Abstract
High-risk human papillomavirus (hrHPV)-induced immortalization and malignant transformation are accompanied by DNA methylation of host genes. To determine when methylation is established during cell immortalization and whether it is hrHPV-type dependent, DNA methylation was studied in a large panel of HPVE6E7-immortalized keratinocyte cell lines. These cell lines displayed different growth behaviors, i.e., continuous growth versus crisis period prior to immortalization, reflecting differential immortalization capacities of the 7 HPV-types (16/18/31/33/45/66/70) studied. In this study, cells were monitored for hypermethylation of 14 host genes (APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2) at 4 different stages during immortalization. A significant increase in overall methylation levels was seen with progression through each stage of immortalization. At stage 1 (pre-immortalization), a significant increase in methylation of hTERT, mir124–2, and PRDM14 was already apparent, which continued over time. Methylation of ROBO3 was significantly increased at stage 2 (early immortal), followed by CYGB (stage 3) and FAM19A4, MAL, PHACTR3, and SFRP2 (stage 4). Methylation patterns were mostly growth behavior independent. Yet, hTERT methylation levels were significantly increased in cells that just escaped from crisis. Bisulfite sequencing of hTERT confirmed increased methylation in immortal cells compared to controls, with the transcription core and known repressor sites remaining largely unmethylated. In conclusion, HPV-induced immortalization is associated with a sequential and progressive increase in promoter methylation of a subset of genes, which is mostly independent of the viral immortalization capacity.
Keywords: cervical cancer, E6, E7, FAM19A4, high-risk HPV, hTERT, human papillomavirus, immortalization, methylation
Abbreviations
- CIN
cervical intraepithelial neoplasia
- HFK
human foreskin keratinocytes
- hrHPV
high-risk human papillomavirus
- LZRS
empty vector
- MIP
methylation independent PCR
- (q)MSP
(quantitative) methylation specific PCR
- SCC
squamous cell carcinoma
- SFM
serum free medium
Introduction
Infection with high-risk human papillomavirus (hrHPV) is the major cause of cervical cancer, as well as a subset of other anogenital cancers and head and neck cancers.1,2 According to their prevalence in cervical cancer, 12 HPV types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59) are classified as high-risk and 8 (HPV26, 53, 66, 67, 68, 70, 73, and 82) as probable or possible high-risk.1
Cervical cancer develops via morphologically recognizable precancerous lesions, also known as cervical intraepithelial neoplasia (CIN). Cervical carcinomas and their closest precursor lesions, i.e., high-grade or transforming CIN, are characterized by elevated expression of the viral oncogenes E6 and E7 in proliferating cells.3 E6 and E7 are known to dysregulate apoptosis, cell cycle control, and the replicative lifespan by interfering, among others, with activities of p53, pRB and hTERT.4,5 This is further accompanied by the induction of genomic instability and epigenetic changes.4 Epigenetic alterations include both histone modifications and DNA methylation, which control the chromatin status and affect gene transcription.6
We and others have previously shown that various established and probable/possible hrHPV types display differential properties in terms of immortalization of primary human foreskin keratinocytes (HFK).7-9 In our studies, the E6/E7 genes of HPV16, 18, 31, and 33 consistently triggered a continuous growth without apparent growth arrest (crisis). Transduction of HFK by E6/E7 of HPV45, 66, and 70, on the other hand, resulted initially in an extended lifespan, but only after a long period of growth reduction or crisis some immortal clones emerged. In the case of HPV45, only HFKs of one out of 3 donors ultimately became immortal. In all cultures, immortalization was characterized by upregulated expression of hTERT, the catalytic subunit of telomerase, and by activation of telomerase. The differential growth behaviors suggest that immortalization induced by HPV16, 18, 31, and 33 either requires less additive epigenetic or genetic events, or that the timing of these events is different, i.e., before normally a crisis period would become manifest. Candidate epigenetic events associated with HPV-induced cervical carcinogenesis include promoter methylation of various host cell genes, often resulting in silencing of the respective genes. Frequently methylated genes in cervical cancer include APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2 (for reviews see refs. 3, 10–12). A subset of the above mentioned genes (APC, CADM1, FAM19A4, hTERT, MAL, mir124–1, mir124–2, mir124–3, PHACTR3, and PRDM14) have previously been tested in HPV16- and HPV18-immortalized HFK cell lines to determine the onset and order of DNA methylation alterations during HPV16 and HPV18-induced transformation.13-18 A progressive increase in methylation levels with passaging of these cell lines was seen and methylation levels were generally higher in cervical cancer cell lines than in in vitro HPV16- or HPV18-immortalized HFKs. Furthermore, for CADM1, hTERT, MAL, mir124, PRDM14, and SFRP2 a functional role of methylation-mediated gene silencing in HPV-induced transformation could be demonstrated.15-20 Whereas promoter hypermethylation of most of the above mentioned genes has been linked to gene silencing, hypermethylation of hTERT has been found to positively correlate to gene expression and activation.18,21-24 This phenomenon may be attributed to methylation-mediated inhibition of the transcriptional repressor CTCF, which can bind to the hTERT gene in the first and second exon.25 Since upregulated hTERT mRNA expression is critical to telomerase activation and HPV-induced immortalization,26 hTERT methylation may represent an important regulatory mechanism of HPV-induced immortalization.
