To the Editor: The epidemiology of cholera, especially in Africa and Asia, has periodically changed in subtle ways (1). The recent cholera epidemic in Haiti, a Caribbean country with no cholera cases in decades, affected >500,000 persons, caused ≈8,000 deaths, and brought this illness to the forefront of Haitian public health concerns (2,3). This life-threatening disease is caused by Vibrio cholerae, a waterborne bacterium with >200 serogroups, 2 of which, O1 and O139, cause epidemic or pandemic cholera. V. cholerae O1 is categorized as classical and El Tor biotypes, which differ biochemically and have different levels of virulence. Classical strains typically cause more severe illness than El Tor strains, which result in mild or moderate and sometimes asymptomatic cases. However, El Tor strains have replaced classical strains as the cause of cholera; the classical biotype is believed to be extinct, and El Tor strains currently prevail. However, the genetic traits specific to classical strains are still present in environmental and clinical V. cholerae isolates. Currently, all clinical strains of V. cholerae in Kolkata produce classical cholera toxin. Such phenotypic and genetic changes in V. cholerae are being monitored worldwide.
Several phenotypic and genetic laboratory tests are used to determine whether isolates are classified as classical or El Tor biotypes. Among phenotypic traits distinguishing the 2 biotypes, sensitivity to polymyxin B (50 U) is considered a reliable indicator and stable phenotype for biotyping. Research has shown that the genome of V. cholerae strains is undergoing cryptic changes that influence the strains’ virulence, rapid transmission, and spread (4). Our previous findings showed El Tor strains with few biotype traits of classical strains (5).
Since the seventh cholera pandemic, which occurred during the 1960s and 1970s and was caused by El Tor strains, the El Tor biotype had been resistant to polymyxin B, a cationic antimicrobial peptide. However, when cholera strains first appeared in patients in Kolkata, India, in June 2012, V. cholerae O1 was found to be sensitive to polymyxin B (6). To determine whether this phenomenon occurred earlier, we tested 255 clinical strains isolated from patients in Kolkata during 2003–2014 and found that, from March 2013, polymyxin B–sensitive El Tor strains had replaced resistant strains (Figure, panel A). The MIC of polymyxin B, determined by Etest (bioMérieux, Marcy l’Etoile, France), confirmed that the El Tor strains were susceptible to this antimicrobial drug (Figure, panel B). In this assay, the El Tor strain (N16961) was highly resistant to polymyxin B (MIC 96 µg/mL), whereas the variant strains in Kolkata showed a drastic reduction in resistance (6,7).
Figure.
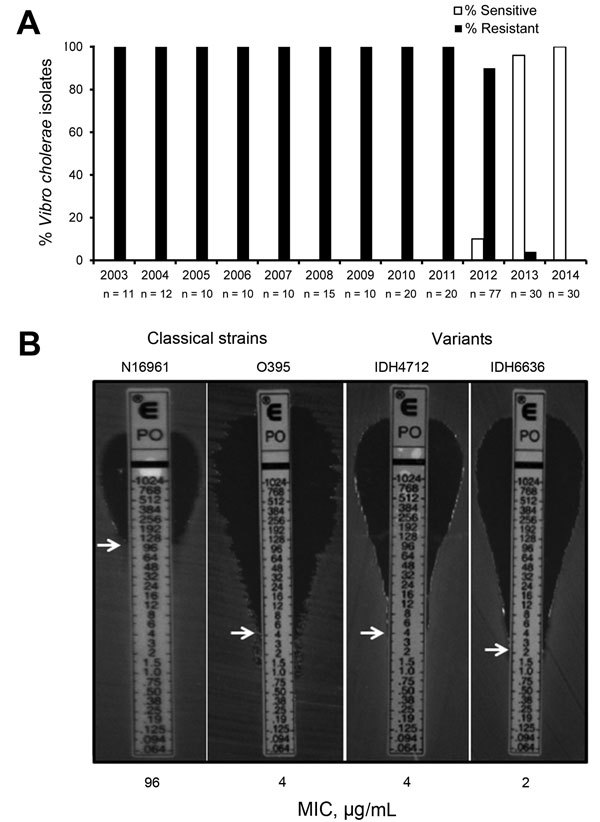
Isolation profile of polymyxin B–sensitive Vibrio cholerae strains in Kolkata, India, 2003–2014. A) Yearly occurrence of polymyxin B sensitivity and resistance in V. cholerae O1 El Tor variant strains isolated from Kolkata patients. During the study period, 255 strains were tested; n values indicate the number of strains tested each year. Polymyxin B–sensitive strains first appeared in Kolkata in June 2012. The first isolate in January 2013 was resistant, but, thereafter, all strains isolated during 2013–2014 were sensitive to polymyxin B, a biotyping marker for classical strains. B) MIC of polymyxin B in El Tor variant strains (classical and El Tor). MICs are indicated by white arrows. Polymyxin B sensitivity, a characteristic of classical strains, was displayed by El Tor variant strains. Data represent 3 biologic repetitions.
