This study examined the appropriateness of common culture systems and their ability to reveal unusual (noncanonical) signal transduction pathways that actually operate in vivo. Novel pathways and their importance in various plastic cell types, culminating in their emerging roles in reprogramming mechanisms, and a number of reprogramming paradigms are also discussed. A new molecular perspective in the field of reprogramming with broad implications in basic, biotechnological, and translational research is introduced.
Keywords: Cellular reprogramming, Cellular transdifferentiation, Induced pluripotent stem cells, Signal transduction, STAT3 transcription factor, Hes3 protein, Pancreatic islets
Abstract
Interest is great in the new molecular concepts that explain, at the level of signal transduction, the process of reprogramming. Usually, transcription factors with developmental importance are used, but these approaches give limited information on the signaling networks involved, which could reveal new therapeutic opportunities. Recent findings involving reprogramming by genetic means and soluble factors with well-studied downstream signaling mechanisms, including signal transducer and activator of transcription 3 (STAT3) and hairy and enhancer of split 3 (Hes3), shed new light into the molecular mechanisms that might be involved. We examine the appropriateness of common culture systems and their ability to reveal unusual (noncanonical) signal transduction pathways that actually operate in vivo. We then discuss such novel pathways and their importance in various plastic cell types, culminating in their emerging roles in reprogramming mechanisms. We also discuss a number of reprogramming paradigms (mouse induced pluripotent stem cells, direct conversion to neural stem cells, and in vivo conversion of acinar cells to β-like cells). Specifically for acinar-to-β-cell reprogramming paradigms, we discuss the common view of the underlying mechanism (involving the Janus kinase-STAT pathway that leads to STAT3-tyrosine phosphorylation) and present alternative interpretations that implicate STAT3-serine phosphorylation alone or serine and tyrosine phosphorylation occurring in sequential order. The implications for drug design and therapy are important given that different phosphorylation sites on STAT3 intercept different signaling pathways. We introduce a new molecular perspective in the field of reprogramming with broad implications in basic, biotechnological, and translational research.
Significance
Reprogramming is a powerful approach to change cell identity, with implications in both basic and applied biology. Most efforts involve the forced expression of key transcription factors, but recently, success has been reported with manipulating signal transduction pathways that might intercept them. It is important to start connecting the function of the classic reprogramming genes to signaling pathways that also mediate reprogramming, unifying the sciences of signal transduction, stem cell biology, and epigenetics. Neural stem cell studies have revealed the operation of noncanonical signaling pathways that are now appreciated to also operate during reprogramming, offering new mechanistic explanations.
Introduction
Progress in biomedical science has been hindered by the all too common difficulty in translating in vitro observations to in vivo systems [1]. This problem applies to both transformed and primary cell culture systems, suggesting a difficulty in modeling the in vivo signaling state of a cell inside a culture dish. Traditionally, highly cancerous or genetically altered cells have been used in research, because they are typically easier to grow than primary cells. Serum is often included in the culture medium as a generic growth stimulator, providing a plethora of undefined nutrients and signal transduction pathway modulators. However, the propensity of primary cells to irreversibly differentiate in these conditions has forced experimentalists to modify how these cells are maintained in vitro, removing serum and using specific mitogens, to preserve their self-renewal state [2]. The value of such culture systems is not merely a practical one allowing for their expansion, they also force cells to grow using particular signaling pathways (that promote self-renewal) at the expense of others (that promote differentiation) and can serve as formidable model systems by providing access to these pathways. Early indications revealed that a variety of immature and differentiated plastic cells use common, noncanonical signaling pathways, with implications in regenerative medicine, cancer, diabetes, and reprogramming technologies.
An example of a signaling pathway that can be identified through this rationale is the STAT3-Ser/Hes3 signaling axis. It was originally characterized in neural stem cells (NSCs) and subsequently shown to regulate additional cell types, including prostate tumor-initiating cells, glioblastoma multiforme cancer stem cells (GBM CSCs), adrenomedullary chromaffin progenitors, and mouse insulinoma cell lines (Fig. 1A) [3–7]. In brief, at the center of the pathway is the phosphorylation of STAT3 on serine residue 727 (STAT3-Ser) [8]. This modification is largely redundant for many cell types but is of great importance to the survival of NSCs. STAT3-Ser is a convergence point for several other stimuli, including fibroblast growth factor (FGF), a noncanonical Notch signaling branch, the angiopoietin2/Tie2 system, and insulin. These lead to STAT3-Ser phosphorylation and subsequent transcriptional activation of Hes3, a transcription factor and passive repressor, with roles that are only now starting to be understood [9–11]. The pathway is opposed by Janus kinase (JAK) activity, a key component of the growth machinery of many cell types [12]. In addition to NSCs, pharmacological inhibitors of JAK promote the survival of human pluripotent stem cells and the developmentally equivalent mouse equivalent epiblast stem cell, further highlighting the stark differences in signal transduction preferences between most cell types studied and stem cell populations [13, 14].
