Abstract
Histone deacetylases (HDAC) play a crucial role in transcriptional regulation and are often deregulated in many cancers. However, global HDAC enzymatic activity has never been investigated in Chronic Lymphocytic Leukemia (CLL). We measured HDAC activity in protein extracts from CD19+ B-cells purified from 114 CLL patients with a median follow-up of 91 months (range: 11–376). HDAC activity was equivalent in CLL and normal B-cells but higher in patients who died during the study than in living patients (152.1 vs. 65.04 pmol; P = 0.0060). Furthermore, HDAC activity correlated with treatment-free survival (TFS; P = 0.0156) and overall survival (OS; P < 0.0001): patients with low HDAC activity (n = 75) had a median TFS and OS of 101 and >376 months, respectively, whereas patients with high HDAC activity (n = 39) had a median TFS and OS of 47 and 137 months, respectively. Multivariate analyses indicated that HDAC activity is an independent predictor of OS (hazard ratio = 7.68; P = 0.0017). Finally, HDAC activity increased after B-cell receptor stimulation using IgM, suggesting a role for microenvironment stimuli (n = 10; P = 0.0371). In conclusion, high HDAC activity in CLL B-cells is associated with shorter TFS and OS and is an independent marker of OS, refining the use of other prognostic factors. This work provides a biological base for the use of HDAC inhibitors in CLL treatment.
Keywords: chronic lymphocytic leukemia, enzymatic activity, HDAC, Sirtuin, prognosis
Abbreviations
- HDAC
Histone deacetylase
- CLL
Chronic lymphocytic leukemia
- TFS
Treatment-free survival
- OS
Overall survival
- LDT
Lymphocyte doubling time
- IgHV
Immunoglobulin heavy-chain variable-region
- ZAP70
Zeta-chain-associated protein kinase 70
- LPL
Lipoprotein lipase
- sCD23
Soluble CD23
- B2M
Beta-2-microglobulin
- SIRT
Sirtuin
- HDACi
HDAC inhibitor
- VPA
Valproic acid
- SAHA
Suberoylanilide hydroxamic acid
- MSC
Mesenchymal stromal cell
- BCR
B cell receptor
- NF-κB
Nuclear factor κB
- FCR
Fludarabine-cyclophosphamide-rituximab
- HR
Hazard ratio
Introduction
In western countries, Chronic Lymphocytic Leukemia (CLL) is a highly prevalent hematological malignancy characterized by clinical heterogeneity.1 Certain patients present severe symptoms, need rapid treatment and have shorter survival duration, while others live for several decades without drastic alteration in their lifetime. Clinical and molecular factors, such as Binet stage, lymphocyte doubling time (LDT), mutational status of the immunoglobulin heavy-chain variable-region (IgHV), zeta-chain-associated protein kinase 70 (ZAP70), lipoprotein lipase (LPL), CD38 expression, serum levels of soluble CD23 (sCD23), and β-2-microglobulin (B2M), are currently used to classify patients into different prognostic subgroups.2 Presently, CLL remains incurable, and further research is needed to understand its physiopathology.
In this context, histone deacetylases (HDAC) are potentially interesting targets. These epigenetic regulators can remove acetyl groups from histone tails, leading to chromatin condensation and gene expression regulation.3 A total of 18 HDAC isoenzymes that are divided into 4 classes have been identified. Class I includes HDAC1, 2, 3 and 8; Class II, includes HDAC4, 5, 6, 7, 9, and 10; Class III contains the sirtuins (SIRT1 to 7); and Class IV only comprises HDAC11.4 In a pathological context, the deacetylation process could be associated with carcinogenesis.4,5 Deregulation of HDAC expression has been observed in several cancers, such as breast, lung, ovarian and prostate cancer, and lymphoma.5 HDAC inhibitors (HDACi), such as valproic acid (VPA) and suberoylanilide hydroxamic acid (SAHA), have thus been considered as new and promising therapeutic agents for the treatment of solid cancers and hematological malignancies.6 Indeed, HDACi were able to induce apoptosis of CLL B-cells,7,8 and clinical trials are currently ongoing.
Recently, we showed, for the first time, that HDAC mRNA expression is globally upregulated in CLL B-cells compared to normal B-cells. Moreover, the mRNA levels of several HDAC isoenzymes have an impact on survival. Specific combinations, such as underexpression of HDAC6 and SIRT3, and overexpression of HDAC7 and HDAC10, were strongly linked to poor prognosis for treatment-free survival (TFS). Whereas overexpression of SIRT5 together with underexpression of SIRT6 was strongly linked to overall survival (OS).9
Increasing evidence suggests a role for the microenvironment in CLL pathogenesis. Our group previously demonstrated that bone marrow mesenchymal stromal cells (MSC) protect CLL but not normal B-cells from apoptosis by contact.10 T-cells also interact with leukemic cells through the CD40-CD40 ligand11 and the B-cell receptor (BCR), which has been shown to promote survival and proliferation of B-cells through upregulation of the nuclear transcription factor κB (NF-κB).12 BCR stimulation is also linked to poor prognosis: responders to IgM stimulation are indeed characterized by a shorter TFS.13
To our knowledge, global HDAC enzymatic activity has never been investigated in CLL by studying the deacetylation power itself. Here, we quantified global HDAC enzymatic activity in CD19+ B-cells obtained from a cohort of 114 patients and found that HDAC enzymatic activity correlated with patients’ clinical outcomes. We also highlighted a potential link between the microenvironment and histone deacetylation.
