Abstract
Background
Genetic and environmental factors both play important roles in the occurrence and progression of diabetic retinopathy (DR). IL-10 592 gene polymorphism is associated with diabetes pathogenesis. This study analyzed the relationship between IL-10 gene promoter-592 loci polymorphism (SNP) in a diabetic model rats with DR.
Material/Methods
Streptozotocin (STZ) was injected through the tail vein to establish a diabetic rat model. The rats were randomly divided into 2 groups for 3 months’ feeding, including 100 rats in the diabetes-positive control group and 100 rats only injected with citric acid buffer as the blank control group. Fundus fluorescein angiography (FFA) was used to observe retinal vascular changes. Polymerase chain reaction-restriction fragment polymorphisms assay (PCR-RFLP) was used to detect IL-10 gene promoter-592 loci polymorphism in DNA samples. Enzyme-linked immunosorbent assay (ELISA) was performed to test serum IL-10 concentration.
Results
Serum IL-10 level in DR rats was 33.18±5.0 pg/mL and in the control rats it was 53.33±4.16 pg/mL in (P<0.01). Diabetes susceptibility with IL-10-592 genotype frequency and gene frequency analysis showed that IL-10-592 genotype frequency and allele frequency were significantly different in the DR group compared with the control group (P<0.01).
Conclusions
IL-10 592 polymorphism was associated with DR susceptibility, suggesting that the gene polymorphism might be a risk factor for DR.
MeSH Keywords: Diabetes Insipidus, Nephrogenic; Diabetic Retinopathy; Rats, Inbred BB
Background
Type 2 diabetes mellitus (T2DM) is an endocrine disorder characterized as high blood glucose. In recent years, T2DM incidence has been rising. Diabetes mellitus patients account for more than 90% of DR cases in the developed countries such as the USA, its incidence increased yearly because of the continuous improvement of living standards and economics in China, with a serious threat to life and health [1,2]. Diabetic retinopathy (DR) is a serious complications of diabetes. A crucial characteristic of DR is microangiopathy [3]. At present, DR is the leading cause of blindness worldwide [3]. Numerous cytokines, such as VEGF, TNF-α, oxidative stress genes, and endothelial nitric oxide synthase, had been found to be correlated with T2DM.
Although the pathogenesis of DR is still not fully elucidated, numerous studies have shown that genetic and environmental factors play considerable roles in the process of DR occurrence and progression [4,5]. T2DM is often accompanied by a variety of change in cytokines, and multiple cytokine-mediated inflammatory responses are often associated with its occurrence [6–9].
A recent study found that interleukin 10 (IL-10) expression in serum is closely related to T2DM incidence [10]. In recent years, the role of IL-10 in T2DM pathogenesis had been preliminarily defined [11,12]. It was found that people and experimental animals with T2DM have inadequate IL-10 secretion and that IL-10-592 gene polymorphism is closely related to T2DM onset [13].
However, whether the polymorphism is associated with DR has not been determined. To provide a basis for DR diagnosis, treatment, and prognosis, this study aimed to investigate the relationship of IL-10 level in serum and IL-10-592 gene polymorphism with DR,
Material and Methods
Animals
We purchased 200 SD rats weighing 230–270 g from Zhengzhou University laboratory animal center. This study was approved by Zhengzhou University animal ethics committee (Register number: 20130618) The rats were maintained at 20–26°C and relative humidity 40–60%, and fed alternately day and night, with alternating light and dark cycles, with natural and mechanical ventilation.
Reagents
Malondialdehyde (MDA, batch number 20131221) and streptozotocin (STZ) were purchased from Sigma (batch number S8260). Superoxide dismutase (SOD, batch number 20131220), and glutathione kit (GSH, batch number 20131118) were bought from Nanjing Jiancheng Bioengineering Institute (China).