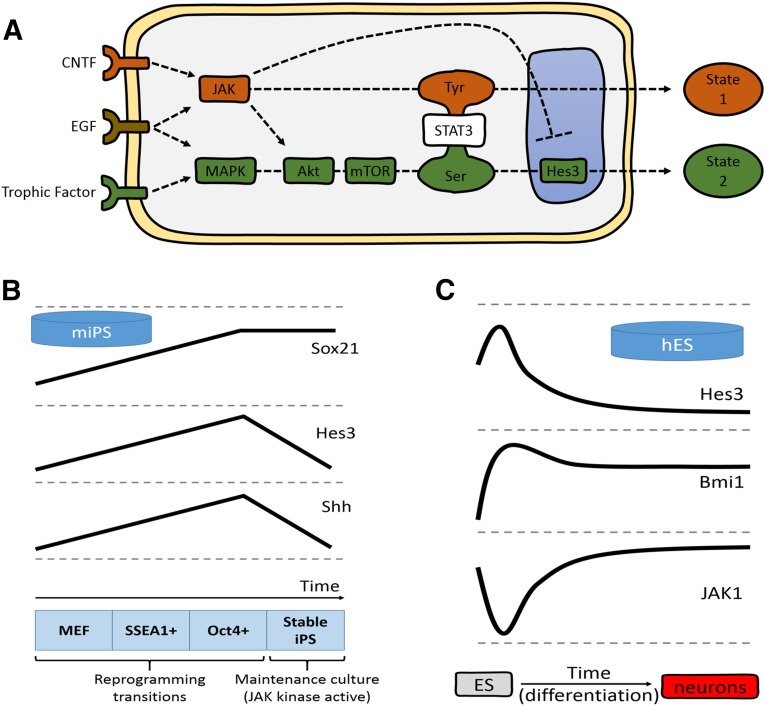
Figure 1.
Noncanonical signaling pathway regulation during reprogramming. (A): Extracellular factors lead to the phosphorylation of STAT3-Tyr via JAK activation or STAT3-Ser via MAPK, Akt, and mTOR activation, and subsequent Hes3 transcription. The two pathways are opposing (e.g., JAK activity in neural stem cells [NSCs] suppresses induction of Hes3). Some cell types (e.g., primary NSCs) are confined to using the STAT3-Ser branch, because the STAT3-Tyr branch leads to their irreversible differentiation. Other cell types (e.g., primary cancer stem cells from glioblastoma multiforme patients and MIN6 cells) grow effectively using either pathway and, through repeated changes in cell culture conditions, can switch their signaling state back and forth. (B): Genes in the STAT3-Ser/Hes3 signaling axis are regulated during mouse fibroblast reprogramming. Sox21, Hes3, and Shh gene expression increases as MEFs transition to SSEA1+ and then to Oct4+ populations during reprogramming to the pluripotent state. Hes3 and Shh are downregulated in resultant stable mouse iPS cells grown in culture conditions that activate JAK (lines not to scale; expression levels at the MEF stage normalized to help visualize patterns and trends). (C): Genes in the STAT3-Ser/Hes3 signaling axis are regulated during neural specification of hES cells. The diagram summarizes the expression patterns of Hes3, Bmi1, and JAK1 over the course of a 77-day protocol to differentiate the human ES cell line WA09 to dorsal telencephalic neuronal fates (lines not to scale; expression levels at day 0 of ES cell stage normalized to help visualize patterns and trends). (B, C): The concepts shown are from gene expression data previously published and reanalyzed for the purposes of the present report [25]. Abbreviations: CNTF, ciliary neurotrophic factor; EGF, epidermal growth factor; hES, human embryonic stem (cell); Hes3, hairy and enhancer of split 3; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; MEF, mouse embryonic fibroblasts; mIPS, mouse induced pluripotent stem (cell); SSEA1, stage-specific embryonic antigen 1; Shh, sonic hedgehog; STAT3, signal transducer and activator of transcription 3.