Results
Population characteristics
The median age of the population was 64 years (range: 34-86). The median TFS was 88 months (range: 0.3–244), whereas the median OS was higher than 376 months (range: 0.4–376). The median follow-up was 91 months (range: 11–376). In this population, prognostic factors, such as IgHV mutational status, ZAP70, CD38, Binet stage, sCD23, B2M, LDT, and cytogenetic abnormalities, were significantly correlated with TFS and OS (P < 0.05), as previously described.2 Complete prognostic data were not available for some patients for which insufficient biological material was available. Binet stages, ZAP70, LPL expression, CD38 expression, and clinical data were available for all 114 patients included in this study. Incomplete data were available for IgHV mutational status (91%), LDT (89%), B2M (97%), sCD23 (96%), and cytogenetic abnormalities (80%). All details on the patient cohort can be found in Table S1 and Figure S1. Of the 63 (100%) patients who received treatment, 23 (36.5%) received alkylating agent-based treatment (i.e., chlorambucil alone and mini-CHOP), 14 (22.2%) received fludarabine-based (without rituximab) treatment (i.e., fludarabine alone and fludarabine-cyclophosphamide), 19 (30.2%) received fludarabine-cyclophosphamide-rituximab (FCR), and 7 (11.1%) received other treatments (i.e., lumiliximab, alemtuzumab, lenalidomide, and ibrutinib). However, the impact of these treatments on OS was not statistically significant (Figure S2). After CD19+ sorting, the mean purity of the 114 CLL and 30 control samples, measured by flow cytometry, was 99.49 ± 0.07% (range: 98.00–99.99) and 97.40 ± 0.71%(range: 92.46–99.81), respectively.
HDAC activity in normal and CLL B-cells
We previously showed that HDAC mRNA levels in CLL samples were generally upregulated compared with those of control groups.9 HDAC activity was compared between CLL and normal CD19+ B-cells. Umbilical cord blood was also used as a source of B-cells, which were CD5+ (mean: 88.56% ± 1.5; range: 80.87–97.63), similarly to CLL B-cells (mean: 98% ± 0.2; range: 91–100). While we observed a trend toward higher HDAC RNA levels, global HDAC activity in leukemic B-cells (97.28 ± 26.39 pmol) was not significantly different from that of normal peripheral blood (57.24 ± 16.68 pmol; P = 0.5741) or umbilical cord blood B-cells (58.26 ± 10.96 pmol; P = 0.1238; Fig. 1).
Figure 1.
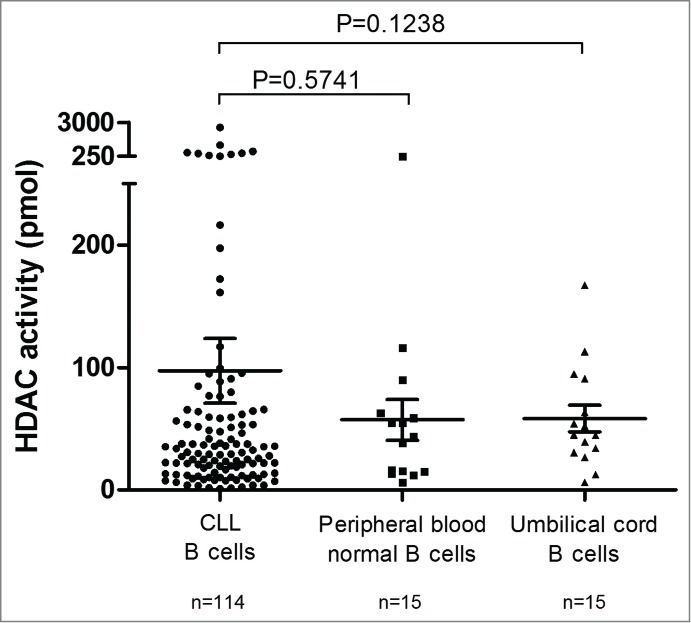
HDAC activity in CLL B-cells, peripheral blood normal B-cells and umbilical cord blood B-cells. Mean HDAC activity for CD19+ purified CLL B-cells (n = 114), CD19+ purified peripheral blood normal B-cells (n = 15) and CD19+ purified umbilical cord blood B-cells (n = 15) is displayed with standard error of the mean (SEM) overlaid with scatter plot. Statistical differences were assessed using the Mann-Whitney non-parametric test.
