4%–20% Tris–glycine. Additional band observed in each preparation appears to be contaminating E. coli Hfq, suggesting that Hfq1 can interact with other Hfqs. In (H), the dotted arrow represents 55 kDa, and not 50 kDa, in size. Hfq1 mutants and chimeras in this figure were assessed at concentrations greater than 8 µM (8–30 µM).
Next, we examined the impact of C-terminal tail length on formation/stability of hexamers, since Hfq1 and Hfq3 are notable for very short tails, and the C-terminal tail of E. coli Hfq abuts the final β-strand of the Hfq monomer (which helps form the intersubunit interface). While a C-terminally truncated E. coli Hfq (approximately the same length as Hfq1/Hfq3) can form hexamers and function in vivo, the hexamer:monomer ratio on a semi-native gel was demonstrated to be reduced.23 Here, Hfq2 truncated to the corresponding length (B. anthracis residue 65) retained hexameric structure on semi-native PAGE [Fig. 6(E), Lane 1 vs. Lane 2; some monomer appears to also be present in Lane 1]. Since His-TEV-Hfq2 is approximately the same size as the potential contaminant of E. coli Hfq (≈11 kDa), the boiled monomer is depicted in comparison to His-TEV-Hfq2 to demonstrate that it is a distinct, quaternary structure-forming product at the expected size.
While Sm2 (an RNA-binding motif common to the Hfq family) is well conserved among B. anthracis Hfqs, the N-terminus and C-terminus of the Hfqs are divergent in sequence (Fig. 1). Therefore, we created two chimeras to examine effects of these differences [Fig. 6(F)]: Hfq 1-2 and Hfq 2-1. His-Hfq 2-1 was monomeric on semi-native PAGE [Fig. 6(D)]. His-Hfq 1-2 purification produced a mixture of bands (potentially contaminants) that did not resolve to monomers upon boiling [Fig. 6(D)]. To enhance production, His-Hfq 1-2 was expressed at 30°C overnight, generating a monomeric product [Fig. 6(G)]. While it is possible that the Hfq chimeras do not fold properly, these results suggest that sequences between Hfq1 residues 17 and 45 are responsible for disrupting hexamer formation. Therefore, we made two additional substitutions in Hfq1 in this region, introducing residues from the Hfq2 sequence: V33R and V37K [Fig. 6(A,B)]. However, the resulting proteins still migrated on semi-native PAGE as monomers [Fig. 6(H)]. Note that for each Hfq1 mutant assessed, protein concentrations were greater than 8 µM; E. coli Hfq is hexameric in solution at only 1 µM.16
Functional properties of B. anthracis Hfqs
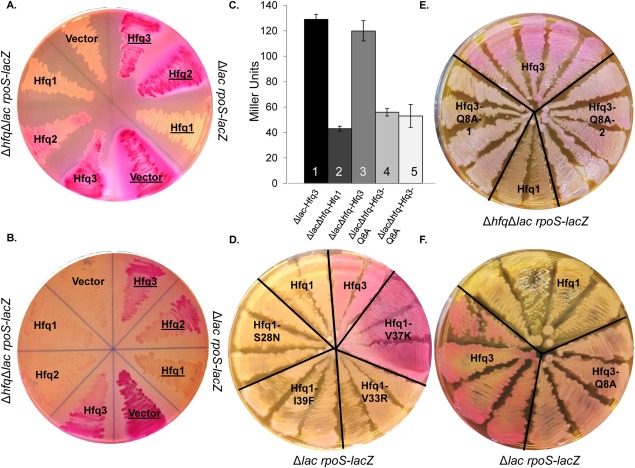
Finally, we sought to identify effects of B. anthracis Hfq sequence and structural differences on in vivo functionality. While a phenotypic role for Hfqs in B. anthracis has not yet been defined, complementation assays in E. coli can be used to explore structure–function relationships. The activity of B. anthracis Hfqs was assessed in E. coli strains carrying rpoS-lacZ translational fusions, where lacZ expression induced by E. coli Hfq activity is manifested as a bright red color on MacConkey agar [Supporting Information Fig. 5(A), #1–6; 130 ± 2 Miller units of β-galactosidase in suspension from cells scraped from plate], as compared with the tan color in the absence of Hfq expression [Supporting Information Fig. 5(A), #7–9; 23 ± 1 Miller units]. When each B. anthracis Hfq was expressed constitutively from the T5-His-Hfq plasmids in an E. coli Δhfq rpoS-lacZ strain [total protein expression depicted in Supporting Information Fig. 5(B)], His-Hfq3 and His-Hfq2 exhibited partial phenotypic complementation of the Δhfq phenotype on the plates, with no complementation by His-Hfq1 [left half of Fig. 7(A), 37°C; and left half of Fig. 7(B), 30°C]. β-galactosidase activities in plate scrapings of His-Hfq2 and His-Hfq3 at 30°C were 35 ± 1 and 53 ± 2 Miller units, respectively (with His-Hfq1 activity at the background level of 20 Miller units). In liquid cultures grown overnight in the presence of IPTG induction of Hfq expression, complementation by His-Hfq3 is also observed in an assay of β-galactosidase activity [Fig. 7(C); Bar #3 vs. Bar #2].
