Abstract
Since chikungunya virus (CHIKV) was introduced into the Americas in 2013, its geographic distribution has rapidly expanded. Of 119 serum samples collected in 2014 from febrile patients in southern Mexico, 79% were positive for CHIKV or IgM against CHIKV. Sequencing results confirmed CHIKV strains closely related to Caribbean isolates.
Keywords: alphavirus, Aedes, arboviruses, arthralgia, chikungunya virus, disease outbreaks, Mexico, Pan American Health Organization, phylogeny, public health, viruses, vector-borne infections
Chikungunya virus (CHIKV), an arbovirus in genus Alphavirus, family Togaviridae, has undergone a rapid geographic expansion during the past decade (1). CHIKV is the causative agent of chikungunya fever, which may be accompanied by severe, debilitating, and even chronic arthralgia. During urban outbreaks, CHIKV uses the highly susceptible and anthropophilic Aedes aegypti and A. albopictus mosquitoes as vectors, which results in high attack rates (1).
The West African and East/Central/South African (ECSA) CHIKV lineages, the 2 most ancient enzootic lineages, primarily circulate in sub-Saharan Africa. Over the past century, the ECSA lineage has given rise to the Asian lineage, found in urban cycles in India and Southeast Asia, and later to the Indian Ocean lineage, which emerged from Kenya into the Indian Ocean Basin in 2004 (1). In late 2013, an outbreak of chikungunya fever caused by an Asian lineage strain began in the Caribbean island of St. Martin (2,3). During 2014, CHIKV spread throughout the Caribbean and into Latin America, causing epidemics in South and Central America, while also causing sporadic autochthonous cases in North America (1). The importation from Angola and local circulation of an ECSA strain was also recently reported in Brazil (4), which now has 2 CHIKV lineages circulating. The total number of suspected cases in the Americas now exceeds 1.6 million and is steadily rising (3,5).
In October 2014, physicians in Chiapas State, Mexico, noticed large numbers of patients reporting febrile illness accompanied by rash and an unusual arthralgia, and chikungunya fever was suspected. Here we report a chikungunya fever outbreak in southern Mexico, involving CHIKV of Asian lineage as the etiologic agent.
The Study
To ascertain the etiologic agent causing an outbreak of febrile illness with symptoms similar to chikungunya fever, we selected 3 sites in Chiapas State, Mexico, for sampling: Tapachula, La Libertad, and Ciudad Hidalgo (Figure 1, panel A). After patients’ informed consent was obtained, blood samples were collected from persons whose condition met the following case definition for possible chikungunya fever: acute onset of fever >38.5°C, accompanied by severe arthralgia not explained by other medical conditions (6). Samples from Tapachula were collected from patients who sought treatment at the Centro Regional de Investigacion en Salud Publica, whereas in La Libertad and Ciudad Hidalgo, researchers surveyed houses to identify potential case-patients. In total, 119 blood samples were collected, and serum was isolated by centrifugation. Six samples were stored in MagnaPure LC buffer (Roche, Nutley, NJ, USA), which inactivates virus particles but preserves the genomic RNA.
Figure 1.
Map of Mexico showing the 3 sites where serum samples were obtained to test for chikungunya virus in Chiapas, Mexico, 2014: Tapachula, La Libertad, and Ciudad Hidalgo.
Viral RNA was extracted from serum samples using the ZR-96 Viral RNA Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. One-step quantitative reverse transcription PCR (qRT-PCR) (7) was performed by using the TaqMan RNA-to-Ct 1-step Kit (Applied Biosystems, San Francisco, CA, USA). Standard plaque assays were performed for the samples positive by qRT-PCR with Vero cells. Anti-CHIKV IgM-capture enzyme-linked immunosorbent assays (ELISAs) (8) were performed by using a chimeric Eilat-CHIKV (9) that contained the nonstructural proteins from Eilat virus and structural proteins from CHIKV, resulting in a structure indistinguishable from that of CHIKV. Plaque-reduction neutralization tests were used to confirm ELISA results. A sample was considered to be CHIKV negative if the sample was not positive by qRT-PCR or IgM ELISA.
Viral RNA from 5 serum samples was sent for Illumina deep sequencing and assembled by using the virus-specific HIVE-Hexagon algorithm (10) and the NGen module in Lasergene Suite version 10 (Bioinformatics Pioneer DNAStar, Inc., Madison, WI, USA ). Single nucleotide polymorphisms were analyzed by using the sequencing profiling tool in the HIVE suite of programs (10). Sequences were aligned in SeaView (11) by using translated proteins for the open reading frames and using nucleotides for the untranslated genome regions, and all gaps were removed. Bayesian phylogenetic inference was performed by using the general time reversible plus invariant sites plus gamma distribution 4 model in MrBayes (12) with 1.5 million iterations to reach congruence. Partial genome sequencing of the E2 and E1 glycoproteins was performed by using traditional Sanger methods on PCR amplicons on an additional 8 samples.