Correlation of HDAC activity with classical prognostic factors and clinical data
We compared HDAC activity in the different prognosis subgroups classified using the previously mentioned prognostic factors. As shown in Table S2, no difference in HDAC activity was assessed among the subgroups, with the exception of the survival subgroups. Indeed, patients who died during the study showed higher deacetylation activity than living patients (152.1 pmol vs. 65.04 pmol; P = 0.0060; Table S2). The patients were then stratified according to low (n = 75) and high (n = 39) global HDAC activity with a cut-off value set by recursive partitioning, maximizing the concordance with OS. We observed that HDAC activity significantly correlated with TFS (P = 0.0156) and OS (P < 0.0001): patients with low HDAC activity had a median TFS and OS of 101 and >376 months, respectively, whereas patients with high HDAC activity had a median TFS of 47 months and a median OS of 137 months (Fig. 2 A-B). To validate HDAC activity prognostic power, we performed a 2-fold cross-validation study. Statistically significant correlation between HDAC activity and OS was observed in 3 independent random iterations (Figure S3). Additional details concerning cross-validation can be found in the Supplemental Material. In order to confirm that the differences in HDAC activity were linked to histone modifications, we compared histone H4 acetylation levels in patients with low (n = 8) and high (n = 8) HDAC activity using flow cytometry, as described by Ronzoni et al.,14 and confirmed higher levels of histone H4 acetylation in patients with low HDAC activity (P = 0.0019; Figure S4).
Figure 2.
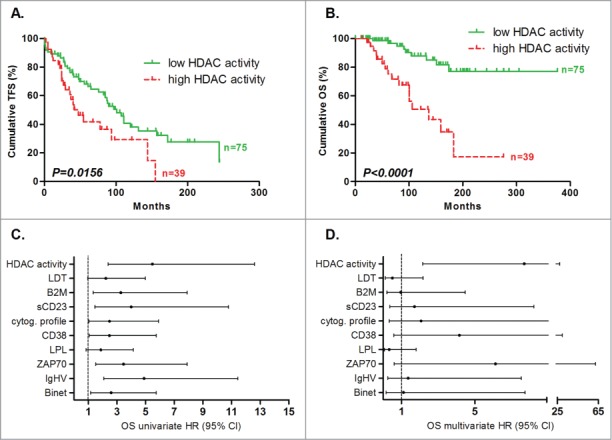
Prognostic power of HDAC activity. Deacetylation power was measured by enzymatic assay and cut-offs were optimized to maximize concordance with overall survival. Statistical differences between curves were calculated using the log-rank test. Statistical details can be found in Table S1. Kaplan-Meier curves showed the TFS (A) and OS (B) prognostic power of global HDAC activity. Cox regression was used to calculate hazard ratio for classical prognostic factors and HDAC activity in univariate (C) and multivariate analyses (D) for OS.
Survival cox regression analysis
Survival analyses with Cox regression were performed to determine the hazard ratio (HR) of prognostic factors. Univariate analyses confirmed that the population was well characterized (in CLL patients, all prognostic factors correlated with clinical data; Table S3). Additionally, we observed that HDAC activity displayed higher HR compared to all tested factors (5.481; 95% CI: 2.383 - 12.603; Fig. 2C). We then used 2 models for multivariate analyses: the “enter method,” which forces the model to include all factors in one equation explaining the survival data, and the “stepwise method,” which selects only the significant factors that provide independent information about survival. Using the enter method to determine OS, HDAC activity was the only prognostic factor, out of the 9 investigated (Binet stage, IgHV mutational status, ZAP70, LPL, CD38, sCD23, B2M, LDT, and HDAC activity), with significant HR (7.68; 95% CI: 2.15 – 27.37; P = 0.0017; Fig. 2D). Following the stepwise regression method, the HR for HDAC activity, ZAP70, and CD38 were 6.85 (95% CI: 2.26 – 20.7; P = 0.0007), 4.15 (95% CI: 1.42 – 12.11; P = 0.0093) and 3.77 (95% CI: 1.19 – 11.91; P=0.0240), respectively, suggesting that these factors are good independent markers of OS (Table S3). However, for TFS, HDAC activity did not reach significance using either method (details about TFS analyses are available in Table S3).
Refinement of classical prognostic factors
We investigated whether HDAC activity could refine other prognostic markers. Patients were divided into poor or good prognosis subgroups and HDAC activity was evaluated in each subgroup. The median OS of patients with low and high HDAC activity belonging to either good or poor prognostic subgroups were all significantly different (P < 0.05), except for the Binet A group (P = 0.0514), not mutated patients (P = 0.1979), and those with LDT <1 year (P = 0.0742). However, the results were close to the limit of significance, most likely due to the low number of patients in these subgroups (Fig. 3). Details about OS analyses in different prognostic subgroups are available in Table S4.
Figure 3.