Figure 7.
In vivo functionalities of B. anthracis Hfqs. (A, B) Effect of expression of His-Hfqs in ΔlacΔhfq rpoS-lacZ E. coli (CEV017-CEV019; left side of plate) or Δlac rpoS-lacZ (hfq+) E. coli (CEV020-022; right side of plate, with labels underlined). Plate (A) was incubated overnight at 37°C, and Plate (B) was incubated overnight at 30°C. (C) β-galactosidase assay of SG30013 or YN585 transformed with His-Hfq1, His-Hfq3, or His-Hfq3-Q8A vectors, grown overnight in LB + 100 µg/mL Amp + 100 µg/mL IPTG at 37°C. Bars represent average of three biological replicates ± standard error. (D) Impact of mutants on His-Hfq1 interference (strains CEV023-026). (E) Impact of Q8A mutation on His-Hfq3 complementation (CEV028). #1 and #2 indicate two independently mutagenized Hfq3-Q8A vector stocks for generation of CEV028; CEV017 and CEV019 are provided for comparison. (F) Impact of Q8A mutation on His-Hfq3 in the hfq+ background (CEV027).
In contrast, in the presence of E. coli hfq, expression of His-Hfq1 not only did not complement, but also had a disruptive effect on E. coli hfq function [right half of Fig. 7(A), 37°C; right half of Fig. 7(B), 30°C]. This effect was retained when the His-Hfq1 had either a Q8A mutation, which has been demonstrated to disrupt E. coli Hfq hexamer formation and RNA binding16,24 [Supporting Information Fig. 5(C)], or any of the S28N, I39F, or V33R mutations [Fig. 7(D)]. While disruption of lacZ expression was not observed for Hfq1-V37K, denaturing gels indicated a low level of Hfq1 expression in this strain [Supporting Information Fig. 5(D)]. The inhibitory effect of Hfq1 may be strengthened by the high level of soluble protein expression in T5-His-Hfq1 strains, as demonstrated in Figure 3.
Finally, we observed two behaviors of a His-Hfq3-Q8A mutation that suggest that functional Hfq3 hexamers are forming in vivo. First, the Q8A mutation reduced the level of complementation by His-Hfq3 on MacConkey agar [Fig. 7(E); protein expression levels of wild-type and Q8A mutant were comparable, Supporting Information Fig. 5(E)]; the same effect was observed by β-galactosidase assay in liquid culture [Fig. 7(c), Bars #4,5 vs. Bar #3]. Second, the Q8A mutation caused observable disruption of lacZ expression by Hfq3 in hfq + E. coli [Fig. 7(F); again, expressed protein levels were similar between wild-type Hfq3 and Q8A mutant, Supporting Information Fig. 5(E)]. The latter result reinforces the hypothesis that the in vivo presence of Hfq monomers exacerbates the dominant negative, disrupting effect.
Discussion
The set of three distinct Hfq proteins in the pathogen B. anthracis provides a unique opportunity to explore structure–function relationships of the Hfq protein family. Here, we provide a detailed description of purification and characterization of B. anthracis Hfq1, 2, and 3. While His-Hfq2 assembled into higher-order structure characteristic of other Hfqs in the family, surprisingly, Hfq1 behaved as a monomer. The interpretation of higher-order structure is complicated by the formation of much larger aggregates. If Hfq1 hexamers can form, as considered during the interpretation of DLS, they would be much less stable than Hfq2 hexamers. Hfq from the Gram-positive relative Clostridium difficile was recently demonstrated to form somewhat unstable hexamers,25 and Hfq from the Gram-negative Borrelia burgdorferi is not a stable hexamer in solution,26 but both are able to complement E. coli Hfq function.