Modeling and grouping
We randomly divided 200 rats into 2 groups after 3 days of feeding, with 100 rats in the control group, and 100 rats in the DR group who were fed a high-fat diet (1% cholesterol, 10% lard, 30% egg, 59% sucrose, 2 mL/rat) and normal breeding. In addition, rats in the DR group received 45 mg/kg STZ via tail vein injection, while the control group received an equal amount of citric acid buffer. Fundus examination was performed every week and the samples were collected after 3 weeks.
Biochemical indicators detection
Related biochemical indicators were detected for the model. The eyes of 3 rats in each group were collected and ground. After centrifugation, the supernatant was collected and SOD, MDA, and GSH levels were detected by the kits.
Retina digestion stretched preparation
The eyes were sampled from 4 rats in each group and fixed in 10% formaldehyde solution. The retina was isolated after 48 h and flushed in water for 24 h. After digestion in 3% trypsin (Serva 1:250) solution at 37°C for 1–3 h, the tissue was moved into distilled water. The transparent retinal vascular net was exposed after removing the internal limiting membrane and residual retinal nerve, then was placed onto a slide and stained by PAS after natural drying.
Blood sample collection
We collected 2 ml of tail venous blood into 2 tubes − 1 with EDTA for DNA extraction and restriction fragment length polymorphism (PCR-RFLP) analysis, and 1 without EDTA for IL-10 detection. The serum was isolated after centrifugation and stored in a refrigerator. Sample DNA was extracted from the white blood cells after removing the red blood cells. DNA samples were diluted to 100 ng/μL for future testing.
IL-10 ELISA detection
ELISA kit (Senxiong Biotech, Shanghai, China) was applied to detect IL-10 expression changes according to the manual. Major steps included: put 20 μl diluted standard product into the corresponding reaction holes to prepare the standard curve; add 20 μl of samples into each hole; after washing the plate 5 times, add 100 μl enzyme reagent; wash the plate 5 times again after incubation at 37°C for 30 min; insert 100 μl of color agent into each hole and incubate the plate at 37°C for 15 min. The reaction was terminated after adding 50 μl of termination liquid. The plate was measured at 450 nm wavelength to get the absorbance value (OD value). The sample concentration was calculated according to the OD value and standard curve.
PCR-RFLP
PCR-RFLP was used to analyze IL-10 gene promoter region 592 loci polymorphism. Oligio 6.0 software was used to design specific primers for IL-10-592 loci amplification based on rat IL-10 DNA sequence reported in GenBank: forward, 5′-GGTGAG AAC TAC GTG ACTCGC-3′; reverse, 5′-CCTTGA TCC CAG TGC CGTAG-3′ (synthesized by Beijing Biomed Technology Development Co., LTD). PCR reaction was performed in 25 μL system, including 100 mmol/L Tris-HCl (pH 8.3, 25°), 500 mmol/L KCl, 25 mmol/L MgCl2, 215 mol/L dNTP, 10 mol/L primer, 100 ng template DNA, and 1 u TaqDNA polymerase. The cycling conditions consisted of an initial single cycle of 5 min at 94°C, followed by 30 cycles of 60 s at 94°C, 45 s at 57°C, and 60 s at 72°C, followed by 5 min at 72°C for extension. PCR product was detected by 1.5% agarose gel electrophoresis and analyzed in JelDoc type 2001 gel electrophoresis imaging system (Bio-Rad).
PCR product restriction enzyme analysis
Restriction enzyme RsaI was used to digest single-nucleotide polymorphism (SNP) loci. Enzyme digestion system, including 10 uL PCR products, 7.0 uL ddH2O, 2.0 uL 10×RsaN buffer, 0.7 uL RsaNse, and 0.2 uL BSA, was incubated at 37° for 12 h. The products were detected by 2.0% agarose gel electrophoresis and analyzed in the JelDoc type 2001 gel electrophoresis imaging system (Bio-Rad).
Statistical analysis
All statistical analyses were performed using SPSS13.1 software (Chicago, IL). Differences between multiple groups were analyzed using the t-test or chi-square test. P<0.05 was considered as a significant difference.