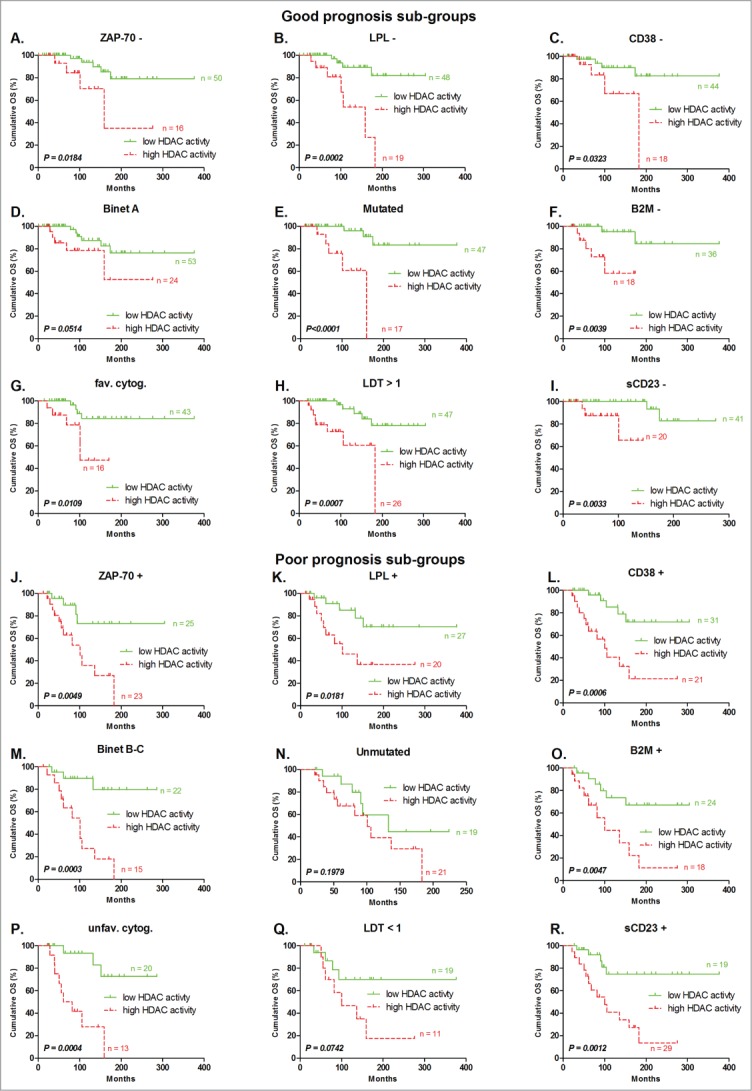
Refinement of prognosis sub-groups by HDAC activity for OS prediction. Patients classified in good prognostic subgroups by classical prognostic factors, including (A) ZAP70 – (n = 66), (B) LPL – (n = 67), (C) CD38 – (n = 62), (D) Binet stage A (n = 77), (E) IgHV mutated (n = 64), (F) B2M – (n = 54), (G) favorable cytogenetic abnormalities (n = 59), (H) lymphocyte doubling time >1 year (n = 73), and (I) soluble CD23 – (n = 61), were divided following their HDAC activity. The same was done for patients classified in poor prognosis subgroups by classical prognostic factors, including (J) ZAP-70 + (n = 48), (K) LPL + (n = 47), (L) CD38 + (n = 52), (M) Binet stage B - C (n = 37), (N) IgHV unmutated (n = 40), (O) B2M + (n = 22), (P) unfavorable cytogenetic abnormalities(n = 33), (Q) lymphocyte doubling time <1 year (n = 30), and (R) soluble CD23 + (n = 48). Statistical differences between curves were calculated using the log-rank test.
HDAC activity and association with HDAC mRNA levels
To further characterize the expression and activity of HDAC genes, we measured mRNA levels of the 18 HDAC isoenzymes in low and high HDAC activity groups using real-time PCR (Fig. 4). mRNA was available from only 75 patients (out of a total of 114): 48 with low HDAC activity (mean: 17.23 ± 1.58 pmol; range: 1.62 – 37.83) and 27 with high HDAC activity (mean: 244.8 ± 77.49 pmol; range 38.12 – 1861). SIRT2 and SIRT6 were the only HDACs with significantly different mRNA levels between the 2 groups. Interestingly, we observed an inverted correlation: the RNA levels were higher in the low HDAC activity group than in the high HDAC activity group (SIRT2: 11.23 vs. 7.63; P = 0.0023 – SIRT6: 5.98 vs. 4.20; P = 0.0301).
Figure 4.
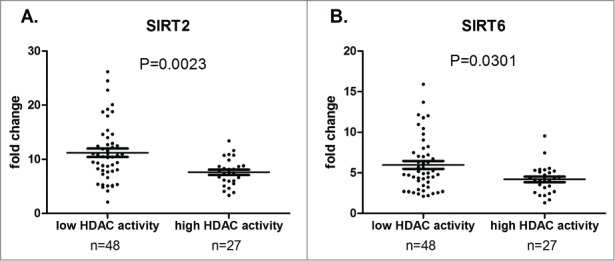
Correlation between HDAC RNA levels and global HDAC enzymatic activity. Mean HDAC mRNA levels (expressed as fold change) is displayed with standard error of the mean (SEM) overlaid with scatter plot for SIRT2 (A) and SIRT6 (B) in low (n = 48) and high (n = 27) HDAC activity groups. Statistical differences were assessed using the Mann-Whitney non-parametric test.