The therapeutic potential of manipulating the components of this pathway has been demonstrated in a series of studies showing powerful protective effects in the brain when pharmacological activators are introduced into the brain in various models of neurodegenerative disease [3, 8, 15–17]. STAT3-Ser phosphorylation mediates carcinogenesis in xenotransplantation models of prostate cancer [6], suggesting that specific inhibitors directed against this site could be useful in treating certain cancer types, in particular those harboring CSC populations. Angiopoietin 2 is a powerful activator of the pathway, pointing toward new functions of angiogenic factors in tissue homeostasis, neurodegenerative disease, and cancer [17, 18]. The value of this pathway in drug discovery is also highlighted by the finding that the efficiency of a γ-secretase inhibitor as an anti-breast cancer drug was predicted by the levels of Hes3 in vivo [19]. That study, in particular has shown the disconnect between in vitro and in vivo effects, validating the idea that great care and thought is needed to appropriately model the signaling state of a cell in vitro. Overall, the STAT3-Ser/Hes3 signaling axis is operational in cell types from several different tissues and might mediate important functions in the context of a wide range of diseases.
The relevance of this pathway to harnessing stem cell technologies for therapeutic benefit is also exhibited by the findings that its activation results in much improved yields and electrophysiological properties of neurons generated from NSCs derived from induced pluripotent stem (iPS) cell sources [20]. More recently, Hes3 was implicated in the direct conversion of adult non-neural cells to the NSC state through a reprogramming method, along with other genes [21]. The STAT3 phosphorylation state is also critical to consider, because the differences between STAT3 tyrosine 705 (STAT3-Tyr) and STAT3-Ser phosphorylation requirements are clearly exhibited in embryonic stem (ES) and NSC systems. Mouse ES cells use STAT3-Tyr; thus, they are cultured in the presence of leukemia inhibitory factor (LIF). Human ES cells, however, do not rely on STAT3-Tyr to remain undifferentiated and are thus cultured in the presence of basic FGF (bFGF). In NSCs, STAT3-Tyr leads to gliogenic differentiation, but STAT3-Ser promotes survival.
In the present report, we discuss the aspects of signaling pathways that involve and/or intercept STAT3 that have been commonly assumed to be mediated by STAT3-Tyr but in reality might also be mediated by STAT3-Ser. We begin by the re-evaluating classic reprogramming paradigms (e.g., mouse embryonic fibroblasts [MEFs] to iPS). We also reanalyze the gene expression data in well-established paradigms of the differentiation of human embryonic stem cells to neurons, further supporting the operation of these pathways in cell conversion decisions. We extend this discussion by examining the conversion of acinar cells to insulin-producing cells, an exciting example of the transdifferentiation that can be induced both in vitro and in vivo. Revisiting the original studies, we reinterpret some of these assumptions and provide alternative interpretations, showing that STAT3-Ser could also be an important mediator in this process. Given the very strong impetus toward understanding the molecular mechanisms driving reprogramming and differentiation, state-of-the-art techniques that are already being applied in experimental therapies, it is essential to explore these new mechanisms of action. We do not aim to give an extensive account of the reprogramming field, which has been expertly provided elsewhere [22].
Is the STAT3-Ser/Hes3 Signaling Axis a Yet Undetected Mediator of Reprogramming?
To date, a common theme with this pathway is a role in the maintenance of the primitive state of cells capable of undergoing massive epigenetic decisions. This prompted us to ponder whether aspects of the pathway should be included in the thought process behind reprogramming. This is a very opportune time to do so, because, in addition to the many studies showing reprogramming using the classic developmental transcription factors, studies demonstrating reprogramming with soluble factors that activate distinct signaling pathways have also been reported. These pathways intercept the STAT3-Ser/Hes3 signaling axis, consistent with its involvement. Furthermore, its involvement appears restricted to particular stages of a cell’s development, with NSCs offering a great example. Pathways that regulate their self-renewal, stimulated by bFGF, and those that regulate differentiation (ciliary neurotrophic factor [CNTF]-driven astrocyte differentiation), in fact, oppose the activity of one another. This is reminiscent of what happens in the case of mouse ES cells, in which LIF-induced STAT3-Tyr activity (a stimulus necessary to maintain pluripotency in these cells) blocks FGF-induced extracellular signal-regulated kinase (Erk) activity (a differentiation signal) [23].