Results
Biochemical indicator detection and retinal morphological observation
SOD activity, MDA, and GSH contents were significantly different in the test group compared with the control group. SOD activity and GSH content were clearly decreased, while MDA increased significantly (P>0.05). In addition, the normal rats exhibited complete retinal vascular net with uniform thickness of retinal capillary diameter and straight direction, but diabetic rats had different degrees of retinal vascular morphology changes with irregular direction, disordered arrangement of capillary net, twisted capillaries, uneven thickness of capillary tube cavities, or section enlargement. Furthermore, nucleus pyknosis, cell number reduction, and endothelial cell hyperplasia were significantly different from controls (Table 1, Figure 1).
Table 1.
SOD, GSH, MDA level comparison.
| Group | SODU (U/mg) | GSH (mg/g) | MDA (mg/g) |
|---|---|---|---|
| Control | 0.65±0.25 | 0.50±0.10 | 0.75±0.20 |
| Model | 0.45±0.10* | 0.32±0.10** | 0.98±0.25** |
P<0. 05;
P<0. 01, compared with control.
Figure 1.
Retinal vascular digestion morphology observation (×400). (A) Control; (B) Model.
Serum IL-10 comparison
Serum IL-10 level in DR rats was 33.18±5.0 pg/mL, while it was 53.33±4.16 pg/mL in the controls (P<0.01) (Table 2). There was no significant difference (P>0.05) between groups, suggesting that IL-10 gene frequency consistently conforms to balance law.
Table 2.
Serum IL-10 comparison.
| Group | IL-10 (pg/ml) | U value | P value |
|---|---|---|---|
| Control | 53.33±4.16 | – | – |
| Model | 33.18±5.0 | 7.355 | 0.006 |
IL-10-592 loci PCR analysis
After PCR amplification, electrophoretic analysis showed that IL-10-592 amplification product was 412 bp. The binding clearly could be used for the subsequent enzyme reaction.
IL-10-592 digestion
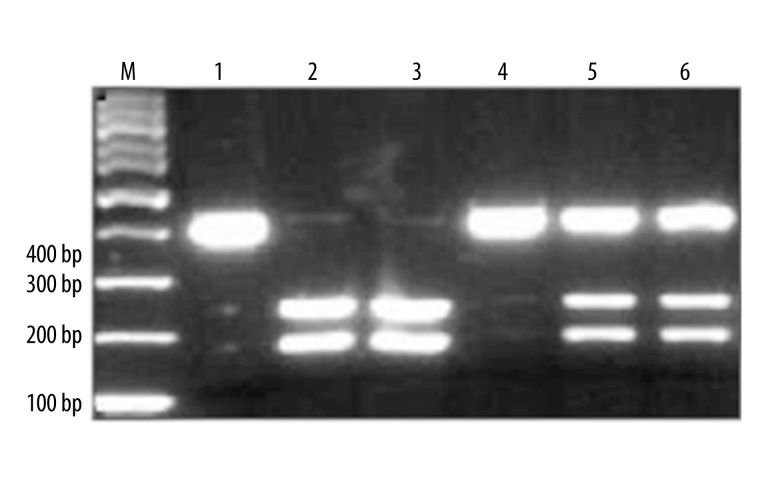
After digested by RsaI, electrophoresis analysis showed that the product could be digested into 2 pieces at 236 bp and 176 bp, while the controls showed no enzyme fragments. IL-10-592 had CyA mutation, leading to fragment enzyme loci. One 412 bp binding could be observed when the CyA mutation genotype was wild-type C/C; 236 bp and 176 bp binding could be found when the genotype was homozygous mutation A/A; 3 bindings at 412, 236, and 176 bp appeared when the genotype was heterozygous mutation A/C (Figure 2).
Figure 2.
PCR-RFLP results of IL-10-592 polymorphism. 1, 4, CC genotype; 2, 3, AA genotype; 5, 6, AC genotype.