The results of our mutagenesis and chimera experiments provide clues as to the structural determinants of hexamer stability. As observed in E. coli, we confirm that truncation of Hfq2 to the end of the β5 strand still permits hexamer formation; therefore, it is other features (besides only tail length)—presumably within the central stretch of 28 amino acids of Hfq1—that prevent stable interactions between monomers. Differences in charge patterns [Fig. 2(B-K)] could also play a role. Functionally, Hfq1 could potentially have a hexamer-disrupting (or other anti-Hfq2) function in B. anthracis. We note that it appears as if E. coli Hfq associates with B. anthracis Hfq1 during purification [Fig. 6(H)]. When mixed as a molar excess of His-Hfq1 and incubated at 25°C or 37°C, purified His-Hfq1 does not disrupt purified Hfq2 hexamers, neither in the presence of 0 or 2M urea nor in the presence of up to 4% SDS (data not shown). However, transient interactions could take place inside the cell, possibly aided by endogenous chaperones, leading to a disruptive phenotype.
In the case of B. anthracis Hfq3, based on its successful complementation of one phenotype of E. coli—which is disrupted by the absence of a functional Q8 residue, known to be involved in hexamer formation and RNA binding—we hypothesize that Hfq3 folds into functional hexamers in vivo, perhaps involving the assistance of chaperones. Indeed, the formation of inclusion bodies and varying results in refolding experiments are consistent with challenges in folding this protein.
While Hfqs of some Gram-positive species, including Listeria monocytogenes and C. difficile, can partially complement Gram-negative Hfq functions,25,27 B. subtilis and S. aureus Hfqs are unable to do so28,29 (we recognize, however, the caveat that these complementation experiments in different species are not performed in parallel). Sequence determinants of differences in function between Hfqs, including between Hfq1 and Hfq3, remain unclear, and simple alignment of Hfq3 and C. difficile Hfq does not identify obvious reasons for their shared functionality in E. coli [Supporting Information Fig. 5(F)]. Therefore, our results set up the potential for the use of B. anthracis Hfqs as a system for screening of mutations to identify residues important in hexamer formation and/or in vivo functionalities. Future work will also expand beyond E. coli phenotypes to examine functional roles for these proteins in B. anthracis, including the potential for disruptive cross-talk and/or heterohexamer formation. Preliminary experiments (with E. coli cell lysates or purified proteins) did not provide evidence of heterohexamer formation between Hfq1 and Hfq2 or between Hfq2 and Hfq3 [Supporting Information Fig. (6)], but further investigation is needed to examine behavior under physiological conditions.
While this article was being finalized, Panda et al.30 reported findings consistent with our observations about differences in hexamer formation between B. anthracis Hfqs. It should be noted that we numbered Hfq1 and 2 according to the designation of Hfq1 in UniProt, while Panda et al. numbered the proteins in the opposite fashion. In addition to the detailed structural findings presented in this article, our work relates quaternary structure to amino acid sequence, as well as relating Hfq structure to in vivo functionality.
Materials and Methods
Sequences and strains
Plasmids, strains, protein sequences and primers are described in Supporting Information Tables I–IV.
RT-PCR
Bacillus anthracis Ames 35 was cultured in LB and lysed with glass beads. RNA was extracted with the Qiagen RNeasy Kit (Valencia, CA), including on-column DNase treatment. RNA was converted to cDNA, and Hfq1, 2, or 3-specific PCR products were generated using the SuperScript III One-Step RT-PCR kit (Life Technologies; Grand Island, NY) and the primers in Supporting Information Table IV. Products were visualized on a 2% agarose gel and absence of product in no-RT PCR reactions was confirmed. No PCR product was observed from Δhfq1hfq2hfq3 B. anthracis (Supporting Information Table II).