At the transcriptional level, recent studies have defined intermediate cell populations during mouse iPS cell generation and identified an early c-Myc/KLF4 wave, followed by a second Oct4/Sox2/KLF4 transcriptional wave required for reprogramming [24]. In addition, we have shown that the SoxB transcription factor family member SOX21 is induced by SOX2 during reprogramming and that SOX21 is required for iPS cell generation [25]. Consistent with our findings, interrogation of the data sets from Polo et al. revealed that Sox21 expression is increased as cells transition toward the iPS cell state. Examination of the genes in the STAT3-Ser/Hes3 signaling axis shows that Hes3 is at low levels in both parental MEFs and stable iPSCs but increases during initial reprogramming, peaking in early Oct4-positive cells (Fig. 1B). Hes3 abruptly decreases as the pluripotent state is locked in; interestingly, the mitogen/morphogen sonic hedgehog (Shh) shares this same expression profile. These data suggest that Hes3 is involved in the critical transition to the stable iPS state during reprogramming. The downregulation of Hes3 in the mouse pluripotent state is consistent with the dependence of mouse ES cells on the canonical JAK/STAT pathway, in contrast to other stem cell populations.
Analysis of RNAseq data sets from human ES cells [26] shows that they express Hes3 in the self-renewing state (as seen with NSCs) and display a marked increase in Hes3 expression during the first steps toward neurectodermal specification (Fig. 1C). The polycomb protein Bmi1 [27–29], an important regulator of stem cell self-renewal, is also increased at this same time point and JAK1 expression is repressed. As these cells further commit and differentiate toward neural fates, Hes3 is rapidly downregulated. In cultures of fetal NSCs, Hes3 overexpression induces production of Shh, a known mitogen for NSCs [8]. Shh is also a positive modulator of BmI1, which leads to the transdifferentiation of mouse fibroblasts to NSC-like cells [30]. Taken together, these data suggest a possible Hes3-Bmi1-Shh axis that could also be involved in the reprogramming of cells to the NSC state.
Epigenetic reprogramming by genetic means has generated new sources for cell replacement strategies [22, 31, 32]. Reprogramming has also been achieved through the use of soluble factors. For example, intraperitoneal administration of a combination of epidermal growth factor (EGF) and CNTF converts acinar cells to β-like cells in vivo with consequences for regulating blood glucose in rodent models of diabetes [33]. These factors were chosen, in part, because of their broad range of functions in cell proliferation and gene regulation and because they have been implicated in various paradigms of regeneration and cell type conversion. This is of particular interest, because it provides a “handle” that is upstream of well-studied signaling pathways, providing both a molecular rationale and additional opportunities for manipulation within these signaling pathways. In a recent study, this logic led to the implementation of genetic overexpression of constitutively activated mitogen-activated protein kinase (MAPK) (caMAPK; specifically, p42/44 also known as ERK1/2) and constitutively activated STAT3 (caSTAT3) in human acinar cells in vitro, which also resulted in the reprogramming of these cells to β-like cells [34].
The simple interpretation of these data is that caMAPK is a surrogate for EGF and that caSTAT3 is a surrogate for CNTF (Fig. 2A). This might well be correct; however, a series of observations suggest the involvement of additional, noncanonical signaling pathways in this reprogramming process and open the possibility that the STAT3-Ser/Hes3 signaling axis might be involved. First, it is important to clarify that caSTAT3 models some, but not all, functions of tyrosine-phosphorylated STAT3. One of these functions is the dimerization of STAT3, which, in caSTAT3 is induced without the need for tyrosine phosphorylation. To achieve this, cysteine residues were engineered on the STAT3 monomer rendering it capable of dimerization through disulfide bonds without the need for tyrosine phosphorylation [35]. caSTAT3 can bind to DNA and activate the transcription of certain genes. However, when stimuli that lead to phosphorylation of STAT3 on tyrosine are used, such as interleukin-6 or v-src, the transcriptional activity of caSTAT3 is greatly enhanced, demonstrating the forced dimerization alone is insufficient in modeling the entire range of functions allocated to tyrosine-phosphorylated STAT3 [35, 36]. Likewise, caSTAT3, in which the tyrosine 705 residue has been mutated to phenylalanine (and cannot, therefore, be phosphorylated), fails to transactivate STAT3 target genes [37]. However, caSTAT3 was found to be serine phosphorylated, suggesting that it might be able to efficiently model the functions of this modification [35]. Therefore, the biological output after transduction with caSTAT3 cannot clearly allocate function to the phosphorylation event of one particular residue.