IL-10-592 genotype and allele frequency comparison
IL-10-592 genotype frequency and allele frequency were significantly different in the DR group compared with controls (P<0.01) (Table 3). No significant difference (P>0.05) existed between groups, indicating that IL-10 gene frequency consistently conforms to balance law.
Table 3.
IL-10-592 genotype and allele frequency comparison (%).
| Group | Cases | Allele frequency | Genotype frequency | |||
|---|---|---|---|---|---|---|
| A | C | AA | AC | CC | ||
| DM | 100 | 16 (16.0) | 55 (55.0) | 29 (29.0) | 43 (43.0) | 57 (56.0) |
| NC | 100 | 9 (9.0) | 50 (50.0) | 41 (41.0) | 34 (34.0) | 66 (66.0) |
| χ2 value | 9.50 | 10.20 | ||||
| P value | 0.002 | 0.005 | ||||
Discussion
Building a DR animal model similar to that in humans is the key to this type of research. This study adopted the animal model according to Smith [14] and tested related markers. Serum IL-10 level was detected and gene polymorphism analysis was performed to confirm its correlation with DR. We found that DR rats had serum IL-10 changes and IL-10-529 gene polymorphism.
Many studies have indicated that T2DM is a chronic, low-grade inflammatory state [6]. Its incidence might be associated with a variety of inflammatory cytokines, which leads to glucose metabolic disorder [15–18]. Among them, studies indicated that diabetic NOD mice presented IL-10 down-regulation, while exogenous IL-10 injection significantly reduced diabetes incidence in NOD mice [19]. Other studies revealed that relatively low serum IL-10 level is often accompanied by high glycosylated hemoglobin (HbA1c) and high blood glucose [20,21]. In this study, we found that serum IL-10 in the DR group was significantly lower than that of normal controls, which is consistent with previous results. This illustrates that low IL-10 can lead to immune dysfunction and eventually trigger DR; such a disorder may cause self-immune tolerance that cannot be effectively maintained. As a result, low serum IL-10 expression in the DR group suggests that IL-10 may reduce diabetes incidence in DR rats, and it may improve DR prognosis.
The transcription starting site of IL-10 is located at the promoter region, which is about 1.3 KB upstream of IL-10. It has a number of positive and negative regulation sequences of promoter. The positive regulatory sequence was between −1100 and −900, while the negative regulatory sequence was between −800 and −300. Three kinds of genotypes were found in the promoter region: −1082 (G/A), −819 (C/T), and −592 (C/A) [22,23]. Studies have found that the IL-10 gene promoter −592 loci polymorphism was associated with diabetes [24–28]. Moreover, it was also indicated that IL-10 gene polymorphism influenced diabetic nephropathy pathogenesis and prevention [29]. Similarly, this study also found that IL-10 592 genotype frequency was significant higher in DR rats, suggesting that IL-10 gene polymorphism may be a risk factor for DR susceptibility and that it is closely related to DR prognosis.
Conclusions
The present study found that IL-10 was associated with DR occurrence. Larger-scale studies including different races are needed to confirm whether it is a susceptible factor. In addition, this study did not thoroughly uncover the related mechanism between IL-10 gene polymorphisms and DR. Because the occurrence of diabetes is often associated with a variety of different genes and genetic factors (e.g., racial heredity and environment), we need studies with larger sample sizes, involving patients of different races and different geographical location to better understand the relationship between polymorphism analysis and disease susceptibility.