It is currently unknown to what extent the changes in methylation are related to the different stages of transformation induced by HPV types other than HPV16 and 18, and whether altered DNA methylation during immortalization is related to the oncogenic capacity of the different HPV types.
Since cells from various passages of HPV16-, 18-, 31-, 33-, 45-, 66, and 70-transduced keratinocytes have been harvested and stored, this offers unique possibilities to relate gene promoter hypermethylation to the immortalization capacities of these HPV types in a longitudinal manner. Here, we analyzed the methylation status of 14 host cell genes, APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2, at various passages pre- and post-immortalization. This included a more in depth methylation analysis of individual CpGs within the hTERT regulatory sequences.
Results
Promoter methylation increases during HPV-induced immortalization
Previously, we have shown that E6/E7 of HPV16, 18, 31, 33, 45, 66, and 70 have differential capacities to immortalize primary HFKs.8 HPV16-, 18-, 31-, and 33-transduced HFKs showed a continuous growth, whereas immortalization of HFKs by HPV45, 66, and 70 was preceded by a long period of crisis. Accordingly, the latter types are considered less oncogenic in terms of in vitro immortalization.
To determine whether methylation of particular genes during the immortalization process is related to the observed variations in growth behavior, methylation was analyzed at 4 different stages of 2 or 3 HPV-transduced HFK donors (donor I-III) per HPV type. These involved the following stages: (1) passage 14–20, pre-immortal cells in their extended life span; (2) passage 25–30, early passage immortal, telomerase positive cells; (3) passage 40–46, intermediate passage immortal, telomerase positive cells and; (4) passage 73–80, late passage immortal, telomerase positive cells. Cultures were considered immortal in case of growth beyond a state where HFKs transduced by E6/E7 of HPV45, 66, and 70 were in crisis. All immortal cultures were characterized by prolonged growth, elevated hTERT mRNA expression, and telomerase activity.8 For stage 1, 17 cell cultures were analyzed, including those of HPV45 transduced HFKs of donor II and III that did not reach stage 2. For stage 2 to 4, 15 cell lines were examined. Early passages of untransduced HFKs (n = 6), HFKs transduced with empty vector (LZRS; n = 3), and HFK of donor I transduced with low-risk HPV11, which did not become immortal, were used as negative controls.8 The cancer cell lines, SiHa and A549 served as positive controls.13,15-18,27,28
Promoter methylation of 14 genes that are frequently methylated in (cervical) cancer,13,15-18,27 i.e., APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2, was determined by qMSP. For hTERT, 2 qMSPs, referred to as hTERT M1 and hTERT M2 that target the promoter region and first intron/second exon, respectively, were tested, given their putative association with methylation-mediated hTERT activation.18,21-25
The promoter regions of APC, mir124–1, mir124–3, and RASSF1A did not show any hypermethylation in the HPV-immortalized cell lines, whereas methylation levels were high in (cervical) cancer cell lines. For the remaining 10 genes, CADM1, CYGB, FAM19A4, hTERT, mir124–2, MAL, PHACTR3, PRDM14, ROBO3, and SFRP2, increased promoter methylation was detected in the HPV-immortalized cells at least at one stage of transformation.