Expression and purification of Hfqs
Hfq expression was driven from T5 or T7 promoters using plasmids as described in Supporting Information Table I, which were transformed into either E. coli M15 (+ plasmid pREP4) or E. coli BL21(λDE3) (or its Δhfq derivative) as described in Supporting Information Table II. M15 strains were cultured in LB + kanamycin (30 µg/mL) + ampicillin (100 µg/mL), and BL21(λDE3) strains were cultured in LB + ampicillin (100 µg/mL). Protein expression was induced overnight at 37°C or 30°C by addition of 1 mM IPTG at OD600 0.8–1.0. Point mutations were created with the Q5 site-directed mutagenesis kit (New England Biolabs; Ipswich, MA). His-tagged Hfq1 and Hfq2 were purified from the soluble fraction of cell lysates by nickel affinity as described in Supporting Information.
For semi-native SDS-PAGE, proteins were mixed with 2× SDS loading dye [Life Technologies; final (SDS) = 4%], separated on precast 16% Tricine gels with 1× Tricine SDS running buffer [Life Technologies; (SDS) = 0.1%], and stained with Coomassie Brilliant Blue. For denaturing SDS-PAGE, samples were boiled for greater than 15 min, or as indicated. For gels completed on the 4%–20% Tris–Glycine system (Life Technologies), the final [SDS] was 1% in loading buffer and 0.1% in running buffer.
Cell-free protein synthesis
Untagged Hfq3 was produced by cell-free synthesis using the PURExpress Protein Synthesis kit (New England Biolabs) as directed.31 DNA template was prepared by PCR and purified with the DNA Clean and Concentrator Kit (Zymo Research; Irvine, CA), and proteins were synthesized for 4 h at 37°C.
Refolding of inclusion bodies
His-Hfq3 was isolated from inclusion bodies and refolded on-column by denaturant dilution. Briefly, expression was driven from E. coli CEV003 overnight at 37°C as described above. After cell lysis with lysozyme and sonication [Lysis buffer: 100 mM Tris–HCl, pH 8.0, 5 mM EDTA, complete protease inhibitor (Roche; Indianapolis, IN)], the pellet was recovered by centrifugation. The pellet was washed (100 mM Tris–HCl, pH 8.0, 5 mM EDTA, 2M urea, 2% Triton X-100) and resuspended in Extraction Buffer [50 mM Tris–HCl, 8M guanidine HCl (GdnHCl), pH adjusted to 8.0], with solubilization for 30 min at room temperature. Insoluble material was removed by centrifugation and supernatant was applied to a nickel column, followed by stepwise reduction of [GdnHCl] into buffer containing 250 mM NaCl (pH 8.0) over ≈30 min, and elution in the 250 mM NaCl buffer (no GdnHCl) + 400 mM imidazole.
Biophysical characterization of proteins
Analytical ultracentrifugation (AUC)
Sedimentation velocity experiments were carried out at 50,000 rpm and 20°C on a Beckman Coulter ProteomeLab XLI following standard protocols.32 Detailed information about data collection and analysis is provided in Supporting Information.
Dynamic light scattering (DLS)
A Zetasizer Nano S from Malvern Instruments was used for estimation of particle radii of His-Hfq1 and His-TEV-Hfq2 as directly eluted from nickel columns, with 2 min pre-equilibration at 25°C and use of the viscosity of water for calculations (0.887 cP).
Bioinformatic and structural analysis
Phylogenetic trees
Hfq protein sequences from Gram-positive and Gram-negative bacteria (as depicted), obtained from GenBank, were utilized to create unrooted phylogenetic trees by Clustal W2 phylogeny [neighbor joining and gap exclusion; Fig. 2(A)] or PhyML 3.0 (maximum likelihood; this alternative tree presented in Supporting Information Fig. 2).33
Protein structural prediction
MODELLER package 9.10 was used to build 3D models of Hfq1 and Hfq3 based on X-ray structures of S. aureus (PDB: 1KQ2 and PDB: 1KQ1, respectively),11 and the E. coli structure (PDB: 1HK9)10 was used to model Hfq2. We used Chimera's default Coulombic Surface Coloring thresholds of ±10 kcal/mol to display surface electrostatic potential on the hexameric models. Additional modeling information is provided in Supporting Information.
In vivo complementation
Two ΔlacZ strains of E. coli, each carrying an rpoS-lacZ fusion, were utilized: one hfq+, and one Δhfq (SG30013 and YN585, Supporting Information Table II). The pQE80-His-Hfq series of plasmids were used for Hfq expression. Strains were streaked to MacConkey (+100 μg/mL ampicillin) plates (BD Difco; Franklin Lakes, NJ) and incubated for ≈16 h at 30°C or 37°C to observe color. We observed that the intensity of color development varied between experiments with different incubation times. To observe Hfq protein expression levels in lysates, cells scraped from plates were lysed in 8M urea before separation on 16% Tricine. β-galactosidase assays were performed essentially as described,34 with samples taken from cultures grown overnight in LB + 100 μg/mL ampicillin + 100 μg/mL IPTG, at 37°C; or, when indicated, cells from plate scrapings resuspended in 1× PBS.