Footnotes
Source of support: This work was supported by the Foundation for Health Science and Technology Innovative Talents of Henan province, China (4107)
References
- 1.Selph S, Dana T, Blazina I, et al. Screening for type 2 diabetes mellitus: a systematic review for the u.s. Preventive services task force. Ann Intern Med. 2015;162:765–76. doi: 10.7326/M14-2221. [DOI] [PubMed] [Google Scholar]
- 2.Liebl A, Khunti K, Orozco-Beltran D, Yale JF. Health economic evaluation of type 2 diabetes mellitus: a clinical practice focused review. Clin Med Insights Endocrinol Diabetes. 2015;8:13–19. doi: 10.4137/CMED.S20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JJ, Wendel LJ, Birkholz ES, et al. The metabolic syndrome and severity of diabetic retinopathy. Clin Ophthalmol. 2015;9:757–64. doi: 10.2147/OPTH.S80355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovic D. Candidate genes for proliferative diabetic retinopathy. Biomed Res Int. 2013;2013:540416. doi: 10.1155/2013/540416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheu WH, Kuo JZ, Lee IT, et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013;22:3165–73. doi: 10.1093/hmg/ddt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hameed I, Masoodi SR, Mir SA, et al. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J Diabetes. 2015;6:598–612. doi: 10.4239/wjd.v6.i4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cilensek I, Mankoc S, Petrovic MG, Petrovic D. GSTT1 null genotype is a risk factor for diabetic retinopathy in Caucasians with type 2 diabetes, whereas GSTM1 null genotype might confer protection against retinopathy. Dis Markers. 2012;32:93–99. doi: 10.3233/DMA-2011-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustavsson C, Agardh E, Bengtsson B, Agardh CD. TNF-alpha is an independent serum marker for proliferative retinopathy in type 1 diabetic patients. J Diabetes Complications. 2008;22:309–16. doi: 10.1016/j.jdiacomp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Cilensek I, Mankoc S, Globocnik Petrovic M, Petrovic D. The 4a/4a genotype of the VNTR polymorphism for endothelial nitric oxide synthase (eNOS) gene predicts risk for proliferative diabetic retinopathy in Slovenian patients (Caucasians) with type 2 diabetes mellitus. Mol Biol Rep. 2012;39:7061–67. doi: 10.1007/s11033-012-1537-8. [DOI] [PubMed] [Google Scholar]
- 10.Insulin in type 2 diabetes: a useful alternative despite limited assessment based on surrogate endpoints. Prescrire Int. 2005;14:187–93. [PubMed] [Google Scholar]
- 11.Brauner H, Luthje P, Grunler J, et al. Markers of innate immune activity in patients with type 1 and type 2 diabetes mellitus and the effect of the anti-oxidant coenzyme Q10 on inflammatory activity. Clin Exp Immunol. 2014;177:478–82. doi: 10.1111/cei.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorand B, Kolb H, Baumert J, et al. Elevated levels of interleukin-18 predict the development of type 2 diabetes: results from the MONICA/KORA Augsburg Study, 1984–2002. Diabetes. 2005;54:2932–38. doi: 10.2337/diabetes.54.10.2932. [DOI] [PubMed] [Google Scholar]
- 13.Acharya AB, Thakur S, Muddapur MV. Effect of scaling and root planing on serum interleukin-10 levels and glycemic control in chronic periodontitis and type 2 diabetes mellitus. J Indian Soc Periodontol. 2015;19:188–93. doi: 10.4103/0972-124X.148644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DK, Korbutt GS, Suarez-Pinzon WL, et al. Interleukin-4 or interleukin-10 expressed from adenovirus-transduced syngeneic islet grafts fails to prevent beta cell destruction in diabetic NOD mice. Transplantation. 1997;64:1040–49. doi: 10.1097/00007890-199710150-00017. [DOI] [PubMed] [Google Scholar]