Influence of BCR stimulation on HDAC activity
The microenvironment is known to play a crucial role in CLL cell survival. B-cells were cultured in the presence or absence of IgM for 3 days, and HDAC activity was measured. Following BCR stimulation, deacetylated substrates were higher (22.87 ± 3.56 pmol vs. 29.34 ± 4.59 pmol; n = 10; P = 0.0371), corroborating the influence of microenvironmental stimuli on HDAC activity, which results in poor prognosis (Fig. 5).
Figure 5.
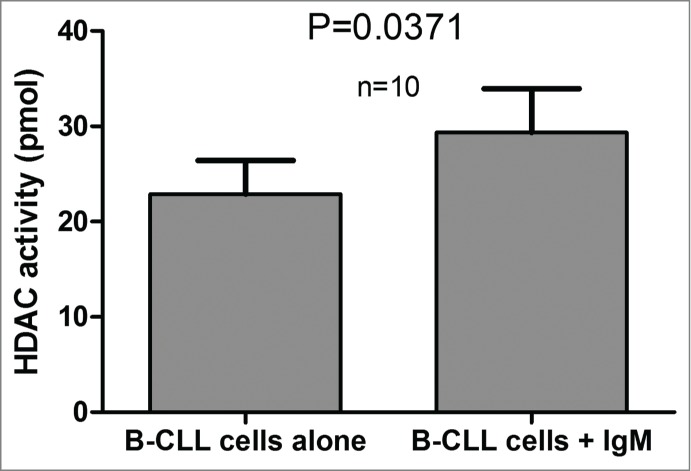
HDAC activity measured with or without BCR stimulation by IgM. HDAC activity values were plotted for 10 CLL patients for samples cultured in the presence of absence of IgM. The significance of the differences was assessed using the Wilcoxon signed rank test.
After culturing CLL B-cells in the presence or absence of IgM, we quantified RNA levels for the 18 HDAC isoenzymes in each condition (Wilcoxon comparison of a minimum of 10 pairs; Figure S5). With the exception of HDAC3 (P = 0.0068), mRNA levels for all isoenzymes were not significantly different between IgM-stimulated and non-stimulated cells (P > 0.05).
Discussion
Targeting HDAC in cancer has become of utmost interest as global loss of monoacetylation and trimethylation on histone H4 is commonly observed in cancer cells.15 Previous studies demonstrate aberrant expression of HDAC in human tumors (as reviewed in5,16). We have previously established the importance of HDAC isoenzymes in prognostic assessment;9 this current study analyzes, for the first time, the relevance of HDAC activity in hematological malignancies.
We demonstrated that HDAC activity is strongly associated with clinical outcome and especially correlated with an increased risk of death in CLL patients. In terms of OS, multivariate cox analyses identified HDAC activity as the sole significant factor (enter method). Using the stepwise method, HDAC activity, together with ZAP70 and CD38, was also identified as a significant prognostic factor. Moreover, protein extracts from CLL B-cells from patients who died during the study produce more deacetylated “Fluor de Lys” substrate compared to protein extracts of living patients. HDAC activity is therefore a predictor of poor prognosis for TFS (P = 0.0156) and, in a stronger manner, for OS (P < 0.0001).
Some patients present discrepancies between their biological status and their clinical evolution. This is why parallel analyses are needed, and various prognostic factors must be assessed to best evaluate the outcome of a patient and to adapt the frequency of their follow-up assessments. We analyzed the impact of classical biological factors (ZAP70, LPL, CD38, IgHV mutational status, sCD23, B2M, and LDT) on TFS and OS in our study population. Concomitantly, we evaluated whether HDAC activity provided additional prognostic information. The results of the multivariate analyses were confirmed by correlation with clinical data; compared with other prognostic factors, HDAC activity presents the highest and most significant χ2 in the log-rank test for OS analysis (Table S1). Moreover, its independence is illustrated by the ability of the variable to refine other prognostic factors.
Interestingly, we observed that in the population with poor prognosis, HDAC activity isolated 2 different populations: one population (high HDAC activity) with a short median OS (<101 months, all groups combined) and another population (low HDAC activity) with a longer median OS (>376.5 months, all groups combined, except unmutated IgHV group ). For low-risk patients, HDAC activity also isolated 2 populations with different prognosis but not in the Binet A population (most likely due to the low number of deaths in this subgroup). These analyses reveal that HDAC activity is a powerful prognostic factor that allows the identification of patients with a worse prognosis or a more rapid evolution defined by the other recently admitted prognostic factors.
The determination of the optimal cut-off is a recurrent problem in all prognostic studies using continuous variables. Here we decided to determine it by recursive partitioning to maximize the correlation with overall survival. We tested other methods to assess an optimal cut-off such as ROC curves maximizing the concordance with IgHV status or simply by choosing the median. Each one depicts different statistical values for the log-rank test in Kaplan-Meier curves but they were all significant for the correlation with OS (P < 0.01). In conclusion, independently of the method used to select a cut-off, HDAC activity remains a strong prognostic factor for OS (Figure S6).