When comparing the average methylation level of all 14 genes, a significant increase was seen with progression through each stage of transformation (P<0.01 at each stage; Fig. 1A). Moreover, methylation levels at stage 1 were significantly increased compared to controls (P<0.01) and the highest methylation levels were detected in cancer cells (Fig. 1A).
Figure 1.
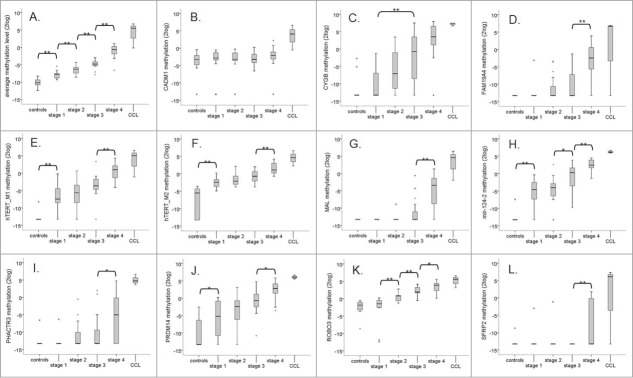
Progressive increase in DNA methylation levels. (A) Box-plot of the average methylation level of all 14 genes (APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2) per stage. (B–L) Boxplots of the methylation levels of the individual genes per stage. Only genes that became methylated in at least one stage are shown (CADM1 (B), CYGB (C), FAM19A4 (D), hTERT M1 (E), hTERT M2 (F), MAL (G), mir124–2 (H), PHACTR3 (I), PRDM14 (J), ROBO3 (K), and SFRP2 (L)). * P < 0.05, ** P < 0.01.
The methylation levels per individual gene at stage 1 to 4 are shown in Figure 1B–L. At stage 1 a significant increase in methylation of hTERT M1 and M2, mir124–2, and PRDM14 compared to controls was already apparent (Fig. 1E, F, H, and J, respectively). Moreover, methylation of all 3 genes showed a progressive increase over time. Methylation of ROBO3 was significantly increased at stage 2 compared to stage 1 (Fig. 1K) and methylation of CYGB was significantly increased at stage 3 compared to stage 1 (Fig. 1C). Methylation of FAM19A4, MAL, PHACTR3 and SFRP2 was significantly increased at stage 4 (Fig. 1D, G, I, and L, respectively). For CADM1 a slight increase in methylation was seen with passaging, which did not reach significance in this relatively small sample set.
Progressive increase in levels of DNA methylation is mostly growth behavior independent
In order to determine whether the progressive increase in DNA methylation levels is related to the growth behavior of the cells, the average methylation level of all 14 genes at the individual stages was compared between cell lines that grew continuously (i.e., HPV16-, 18-, 31-, and 33-immortalized cells) and cell lines that encountered a crisis period prior to immortalization (i.e., HPV45-, 66-, and 70-immortalized cells). As shown in Figure 2A, a borderline significant difference between both groups is seen at stage 2 (p=0.059), which is just after immortalization. No clear differences were seen at stage 1, 3 and 4. Further comparison of individual gene methylation levels at stage 2 between cells with and without growth crisis, revealed that methylation of hTERT at the M2 region and methylation of PHACTR3 were both significantly increased in cells that underwent a crisis compared to those that grew continuously (Fig. 2B and C).
Figure 2.