To confirm additional changes in biotype attributes in the variant Kolkata isolates during 2003–2014, we used the Voges-Proskauer test to determine production of acetylmethyl carbinol and found that the tested strains produced acetoin and were positive for chicken erythrocytes agglutination. The rtsC gene encoding the activator protein, which is absent from classical biotype strains but present in El Tor strains, was found in all the tested strains of the El Tor biotype. Biotype-specific CTX prophage repressor rstR was amplified with the El Tor–specific primers, indicating presence of El Tor rstR. The tcpA gene has distinct alleles specific to classical and El Tor biotypes of O1. Our study showed that all strains yielded amplicons with the El Tor-tcpA–specific primers but not with the classical-tcpA–specific primers. However, these strains had a single-base substitution at the 266-nt position of tcpA, also present in variant strains from Haiti. Furthermore, Vibrio seventh pandemic (VSP) gene clusters VSP I and VSP II are unique to El Tor strains of the seventh pandemic. We found presence of VSP I and II encoding genes in all our tested strains, indicating that the strains are El Tor, but with specific classical traits.
We also checked the strains’ sensitivity to many antimicrobial drugs: tetracycline, trimethoprim/sulfamethoxazole, streptomycin, erythromycin, gentamicin, ciprofloxacin, and azithromycin, and all strains were sensitive to all drugs except trimethoprim/sulfamethoxazole and streptomycin. All strains isolated during 2013–2014 were fully resistant to trimethoprim/sulfamethoxazole and streptomycin, but 55% of strains isolated before 2012 were sensitive to these drugs.
Genes encoding lipid IVA acyltrasferase (msbB), biofilm formation, antimicrobial peptide resistance (carR), and 3 aminoacyl lipid modification (almEFG) have been shown to contribute to polymyxin resistance in V. cholerae (6–8). Analysis of these genes from the newly emerged polymyxin-B–sensitive strains may provide additional useful information. We found that these strains contained Haitian variant ctxB (ctxB7) similar to the classical cholera toxin. Our earlier studies identified many new attributes of Haitian V. cholerae variant strains in Kolkata since 2003 (9,10).
We report the emergence of El Tor strains producing classical cholera toxin. These strains have lost an El Tor biotype marker and acquired a vital classical biotype characteristic, a change that has probably altered the regulatory mechanisms of lipid A modification machinery in V. cholerae (6–8). This change is a major event in the history of cholera after 1961, when El Tor strains first appeared. The recent changes in V. cholerae O1 strains should be carefully monitored to determine their clinical and epidemiologic implications.
Acknowledgments
This study was supported in part by the Indian Council of Medical Research, Government of India; the Japan Initiative for Global Research Network on Infectious Diseases of the Japan Agency for Medical Research and Development; and the National Institute of Infectious Diseases, Japan.
P.S. received a Junior Research Fellowship from the Council of Scientific and Industrial Research, India, during the study.
Footnotes
Suggested citation for this article: Samanta P, Ghosh P, Chowdhury G, Ramamurthy T, Mukhopadhyay AK. Sensitivity to polymyxin B in El Tor Vibrio cholerae O1 strain, Kolkata, India [letter]. Emerg Infect Dis. 2015 Nov [date cited]. http://dx.doi.org/10.3201/eid2111.150762
References
- 1.Safa A, Nair GB, Kong RYC. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. 10.1016/j.tim.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 2.Piarroux R, Barrais R, Faucher B, Haus R, Piarroux M, Gaudart J, et al. Understanding the cholera epidemic, Haiti. Emerg Infect Dis. 2011;17:1161–8. 10.3201/eid1707.110059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, et al. Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–9. 10.3201/eid1711.110805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robins WP, Mekalanos JJ. Genomic science in understanding cholera outbreaks and evolution of Vibrio cholerae as a human pathogen. Curr Top Microbiol Immunol. 2014;379:211–29. 10.1007/82_2014_366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychoudhuri A, Patra T, Ghosh K, Ramamurthy T, Nandy RK, Takeda Y, et al. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis. 2009;15:131–2. 10.3201/eid1501.080543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, Trent MS. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio. 2014;5:e02283-–14.. 10.1128/mBio.02283-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilecen K, Fong JC, Cheng A, Jones CJ, Zamorano-Sánchez D, Yildiz FH. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun. 2015;83:1199–209. 10.1128/IAI.02700-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson JS, Yoo HJ, Hakansson K, DiRita VJ. Polymyxin B resistance in El Tor Vibrio cholerae requires lipid acylation catalyzed by MsbB. J Bacteriol. 2010;192:2044–52. 10.1128/JB.00023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh P, Naha A, Basak S, Ghosh S, Ramamurthy T, Koley H, et al. Haitian variant tcpA in Vibrio cholerae O1 El Tor strains in Kolkata, India. J Clin Microbiol. 2014;52:1020–1. 10.1128/JCM.03042-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS ONE. 2014;9:e112973. 10.1371/journal.pone.0112973 [DOI] [PMC free article] [PubMed] [Google Scholar]