- 15.Shao L, Zhang P, Zhang Y, Ma A. Inflammatory unbalance of TLR3 and TLR4 in PCI patients with or without type 2 diabetes mellitus. Immunol Lett. 2014;161:81–88. doi: 10.1016/j.imlet.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Cheung KK, Luk AO, So WY, et al. Testosterone level in men with type 2 diabetes mellitus and related metabolic effects: A review of current evidence. J Diabetes Investig. 2015;6:112–23. doi: 10.1111/jdi.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barzilay JI, Abraham L, Heckbert SR, et al. The relation of markers of inflammation to the development of glucose disorders in the elderly: the Cardiovascular Health Study. Diabetes. 2001;50:2384–89. doi: 10.2337/diabetes.50.10.2384. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Chow IT, Sosinowski T, et al. Autoreactive T cells specific for insulin B: 11–23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci USA. 2014;111:14840–45. doi: 10.1073/pnas.1416864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derosa G, Cicero AF, D’Angelo A, et al. Effects of 1 year of treatment with pioglitazone or rosiglitazone added to glimepiride on lipoprotein (a) and homocysteine concentrations in patients with type 2 diabetes mellitus and metabolic syndrome: a multicenter, randomized, double-blind, controlled clinical trial. Clin Ther. 2006;28:679–88. doi: 10.1016/j.clinthera.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.van Exel E, Gussekloo J, de Craen AJ, et al. Leiden 85 Plus Study. Low production capacity of interleukin-10 associates with the metabolic syndrome and type 2 diabetes: the Leiden 85-Plus Study. Diabetes. 2002;51:1088–92. doi: 10.2337/diabetes.51.4.1088. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama M, McDaniel K, Fitzgerald-Miller L, et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc Natl Acad Sci USA. 2015;112:4429–34. doi: 10.1073/pnas.1502967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eskdale J, Kube D, Gallagher G. A second polymorphic dinucleotide repeat in the 5′ flanking region of the human IL10 gene. Immunogenetics. 1996;45:82–83. doi: 10.1007/s002510050174. [DOI] [PubMed] [Google Scholar]
- 23.Eskdale J, Gallagher G. A polymorphic dinucleotide repeat in the human IL-10 promoter. Immunogenetics. 1995;42:444–45. doi: 10.1007/BF00179416. [DOI] [PubMed] [Google Scholar]
- 24.Temple SE, Lim E, Cheong KY, et al. Alleles carried at positions −819 and −592 of the IL10 promoter affect transcription following stimulation of peripheral blood cells with Streptococcus pneumoniae. Immunogenetics. 2003;55:629–32. doi: 10.1007/s00251-003-0621-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhu LJ, Liu ZH, Zeng CH, et al. Association of interleukin-10 gene −592 A/C polymorphism with the clinical and pathological diversity of lupus nephritis. Clin Exp Rheumatol. 2005;23:854–60. [PubMed] [Google Scholar]
- 26.Ezzidi I, Mtiraoui N, Kacem M, et al. Interleukin-10-592C/A, −819C/T and −1082A/G promoter variants affect the susceptibility to nephropathy in Tunisian type 2 diabetes (T2DM) patients. Clin Endocrinol (Oxf) 2009;70:401–7. doi: 10.1111/j.1365-2265.2008.03337.x. [DOI] [PubMed] [Google Scholar]
- 27.Tegoshi H, Hasegawa G, Obayashi H, et al. Polymorphisms of interferon-gamma gene CA-repeat and interleukin-10 promoter region (−592A/C) in Japanese type I diabetes. Hum Immunol. 2002;63:121–28. doi: 10.1016/s0198-8859(01)00363-9. [DOI] [PubMed] [Google Scholar]
- 28.Mohebbatikaljahi H, Menevse S, Yetkin I, Demirci H. Study of interleukin-10 promoter region polymorphisms (−1082A/G, −819T/C and −592A/C) in type 1 diabetes mellitus in Turkish population. J Genet. 2009;88:245–48. doi: 10.1007/s12041-009-0034-x. [DOI] [PubMed] [Google Scholar]
- 29.Mtiraoui N, Ezzidi I, Kacem M, et al. Predictive value of interleukin-10 promoter genotypes and haplotypes in determining the susceptibility to nephropathy in type 2 diabetes patients. Diabetes Metab Res Rev. 2009;25:57–63. doi: 10.1002/dmrr.892. [DOI] [PubMed] [Google Scholar]