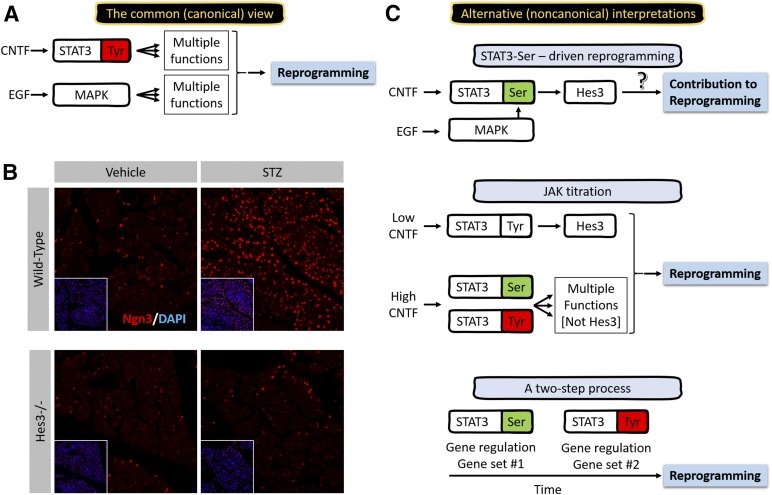
Figure 2.
Possible noncanonical signaling pathway involvement in reprogramming through modulators of STAT3 and MAPK. (A): Common view for the mechanism of reprogramming in acinar-to-β-cell reprogramming downstream of CNTF and EGF. STAT3-Tyr and activated MAPK induce vast transcriptional changes leading to fate specification changes. (B): Hes3 as a regulator of Ngn3 in the context of endocrine pancreas regeneration. A lack of Ngn3 expression induction in Hes3-null (Hes3−/−) mice 5 months after a low-dose streptozotocin regimen (5 consecutive daily injections at 50 mg/kg in phosphate-buffered saline [PBS]; vehicle controls received only PBS). (C): STAT3-Ser as a putative mediator of reprogramming of acinar-to-β cell conversion. Three possible alternative interpretations for the mechanism of action of CNTF- and EGF-induced reprogramming that involve the STAT3-Ser/Hes3 signaling axis. (Both CNTF and EGF lead to the phosphorylation of STAT3-Ser and STAT3-Tyr; the diagrams highlight the particular phosphorylation event that might be driving a given function. It is not meant to suggest that only one residue is phosphorylated. Also, a predominant function of STAT3-Tyr phosphorylation is the dimerization of STAT3. For this reason, and for simplicity, the diagrams depict STAT3-Tyr phosphorylation to also represent STAT3 dimerization). (B): Image width: 534 μm. Abbreviations: CNTF, ciliary neurotrophic factor; DAPI, 4′,6-diamidino-2-phenylindole; EGF, epidermal growth factor; Hes3, hairy and enhancer of split 3; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; STAT3, signal transducer and activator of transcription 3; STZ, streptozotocin.
A clue to the possible involvement of the STAT3-Ser/Hes3 signaling axis in reprogramming comes from observations of the induction of Ngn3 in pancreatic cells. These findings imply the onset of de- and transdifferentiation events, for example, in the conversion of acinar to β cells [38, 39]. Specifically, transduction with caMAPK and caSTAT3 induced Ngn3 only in acinar cells and not in β cells, although both cell types express Ngn3 [34]. In accordance with the involvement of the components of the STAT3-Ser/Hes3 signaling axis in Ngn3 induction in these different cell populations, Hes3-null mice fail to induce Ngn3 in the regenerating pancreas after streptozotocin (STZ) damage. Wild-type mice exhibit Ngn3 expression in both pancreatic islet β cells and acinar cells. At 5 months after STZ-induced damage, Ngn3 expression in acinar cells is strongly upregulated, with no detectable changes in β cells (Fig. 2B). Like wild-type mice, nondamaged Hes3-null mice also exhibit Ngn3 expression in both β and acinar cells. However, after STZ damage, the upregulation of Ngn3 is greatly attenuated. These results suggest the involvement of Hes3 in the induction of Ngn3 in acinar cells and therefore support the hypothesis that caMAPK and caSTAT3 operate through the STAT3-Ser/Hes3 signaling axis in this context.