Over 100 serum samples were obtained from persons seeking treatment for chikungunya fever–like illness during October–December 2014 in 3 locations in Chiapas, Mexico (Tapachula, La Libertad, Ciudad Hidalgo) (Figure 1). These samples were analyzed by qRT-PCR and IgM-capture ELISA to detect viremia and IgM, respectively. No overlap occurred between the samples that were positive for CHIKV by qRT-PCR or those positive by IgM, demonstrating the importance of the humoral response to viral clearance. With few exceptions, viremia was detectable up to 3 days after fever onset (Figure 2, panel A), after which most samples were IgM positive. All age groups were equally likely to be infected, as expected during infectious disease outbreaks involving a naive population (13).
Figure 2.
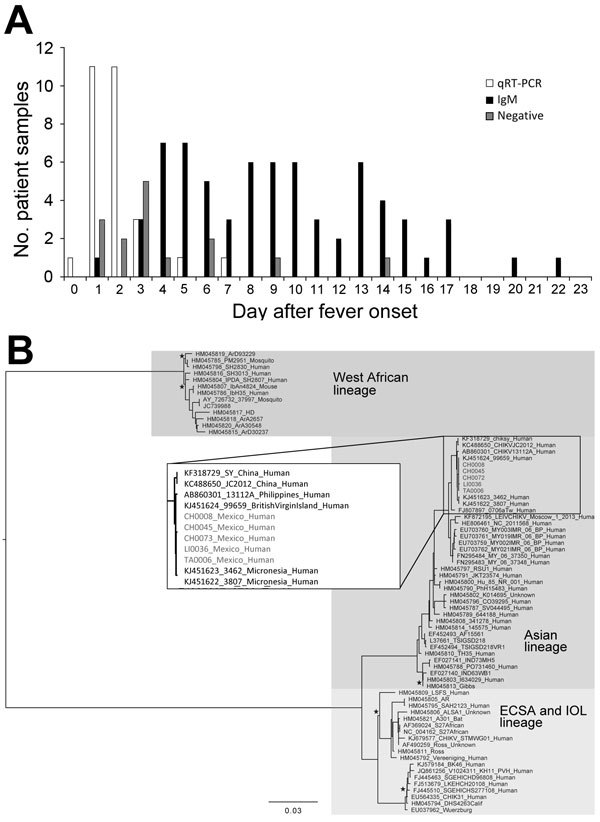
A) Number of serum samples positive for chikungunya virus (CHIKV) by reverse transcription quantitative PCR (qRT-PCR), for CHIKV IgM by ELISA, and negative for CHIKV by both methods, arranged according to day after fever onset. B) Phylogenetic tree generated by Bayesian analysis showing the relationship of the complete genomic sequences of 5 chikungunya virus isolates from Mexico and representative sequences from the GenBank library. All nodes showed posterior probabilities of >0.9, except those indicated with a star. The inset shows the closest relatives of the 5 isolates. ECSA, East/Central/South African, IOL, Indian Ocean lineage. Scale bars indicates nucleotide substitutions per site.
Plaque assays were performed to determine serum virus titers (Table); 3 qRT-PCR samples from Tapachula could not be assayed because of sample limitations. Mean viremia level was similar among the 3 sampled locations and ranged from <2 log10 to 4.2 log10 PFU/mL.
Table. Proportions of CHIKV viremia and IgM in 119 serum samples collected at 3 collection sites in Chiapas, Mexico, October–December 2014*.
| Collection site | No. serum samples | % qRT-PCR positive serum samples (no.) | Mean serum virus titer, log10 PFU/mL (±SD) | % IgM-positive serum samples† (no.) |
|---|---|---|---|---|
| La Libertad | 43 | 20 (9) | 3.26 (0.57) | 51.2 (22) |
| Ciudad Hidalgo | 63 | 23.8 (15) | 3.36 (0.56) | 68.2 (43) |
| Tapachula | 13 | 30.8 (4) | 3.66 | 7.7 (1) |
*CHIKV, chikungunya virus; qRT-PCR, quantitative reverse transcription PCR. †Six patient samples that were negative for CHIKV by qRT-PCR were unable to be used for plaque assays or ELISAs because they were stored in MagnaPure Buffer (Roche, Nutley, NJ, USA).
Five serum samples from diverse locations and collection dates were selected for Illumina sequencing (GenBank accession nos. KT327163–KT327167). The strains circulating in Chiapas were most closely related to Asian lineage strains first detected in the Caribbean (represented by a British Virgin Islands isolate) and now presumed to be circulating in much of Latin America (Figure 2, panel B). Curiously, no novel mutations appear to have been fixed in the year since the British Virgin Islands isolate was collected.