HDAC isoform expression profiles between low and high HDAC activity showed that mRNA of SIRT2 and SIRT6 was higher when the global HDAC activity was low. This observation is in agreement with our previous data showing that patients with high expression of these isoenzymes present higher median OS.9 This good prognosis feature has been demonstrated in previous studies: SIRT6 induces apoptosis in cancer but not in normal cells,17 and overexpression of SIRT2 mediates a delay in cellular proliferation.18 The link between RNA and protein activity is still hypothetical because many steps, such as posttranscriptional or posttranslational modifications, could interfere with a proportional correlation. However, these results lead us to hypothesize that the poor prognosis feature of global HDAC activity was not due to these sirtuins.
Functional analysis revealed that BCR stimulation with IgM influences HDAC activity. It is unlikely that this increase was due to upregulation of transcription unless HDAC3 was highly involved in global deacetylase activity, as HDAC3 was the sole isoenzyme displaying higher mRNA levels in stimulated cells (7.647 ± 2.502 vs. 22.98 ± 6.513; P = 0.0068). Microenvironment influence could act on posttranslational regulation, such as by protein activation during intracellular signaling. Indeed, HDAC5 and 7 were shown to be phosphorylated and exported from the nucleus following ligation of the BCR, which involves protein kinase D.19
It is worth noting, as mentioned in the protocol manual, that the Fluor-de-Lys®-Green HDAC fluorometric activity assay kit works with Class I, Class IIb, Class III, and Class IV HDACs. Class IIa has been reported to have weak deacetylase activity,20 corroborating the fact that the kit might not successfully work with it. Instead, the isoenzymes of this class may act as histone “readers” or scaffold proteins rather than as effective deacetylases.21 HDAC activity described in our work is most likely due to Class I, IIb or IV HDACs. Inhibition tests using valproic acid, SAHA, and tubastatin A HCl partially confirmed this hypothesis, as HDAC activity decreased significantly in the presence of these drugs (data not shown). Unfortunately, lack of specific HDAC inhibitors does not allow the determination of the individual activity of each isoenzyme.
Class I includes HDAC1, 2, 3 and 8. These proteins generally have nuclear localization, are ubiquitous, and deacetylate histones as well as DNA-binding nuclear receptors and transcription factors.22 HDAC6 and HDAC10 belong to Class IIb and are mostly cytoplasmic. HDAC6 does not deacetylate histones, but rather acts on structural proteins, such as tubulin23 or chaperone proteins, such as Hsp90.24 However, we demonstrated a link between global HDAC activity and histone H4 acetylation (Figure S4), suggesting that CLL prognosis is associated with histone epigenetic regulation.
HDAC activity reflects more than just a disrupted epigenetic control of gene transcription and other cellular events. Motility or protein degradation, involving HDAC, may contribute to the pathogenesis of CLL. This work encourages further research in the field of HDAC inhibitors, giving a biological rational for their potential use in the clinic.
Material and Methods
Patients and samples
This study was approved by the Bordet Institute Ethics Committee and conducted according to the principles expressed in the Declaration of Helsinki. All samples were collected at the time of diagnosis before any treatment and after written informed consent was obtained from the 114 CLL patients who presented with a typical CD19+CD5+CD23+ phenotype. TFS and OS were calculated from the time of diagnosis until the date of first treatment and the date of death, respectively. All of the deaths were CLL-related. Two kinds of control samples were used: normal CD19+ purified B-cells from peripheral blood of 15 age-matched healthy volunteers (mean, 69 years old; range: 54–90) and CD19+ purified B-cells from 15 umbilical cord blood samples.
Assessment of classic prognostic factors
ZAP70 and LPL were measured by real-time PCR, as previously described.25 CD38 expression was assessed by flow cytometry, sCD23 and B2M by ELISA, and IgHV gene mutational analysis was performed using the IGH Somatic Hypermutation Assay v2.0 (Invivoscribe; La Ciota, France). LDT was assessed according to Montserrat et al.26 Classical cytogenetics by standard karyotype analysis and additional interphase FISH were performed to screen for the most common aberrations using the Chromoprobe Multiprobe® - CLL System (Cytocell, Amplitech; Compiegne, France). Patients were then classified according to Döhner et al. and Cuneo et al. recommendations: patients with del(17p), del(11q), del(6q), trisomy 12, or a complex karyotype were classified in the “unfavorable cytogenetic abnormalities” subgroup, while patients with a normal karyotype, with del(13q) or other abnormalities, were classified in “favorable cytogenetic abnormalities” subgroup.27,28
Protein extraction, HDAC activity quantification and normalization, and histone H4 acetylation assessment
Purified B-cells were obtained by CD19+ sorting using a magnetic bead system (MidiMACS, Miltenyi Biotec; Bergish Gladbash, Germany) after the isolation of mononuclear cells by density gradient centrifugation (Linfosep, Biomedics; Madrid, Spain). The mean purities of the 114 CLL and the 30 control samples, measured by flow cytometry, were 99.49 ± 0.07% (range: 98.00–99.99) and 97.40 ± 0.71% (range: 92.46–99.81), respectively. Ten million CD19+ cells were then lysed in 400 μl of cell lysis buffer (Cell Signaling, Bioké; Leiden, Netherlands) for 20 minutes on ice. After thrice quick deep-freezing in liquid nitrogen to complete lysis, samples were centrifuged at 14,000 g to eliminate cellular debris. The supernatant containing proteins was recovered and stored at -80°C until HDAC activity measurement.