Another clue is that the order of viral transduction matters. For reprogramming to work efficiently, caMAPK must be transduced before caSTAT3. Transduction of both constructs simultaneously resulted in lower efficiency. Transduction of caSTAT3 before caMAPK resulted in even lower efficiency. Each gene alone was insufficient for reprogramming. We present three alternative interpretations of these findings (Fig. 2C).
Alternative Interpretation 1: STAT3-Ser-Driven Reprogramming
It is possible that, in part, the effects of this reprogramming method involve STAT3-Ser phosphorylation. This would explain why the order of caMAPK first and caSTAT3 second is so important in the efficiency of reprogramming. caMAPK leads to powerful STAT3-Ser phosphorylation via several pathways [40]. Therefore, an initial overexpression of caMAPK would ensure that a subsequent overexpression of caSTAT3 would immediately result in high amounts of STAT3 that would be dimerized and serine phosphorylated. In contrast, the reverse order would result in high amounts of STAT3 that would be dimerized, with possibly submaximal serine phosphorylation, leading to potentially different gene regulation. It is also possible that caMAPK might contribute to reprogramming by elevating serine phosphorylation of endogenous STAT3. This could be tested with experiments in which serine-phosphomimetic constructs of STAT3 that cannot be tyrosine phosphorylated can be assessed for their potential to contribute to reprogramming.
Alternative Interpretation 2: JAK Titration
Although JAK activity is predominantly seen as a means of elevating STAT3-Tyr phosphorylation, JAK also leads to STAT3-Ser phosphorylation [40]. The distinction between these two phosphorylation events can be regulated by the levels of cytokine activity that lead to JAK activation [8]. For example, low levels of CNTF in NSC cultures result in the elevation of STAT3-Ser phosphorylation with no indication of an effect on STAT3-Tyr phosphorylation. Higher CNTF concentrations lead to abrupt increases in STAT3-Tyr phosphorylation [5]. Therefore, CNTF levels can be used either to promote STAT3-Ser phosphorylation in the absence of detectable STAT3-Tyr phosphorylation (and, therefore, promote NSC self-renewal and increase cell numbers) or to promote both serine and tyrosine phosphorylation (and, therefore, inhibit self-renewal and induce differentiation). In another example, the use of a JAK inhibitor in GBM CSCs can significantly increase the ratio of STAT3-Ser to STAT3-Tyr phosphorylation, and as a consequence, the expression of Hes3. This leads to changes in various properties of these cells, including the ability to achieve higher terminal cell densities in vitro [5]. In this scenario, specific levels of JAK activity—which can be regulated using a JAK inhibitor [34, 41]—might result in the stimulation of STAT3-Ser, in the absence of STAT3-Tyr, and promote reprogramming. Therefore, although the tyrosine site might be very important in this reprogramming paradigm, it is also possible that the serine site is also an important player in reprogramming.
Alternative Interpretation 3: A Two-Step Process
It is also possible that the serine and tyrosine phosphorylation of STAT3 represent two distinct events within the reprogramming process and that both are necessary for reprogramming. It is conceivable that the serine event must precede the tyrosine event, as suggested by the observations regarding the importance of order for caMAPK and caSTAT3.
STAT3-Ser/Hes3 in Cancer—Clues to a Role in Reprogramming
Key reprogramming factors are oncogenes, highlighting the similarities between developmental programs and transformation. Hes3 might belong to this category. Direct evidence for this comes from work showing that Hes3 RNA interference opposes the growth of putative CSCs from glioblastoma multiforme biopsies [5], and supporting its putative role in breast cancer, where in vivo Hes3 expression levels correlated with the efficacy of a γ-secretase inhibitor [19]. Other key components of the STAT3-Ser/Hes3 signaling axis are also implicated in carcinogenesis. For example, STAT3-Ser phosphorylation in the absence of STAT3-Tyr phosphorylation has also been demonstrated to drive prostate carcinogenesis [6]. Therefore, the STAT3-Ser/Hes3 signaling axis might represent an oncogenic, noncanonical branch of the Notch signaling pathway, providing an explanation of the dual properties of Notch signaling as both an oncogene and a tumor suppressor [42, 43].