Although the genomic sequences confirmed that the circulating virus in Chiapas belonged to the Asian lineage, which is primarily transmitted by A. aegypti mosquitoes (1), we nevertheless examined the sequences for mutations known to adapt CHIKV for transmission by A. albopictus mosquitoes. Because both mosquito species are found in Chiapas (14), adaptation of the CHIKV strain circulating in Mexico to A. albopictus mosquitoes could place temperate regions of the eastern United States and millions of naive persons at risk for infection. None of the E2 or E1 substitutions previously reported to increase fitness in A. albopictus mosquitoes was observed in 8 additional samples analyzed by Sanger sequencing (GenBank accession nos. KT247378–KT247385) (15). One sample had a nonsynonymous mutation, in comparison to the January 2014 British Virgin Islands isolate (GenBank accession no. KJ451624) that encoded E1-V7M, which had not previously been described.
Conclusions
We found that 79% of febrile illness cases with polyarthralgia in Chiapas State during late 2014 were caused by CHIKV. Our sequencing of CHIKV genomes confirmed spread of an Asian lineage strain from the Caribbean and suggested that although CHIKV has circulated in the Americas since 2013, no adaptive mutations have occurred. However, continued screening for vector-adaptive mutations will be critical, especially now that strains of the ECSA lineage, which gave rise to the Indian Ocean lineage, have been introduced into Brazil (4).
Acknowledgments
We thank Rose Coleman for her assistance with qRT-PCRs and Albert Auguste for critiques of the manuscript.
This research was supported by the Institute for Human Infections and Immunity and the McLaughlin Fellowship Fund at the University of Texas Medical Branch. CHIKV genomes were sequenced through funding provided by the US Department of Homeland Security through contract no. HSHQDC-13-C-B0009 “Capturing Global Biodiversity of Pathogens by Whole Genome Sequencing.”
Biography
Ms. Kautz is a graduate student in the Microbiology and Immunology Program at the University of Texas Medical Branch. Her primary research interests include arbovirus emergence and evolution.
Footnotes
Suggested citation for this article: Kautz TF, Díaz-González EE, Erasmus JH, Malo-García IR, Langsjoen RM, Patterson EI, et al. Chikungunya virus as cause of febrile illness outbreak, Chiapas, Mexico, 2014. Emerg Infect Dis. 2015 Nov [date cited]. http://dx.doi.org/10.3201/eid2111.140546
These first authors contributed equally to this article.
References
- 1.Weaver SC, Forrester NL. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res. 2015;120:32–9 . 10.1016/j.antiviral.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 2.Van Bortel W, Dorleans F, Rosine J, Blateau A, Rousset D, Matheus S, et al. Chikungunya outbreak in the Caribbean region, December 2013 to March 2014, and the significance for Europe. Euro Surveill. 2014;19:••• . [DOI] [PubMed] [Google Scholar]
- 3.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. 10.1016/S0140-6736(14)60185-9 [DOI] [PubMed] [Google Scholar]
- 4.Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. 10.1186/s12916-015-0348-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan American Health Organization. Number of reported cases of Chikungunya fever in the Americas—epidemiological week 31 (August 7, 2015) [cited 2015 Aug 21]. http://www.paho.org/hq/index.php?option=com_docman&task=doc_download&Itemid=270&gid=31087&lang=en
- 6.European Centre for Disease Prevention and Control. Fact sheet for health professionals: chikungunya virus (updated 2015. Feb 27) [cited 2015 Aug 15]. http://ecdc.europa.eu/en/healthtopics/chikungunya_fever/factsheet-health-professionals/Pages/factsheet-for-health-professionals.aspx
- 7.Powers AM, Roehrig JT. Alphaviruses. Methods Mol Biol. 2010;665:17–38. 10.1007/978-1-60761-817-1_2 [DOI] [PubMed] [Google Scholar]
- 8.Beaty BJ, Calisher CC, Shope RE. Arboviruses. In: Schmidt NJ, Emmons RW, editors. Diagnostic procedures for viral rickettsial and chlamydial infections. 6th edition. Washington (DC): American Public Health Association; 1989. p. 797–855. [Google Scholar]
- 9.Nasar F, Gorchakov RV, Tesh RB, Weaver SC. Eilat virus host range restriction is present at multiple levels of the virus life cycle. J Virol. 2015;89:1404–18. 10.1128/JVI.01856-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonyan V, Mazumder R. High-performance integrated virtual environment (HIVE) tools and applications for big data analysis. Genes (Basel). 2014;5:957–81. [DOI] [PMC free article] [PubMed]
- 11.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 12.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 13.Hartfield M, Alizon S. Epidemiological feedbacks affect evolutionary emergence of pathogens. Am Nat. 2014;183:E105–17. 10.1086/674795 [DOI] [PubMed] [Google Scholar]
- 14.Arredondo-Jiménez JI, Valdez-Delgado KM. Aedes aegypti pupal/demographic surveys in southern Mexico: consistency and practicality. Ann Trop Med Parasitol. 2006;100(Suppl 1):S17–32. 10.1179/136485906X105480 [DOI] [PubMed] [Google Scholar]
- 15.Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. [DOI] [PMC free article] [PubMed]