HDAC activity was assessed with the commercial kit “Fluor de Lys” (Enzo Life Sciences BVBA; Antwerpen, Belgium) according to the manufacturer's instructions. Briefly, 15 μl of protein extracts was incubated with 200 μM of Fluor de Lys substrate for 10 min, allowing the removal of acetyl groups from this compound. The sample was then excited at 485 nm to induce a light emission measured at 530 nm by a fluorometer (Organon Teknika; Turnhout, Belgium) after the addition of a developer. To allow sirtuins to be efficient, all reactions were performed with 100 μM of NAD+. The developer contains trichostatin A and nicotinamide to stop the deacetylation process during the measuring.
Cell count or protein concentration is widely accepted as a method to normalize enzyme activity. However, in our experience, both measures exhibit poor reproducibility and low accuracy. Therefore, in the present study, we normalized HDAC activity by determining GAPDH concentration of each sample by ELISA (Bioo Scientific, Sanbio; Uden, Netherlands). All results are expressed as pmol of deacetylated substrate per μg of GAPDH (pmol).
Histone H4 acetylation was measured as described by Ronzoni et al.14 Additional details can be found in Supplemental Material.
Cell cultures and BCR stimulation
We coated plates with 9 μg of IgM (Jackson ImmunoResearch; Suffolk, UK) in PBS containing 0.5% BSA and cultured 10 × 106 of B-CLL cells for 3 days. For this purpose, CLL cells were purified by negative selection (B CLL selection kit, Miltenyi) to avoid B-cell activation.
RNA quantification
Total RNA was extracted from purified CD19+ cells in a single step using TriPure Isolation Reagent (Roche Applied Science). cDNA was generated from 500 ng of RNA using qScript cDNA SuperMix (Quanta Biosciences; Gaithersburg, MD, USA) according to the manufacturer's protocol. HDAC mRNA levels (18 isoenzymes) were quantified by real-time PCR using custom TaqMan® low-density arrays (TLDA, Life Technologies-Applied Biosystems; Carlsbad, CA, USA) as previously described.9 Additional details can be found in Supplemental Material.
Statistical analysis
Cut-off values were determined by recursive partitioning to maximize the concordance with OS allowing the division of our cohort into high and low activity groups. All mean comparisons were performed using a Mann-Whitney test. TFS and OS analysis were plotted with Kaplan-Meier curves, and differences between groups in terms of prognosis were assessed with a log-rank test. To assess the independent feature of prognostic factors (including HDAC activity) in regards to TFS and OS, we performed Cox regression with binarized data. All tests were 2-sided. An effect was considered to be statistically significant at P < 0.05. Results were analyzed using the GraphPad Prism 5.0 (Graph-Pad Software) or SPSS 18.0.0 software. A 2-fold cross-validation study was performed (3 times) to assess the reproducibility of OS prognostic power of HDAC activity.
Supplementary Material
Author Contributions
M.V.D. performed the research, analyzed the data, made the figures and tables, and wrote the manuscript. E.C. participated to the revision of the manuscript. N.M., D.B. and P.M. helped provide the patient samples and data. B.D. and H.E.H. performed IgHV mutational status assessment. L.L. and B.S. performed, supervised and designed the research and revised the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the Fonds IRIS-Recherche, the Télévie Fund, the David and Alice Van Burren Fund, and the Plan National Cancer of the Belgian Ministry of Health.