In line with a dynamic role for Hes3 in regulating the epigenetic state of a cell, Hes3 expression and subcellular localization are themselves dynamically and, in certain cases, reversibly, regulated. Hes3 expression and subcellular localization can also provide clues to its function and reveal information of the state of a cell. GBM CSCs and a mouse insulinoma cell line (MIN6) can be efficiently cultured under conditions that support nuclear Hes3 expression or prevent it [5, 7]. By changing the culture conditions, the cells can be repeatedly switched from one state to the other. For example, serum-free defined cultured conditions allow the nuclear expression of Hes3. Under these conditions, the cells grow independently of JAK activity. In fact, inclusion of a JAK inhibitor promotes GBM CSC growth. Primary, bona fide NSC cultures also exhibit Hes3 expression, which is lost when the cells are induced to differentiate, similar to GBM CSCs. In contrast, however, primary NSCs do not exhibit reversibility in their differentiation and Hes3 expression states, a property that has been proposed to characterize the CSC population [44]. It could be of use in assays determining the NSC state to include measurements of the irreversibility of the differentiation of these cells.
Conclusion
The STAT3-Ser/Hes3 signaling axis has been implicated in a number of phenomena, including improved culture conditions for plastic cell types, their activation in vivo in the context of neurodegenerative disease, as a target for brain CSCs, and as a mediator of pancreatic β-cell function and survival. More recently, the pathway has also been implicated in aspects of reprogramming to the NSC state. An understanding of this signaling pathway will provide possible molecular explanations for its function in the context of reprogramming. The possibility that Hes3 could prove to be an important player in different aspects of epigenetic reprogramming as manifested in the iPS and CSC fields warrants more investigation.
Acknowledgments
This work was supported in part by the Helmholtz Alliance ICEMED (Imaging and Curing Environmental Metabolic Diseases) through the Initiative and Network Fund of Helmholtz Association Grant 051_40001; Deutsche Forschungsgemeinschaft Grant SFB 655, “Cells Into Tissues,” Project A24; and a grant from the Else Kroener Fresenius Stiftung.
Author Contributions
S.W.P. and A.A.-T.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.G.C., C.C., and J.M.: collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; G.C.: data analysis and interpretation, manuscript writing, final approval of manuscript; S.R.B.: financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; R.D.M.: financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
S.W.P. and A.A.-T. are uncompensated coinventors on patent applications with Technische Universitaet Dresden. J.M. has compensated employment and uncompensated equity interest in DxNow. The other authors indicated no potential conflicts of interest.
References
- 1.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 2.Cattaneo E, McKay R. Proliferation and differentiation of neuronal stem cells regulated by nerve growth factor. Nature. 1990;347:762–765. doi: 10.1038/347762a0. [DOI] [PubMed] [Google Scholar]
- 3.Poser SW, Park DM, Androutsellis-Theotokis A. The STAT3-Ser/Hes3 signaling axis: An emerging regulator of endogenous regeneration and cancer growth. Front Physiol. 2013;4:273. doi: 10.3389/fphys.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masjkur J, Levenfus I, Lange S, et al. A defined, controlled culture system for primary bovine chromaffin progenitors reveals novel biomarkers and modulators. Stem Cells Translational Medicine. 2014;3:801–808. doi: 10.5966/sctm.2013-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park DM, Jung J, Masjkur J, et al. Hes3 regulates cell number in cultures from glioblastoma multiforme with stem cell characteristics. Sci Rep. 2013;3:1095. doi: 10.1038/srep01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin HR, Kim HJ, Kim JY, et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68:7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masjkur J, Arps-Forker C, Poser SW, et al. Hes3 is expressed in the adult pancreatic islet and regulates gene expression, cell growth, and insulin release. J Biol Chem. 2014;289:35503–35516. doi: 10.1074/jbc.M114.590687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Androutsellis-Theotokis A, Leker RR, Soldner F, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 9.Lobe CG. Expression of the helix-loop-helix factor, Hes3, during embryo development suggests a role in early midbrain-hindbrain patterning. Mech Dev. 1997;62:227–237. doi: 10.1016/s0925-4773(97)00665-5. [DOI] [PubMed] [Google Scholar]