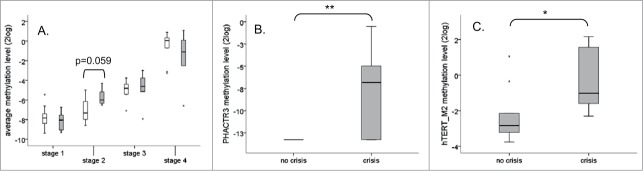
Progressive increase in levels of DNA methylation is mostly growth behavior independent. (A) Boxplot of the average methylation level of all 14 genes (APC, CADM1, CYGB, FAM19A4, hTERT (M1 and M2), MAL, mir124–1, mir124–2, mir124–3, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2) per stage divided between cell lines that grew continuously in culture (white: HPV16, 18, 31 and 33) and cell lines that encounter a crisis period prior immortalization (gray: HPV45, 66 and 70). Significantly differential methylation between cell lines without and with a crisis period prior immortalization was observed for PHACTR3 (B), and hTERT M2 (C) at stage 2. * P < 0.05, ** P < 0.01.
Increased hTERT methylation in HPV-immortalized cells
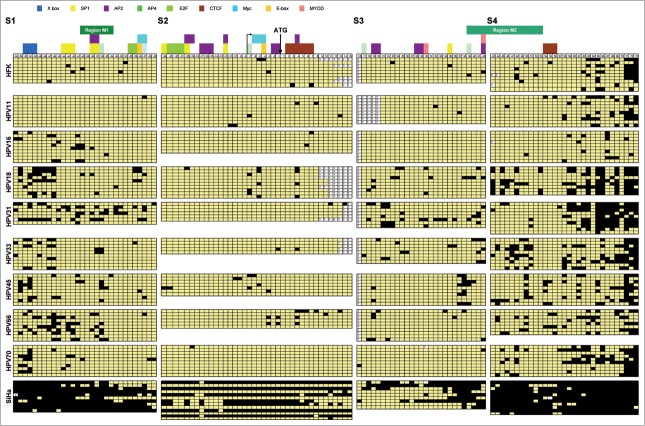
As described above, methylation of hTERT represented one of the earliest methylation events in the majority of HPV-transduced HFKs and methylation at the M2 region was significantly increased in cells that just escaped crisis (i.e., at stage 2 in HPV45-, 66-, and 70- immortalized cells) compared to cells that grew continuously (i.e., HPV16-, 18-, 31-, and 33-immortalized cells). This difference leveled out at stage 3 and 4. The M2 region was included as it is located next to a CTCF binding site implicated in negative hTERT transcription regulation. Methylation of CTCF binding sequences has been shown to inhibit CTCF binding, thereby contributing to hTERT upregulation and telomerase activation.21,22,25 To determine in more detail which transcription factor binding sites in hTERT are targeted by DNA methylation in the HPV-immortalized cells, cells (stage 3) were subjected to bisulfite sequencing on 4 overlapping regions of the hTERT promoter and gene: −476 to −185 bp (region S1), -209 to +96 bp (region S2), +90 to +338 bp (region S3), and +319 to +600 bp (region S4) (top Fig. 3). The methylation patterns in hTERT-positive HPV-immortalized cells were compared to HFKs and HPV11-transduced HFKs which we previously showed to be hTERT mRNA negative,8 and hTERT mRNA positive SiHa cervical cancer cells, in which the hTERT promoter is known to be highly methylated.18
Figure 3.
Bisulfite sequencing results of the hTERT CpG island. Overview of the CpG methylation results of individual cloned PCR products of untransduced HFKs, HPV11- and stage 3 HPV16-, 18-, 31-, 33-, 45-, 66- and 70-transduced HFKs of donor I of 4 regions: S1 spanning −476 to −185 bp, S2 spanning −209 to +96 bp, S3 spanning +90 to +338 bp and S4 spanning +319 to 600 bp relative to the ATG. Numbers refer to the respective CpG and their position relative to the ATG. The position of known and putative transcription factor binding sites are plotted on top of the corresponding CpG. Regions analyzed by qMSP (M1 and M2) are plotted on top of the appropriate CpG in green. Black indicates methylation positive; yellow indicates methylation negative; ? indicates no sequencing results available.