References
- 1.Hamblin T. Chronic lymphocytic leukaemia: one disease or two? Ann.Hematol 2002; 81:299-303 [DOI] [PubMed] [Google Scholar]
- 2.Van Bockstaele F, Verhasselt B, Philippe J. Prognostic markers in chronic lymphocytic leukemia: a comprehensive review. Blood Rev 2009; 23:25-47; PMID:18599169; http://dx.doi.org/ 10.1016/j.blre.2008.05.003 [DOI] [PubMed] [Google Scholar]
- 3.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu.Rev.Biochem 1998; 67:545-579; http://dx.doi.org/ 10.1146/annurev.biochem.67.1.545 [DOI] [PubMed] [Google Scholar]
- 4.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett 2009; 277:8-21; PMID:18824292; http://dx.doi.org/ 10.1016/j.canlet.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 5.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett 2009; 280:168-176; PMID:19103471; http://dx.doi.org/ 10.1016/j.canlet.2008.10.047 [DOI] [PubMed] [Google Scholar]
- 6.Secrist JP, Zhou X, Richon VM. HDAC inhibitors for the treatment of cancer. Curr Opin Investig Drugs 2003; 4:1422-1427; PMID:14763127 [PubMed] [Google Scholar]
- 7.Stamatopoulos B, Meuleman N, De Bruyn C et al. Antileukemic activity of valproic acid in chronic lymphocytic leukemia B cells defined by microarray analysis. Leukemia 2009; 23:2281-2289; PMID:19710697; http://dx.doi.org/ 10.1038/leu.2009.176 [DOI] [PubMed] [Google Scholar]
- 8.Stamatopoulos B, Meuleman N, De Bruyn C et al. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis, down-regulates the CXCR4 chemokine receptor and impairs migration of chronic lymphocytic leukemia cells. Haematologica 2010; 95:1136-1143; PMID:20145270; http://dx.doi.org/ 10.3324/haematol.2009.013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Damme M, Crompot E, Meuleman N et al. HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics. 2012; 7:1403-1412; PMID:23108383; http://dx.doi.org/ 10.4161/epi.22674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood 1998; 91:2387-2396; PMID:9516138 [PubMed] [Google Scholar]
- 11.Scielzo C, Apollonio B, Scarfo L et al. The functional in vitro response to CD40 ligation reflects a different clinical outcome in patients with chronic lymphocytic leukemia. Leukemia 2011; 25:1760-1767; PMID:21709686; http://dx.doi.org/ 10.1038/leu.2011.149 [DOI] [PubMed] [Google Scholar]
- 12.Herishanu Y, Polliack A. B-cell receptor signaling in chronic lymphocytic leukemia leans on Lyn. Leuk Lymphoma 2013; 54:1125-1126; PMID:23270582; http://dx.doi.org/ 10.3109/10428194.2012.754097 [DOI] [PubMed] [Google Scholar]
- 13.Vlad A, Deglesne PA, Letestu R et al. Down-regulation of CXCR4 and CD62L in chronic lymphocytic leukemia cells is triggered by B-cell receptor ligation and associated with progressive disease. Cancer Res 2009; 69:6387-6395; PMID:19654311; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-4750 [DOI] [PubMed] [Google Scholar]
- 14.Ronzoni S, Faretta M, Ballarini M, Pelicci P, Minucci S. New method to detect histone acetylation levels by flow cytometry. Cytometry A 2005; 66:52-61; PMID:15915507; http://dx.doi.org/ 10.1002/cyto.a.20151 [DOI] [PubMed] [Google Scholar]
- 15.Fraga MF, Ballestar E, Villar-Garea A et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005; 37:391-400; PMID:15765097; http://dx.doi.org/ 10.1038/ng1531 [DOI] [PubMed] [Google Scholar]
- 16.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014; 124:30-39; PMID:24382387; http://dx.doi.org/ 10.1172/JCI69738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011; 10:3153-3158; PMID:21900744; http://dx.doi.org/ 10.4161/cc.10.18.17435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle 2007; 6:1011-1018; PMID:17457050; http://dx.doi.org/ 10.4161/cc.6.9.4219 [DOI] [PubMed] [Google Scholar]
- 19.Matthews SA, Liu P, Spitaler M et al. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol.Cell Biol 2006; 26:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischle W, Dequiedt F, Hendzel MJ et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 2002; 9:45-57; PMID:11804585; http://dx.doi.org/ 10.1016/S1097-2765(01)00429-4 [DOI] [PubMed] [Google Scholar]
- 21.Lahm A, Paolini C, Pallaoro M et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A 2007; 104:17335-17340; PMID:17956988; http://dx.doi.org/ 10.1073/pnas.0706487104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dokmanovic M, Perez G, Xu W et al. Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol.Cancer Ther 2007; 6:2525-2534; http://dx.doi.org/ 10.1158/1535-7163.MCT-07-0251 [DOI] [PubMed] [Google Scholar]
- 23.Hubbert C, Guardiola A, Shao R et al. HDAC6 is a microtubule-associated deacetylase. Nature 2002; 417:455-458; PMID:12024216; http://dx.doi.org/ 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- 24.Kovacs JJ, Murphy PJ, Gaillard S et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 2005; 18:601-607; PMID:15916966; http://dx.doi.org/ 10.1016/j.molcel.2005.04.021 [DOI] [PubMed] [Google Scholar]
- 25.Stamatopoulos B, Meuleman N, Haibe-Kains B et al. Quantification of ZAP70 mRNA in B cells by real-time PCR is a powerful prognostic factor in chronic lymphocytic leukemia. Clin.Chem. 2007; 53:1757-1766; [DOI] [PubMed] [Google Scholar]
- 26.Montserrat E, Sanchez-Bisono J, Vinolas N, Rozman C. Lymphocyte doubling time in chronic lymphocytic leukaemia: analysis of its prognostic significance. Br.J Haematol 1986; 62:567-575; http://dx.doi.org/ 10.1111/j.1365-2141.1986.tb02969.x [DOI] [PubMed] [Google Scholar]
- 27.Dohner H, Stilgenbauer S, Benner A et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N.Engl J Med 2000; 343:1910-1916; http://dx.doi.org/ 10.1056/NEJM200012283432602 [DOI] [PubMed] [Google Scholar]
- 28.Cuneo A, Rigolin GM, Bigoni R et al. Chronic lymphocytic leukemia with 6q- shows distinct hematological features and intermediate prognosis. Leukemia 2004; 18:476-483; PMID:14712287; http://dx.doi.org/ 10.1038/sj.leu.2403242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