- 10.Hirata H, Tomita K, Bessho Y, et al. Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 2001;20:4454–4466. doi: 10.1093/emboj/20.16.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imayoshi I, Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Levy DE, Darnell JE., Jr STATs: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 13.Chenoweth JG, McKay RD, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- 14.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 15.Ohta S, Misawa A, Fukaya R, et al. Macrophage migration inhibitory factor (MIF) promotes cell survival and proliferation of neural stem/progenitor cells. J Cell Sci. 2012;125:3210–3220. doi: 10.1242/jcs.102210. [DOI] [PubMed] [Google Scholar]
- 16.Androutsellis-Theotokis A, Rueger MA, Mkhikian H, et al. Signaling pathways controlling neural stem cells slow progressive brain disease. Cold Spring Harb Symp Quant Biol. 2008;73:403–410. doi: 10.1101/sqb.2008.73.018. [DOI] [PubMed] [Google Scholar]
- 17.Androutsellis-Theotokis A, Rueger MA, Park DM, et al. Targeting neural precursors in the adult brain rescues injured dopamine neurons. Proc Natl Acad Sci USA. 2009;106:13570–13575. doi: 10.1073/pnas.0905125106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Androutsellis-Theotokis A, Rueger MA, Park DM, et al. Angiogenic factors stimulate growth of adult neural stem cells. PLoS One. 2010;5:e9414. doi: 10.1371/journal.pone.0009414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang CC, Pavlicek A, Zhang Q, et al. Biomarker and pharmacologic evaluation of the γ-secretase inhibitor PF-03084014 in breast cancer models. Clin Cancer Res. 2012;18:5008–5019. doi: 10.1158/1078-0432.CCR-12-1379. [DOI] [PubMed] [Google Scholar]
- 20.Salewski RP, Buttigieg J, Mitchell RA, et al. The generation of definitive neural stem cells from PiggyBac transposon-induced pluripotent stem cells can be enhanced by induction of the NOTCH signaling pathway. Stem Cells Dev. 2013;22:383–396. doi: 10.1089/scd.2012.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassady JP, D’Alessio AC, Sarkar S, et al. Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Rep. 2014;3:948–956. doi: 10.1016/j.stemcr.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JW, Hsu YC, Kao CY, et al. Leukemia inhibitory factor-induced Stat3 signaling suppresses fibroblast growth factor 1-induced Erk1/2 activation to inhibit the downstream differentiation in mouse embryonic stem cells. Stem Cells Dev. 2013;22:1190–1197. doi: 10.1089/scd.2012.0229. [DOI] [PubMed] [Google Scholar]
- 24.Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmichev AN, Kim SK, D’Alessio AC, et al. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr Biol. 2012;22:1705–1710. doi: 10.1016/j.cub.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 26.van de Leemput J, Boles NC, Kiehl TR, et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 28.Molofsky AV, He S, Bydon M, et al. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon JH, Heo JS, Kim JS, et al. Reprogramming fibroblasts into induced pluripotent stem cells with Bmi1. Cell Res. 2011;21:1305–1315. doi: 10.1038/cr.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez Alvarado A, Yamanaka S. Rethinking differentiation: Stem cells, regeneration, and plasticity. Cell. 2014;157:110–119. doi: 10.1016/j.cell.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baeyens L, Lemper M, Leuckx G, et al. Transient cytokine treatment induces acinar cell reprogramming and regenerates functional beta cell mass in diabetic mice. Nat Biotechnol. 2014;32:76–83. doi: 10.1038/nbt.2747. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Lemper M, Leuckx G, Heremans Y, et al. Reprogramming of human pancreatic exocrine cells to β-like cells. Cell Death Differ. 2015;22:1117–1130. doi: 10.1038/cdd.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Shaw PE. Elevated activity of STAT3C due to higher DNA binding affinity of phosphotyrosine dimer rather than covalent dimer formation. J Biol Chem. 2006;281:33172–33181. doi: 10.1074/jbc.M606940200. [DOI] [PubMed] [Google Scholar]
- 37.Liddle FJ, Alvarez JV, Poli V, et al. Tyrosine phosphorylation is required for functional activation of disulfide-containing constitutively active STAT mutants. Biochemistry. 2006;45:5599–5605. doi: 10.1021/bi0525674. [DOI] [PubMed] [Google Scholar]
- 38.Pagliuca FW, Melton DA. How to make a functional β-cell. Development. 2013;140:2472–2483. doi: 10.1242/dev.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Brown J, Kanarek A, et al. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 41.Baeyens L, Bonné S, German MS, et al. Ngn3 expression during postnatal in vitro beta cell neogenesis induced by the JAK/STAT pathway. Cell Death Differ. 2006;13:1892–1899. doi: 10.1038/sj.cdd.4401883. [DOI] [PubMed] [Google Scholar]
- 42.Lobry C, Oh P, Mansour MR, et al. Notch signaling: Switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59:168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]