Supporting Information
Additional methods and figures (Supporting Information Figs. 1–6), as cited in the text of Results and Materials and Methods, are provided in the Supporting Information section. Detailed information about plasmids (Supporting Information Table I), strains (Supporting Information Table II), protein sequences (Supporting Information Table III), and primers used in the study (Supporting Information Table IV), as well as a table of biochemical properties of the primary Hfq constructs used in this article (Supporting Information Table V), are provided in this section.
Acknowledgments
The authors thank Rasem Fattah for his assistance with the mass spectrometry data collection.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
References
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, Dupuy B, Martin-Verstraete I, Soutourina O. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J Bacteriol. 2014;196:3234–3248. doi: 10.1128/JB.01923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- McKenzie A, Pomerantsev AP, Sastalla I, Martens C, Ricklefs SM, Virtaneva K, Ansick S, Porcella SF, Leppla SH. Transcriptome analysis identifies Bacillus anthracis genes that respond to CO2 through an AtxA-dependent mechanism. BMC Genom. 2014;15:229. doi: 10.1186/1471-2164-15-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CG, Sousa SA, Grilo AM, Feliciano JR, Leitão JH. The second RNA chaperone, Hfq2, is also required for survival under stress and full virulence of Burkholderia cenocepacia J2315. J Bacteriol. 2011;193:1515–1526. doi: 10.1128/JB.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Panja S, Woodson SA. Hexamer to monomer equilibrium of E. coli Hfq in solution and its impact on RNA annealing. J Mol Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- Eisenberg H. Protein and nucleic acid hydration and cosolvent interactions: establishment of reliable baseline values at high cosolvent concentrations. Biophys Chem. 1994;53:57–68. doi: 10.1016/0301-4622(94)00076-x. [DOI] [PubMed] [Google Scholar]
- Ebel C, Eisenberg H, Ghirlando R. Probing protein-sugar interactions. Biophys J. 2000;78:385–393. doi: 10.1016/S0006-3495(00)76601-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Beich-Frandsen M, Konarev PV, Shang W, Vecerek B, Kontaxis G, Hämmerle H, Peterlik H, Svergun DI, Bläsi U, Djinović-Carugo K. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 2012;40:8072–8084. doi: 10.1093/nar/gks510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, Nikulin A. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:760–764. doi: 10.1107/S1744309110017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murina VN, Melnik BS, Filimonov VV, Uhlein M, Weiss MS, Müller U, Nikulin AD. Effect of conserved intersubunit amino acid substitutions on Hfq protein structure and stability. Biochemistry (Mosc) 2014;79:469–477. doi: 10.1134/S0006297914050113. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Napetschnig J, Afonyushkin T, Ecker K, Vecerek B, Moll I, Kaberdin VR, Bläsi U. Functional effects of variants of the RNA chaperone Hfq. Biochem Biophys Res Commun. 2004;323:1017–1023. doi: 10.1016/j.bbrc.2004.08.190. [DOI] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol. 2013;425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet J, Gracia C, Fontaine F, Hajnsdorf E. Clostridium difficile Hfq can replace Escherichia coli Hfq for most of its function. RNA. 2014;20:1567–1578. doi: 10.1261/rna.043372.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Abel CA, Feig AL, Samuels DS. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2010;78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–916. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Bouloc P, Yang Q, Bossi L, Figueroa-Bossi N. Lack of interchangeability of Hfq-like proteins. Biochimie. 2012;94:1554–1559. doi: 10.1016/j.biochi.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Panda G, Tanwer P, Ansari S, Khare D, Bhatnagar R. Regulation and RNA binding properties of Hfq-like RNA chaperones in Bacillus anthracis. Biochim Biophys Acta. 2015;1850(9):1661–1668. doi: 10.1016/j.bbagen.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. PURE technology. Methods Mol Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci. 2013 doi: 10.1002/0471140864.ps2012s71. Chapter 20:Unit 20.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information