As shown in Figure 3 and Table 1 an increase in methylated CpGs in hrHPV-immortalized cells compared to controls is particularly evident for region S1. Methylation in this region ranged from 7% to 22% in HPV-immortalized cells compared to 3% in HFKs and 0% in HPV11-transduced HFKs. In regions S2 and S3, relatively few CpGs were methylated in both HPV-immortalized cells and controls. Only in SiHa cells these regions were heavily methylated. More pronounced differences were observed in region S4, which is within the gene body. In HPV18-, 31-, 33-, 45-, 66-, and 70-, but not in HPV16-immortalized cells, methylation in S4 was higher than in HFKs and HPV11-HFKs. The 3’ part of region S4 was densely methylated in all cells analyzed. No major differences were evident between less oncogenic HPV types (HPV45, 66, and 70), compared to cells immortalized by the more transforming HPV types 16, 18, 31, and 33. However, among the different cell lines, least methylation was seen in the HPV16-immortalized cells in all 4 regions.
Table 1.
Percentage of hTERT methylation in primary human keratinocytes (HFK), HPV E6/E7 containing HFKs and SiHa cells
| % Methylation (total number of clones analyzed) |
||||
|---|---|---|---|---|
| S1 (−476/−185 bp) | S2 (−209/+96 bp) | S3 (+90/+338 bp) | S4 (+319/+600 bp) | |
| Cell lines | ||||
| HFK | 3 (8) | 2 (9) | 2 (8) | 15 (10) |
| +HPV11 | 0 (10) | 2 (10) | 1 (9) | 15 (10) |
| +HPV16 | 7 (10) | 1 (7) | 0 (10) | 5 (10) |
| +HPV18 | 15 (10) | 4 (10) | 6 (10) | 37 (9) |
| +HPV31 | 22 (7) | 0 (6) | 14 (8) | 32 (10) |
| +HPV33 | 10 (8) | 1 (5) | 8 (8) | 28 (10) |
| +HPV45 | 9 (10) | 4 (7) | 7 (10) | 19 (10) |
| +HPV66 | 13 (14) | 5 (6) | 4 (10) | 18 (8) |
| +HPV70 | 7 (10) | 0 (10) | 1 (10) | 25 (10) |
| SiHa | 82 (10) | 38 (13) | 26 (9) | 92 (11) |
With respect to the specific transcription factor binding sites previously associated with hTERT regulation in HPV-containing cells, i.e., the X-box motif, E-box motif, and CTCF binding sites,25,29-32 minor differences between individual cell lines were observed. The X-box motif in promoter region S1 showed increased methylation in HPV18-, 31-, 33-, 45-, 66-, and 70-immortalized cells compared to the controls. However, in HPV16-immortalized cells this site was largely unmethylated. The two E-boxes (c-myc binding sites) were mostly unmethylated, except for a few methylated CpGs in the distal site in HPV18-, and 45-immortalized cells.
Both CTCF-sites, located within the gene body in region S2 and S4, were mostly unmethylated, whereas CpGs flanking the CTCF site in S4 were methylated more frequently.
In conclusion, compared to HFK control and HPV11-containing cells, most HPV-immortalized cells showed increased hTERT methylation (with slight differences between the HPV-types). Importantly, the bisulfite sequencing results correspond with the methylation levels determined by qMSP (Fig. 1). More methylation was observed in the S3/S4 region where qMSP M2 is located compared to region S1 containing the qMSP M1 region. Moreover, in contrast to qMSP analysis showing significantly increased hTERT M2 methylation at stage 2 in cells that underwent a crisis period, both bisulfite sequencing and qMSP showed no relation to growth behavior at stage 3.
Discussion
The efficiency of keratinocyte immortalization by HPV is HPV-type dependent and may result from differential necessities of supplementary (epi)genetic host cell alterations.8 Longitudinal analysis of multiple HPV16-, 18-, 31-, 33-, 45-, 66-, and 70-immortalized keratinocyte cell lines revealed a sequential and progressive increase in host gene promoter methylation with passaging. These genes include both (candidate) tumor suppressor genes that are known to become silenced by DNA methylation in the context of hrHPV, such as CADM1, MAL, mir124, PRDM14 and SFRP2,15-17,19,20 and hTERT, for which methylation has been associated with gene activation.18,21-25
Methylation of hTERT, mir124–2, and PRDM14 represented the earliest methylation events. The methylation levels of these gene were already significantly increased at the pre-immortal stage (stage 1) compared to controls. Subsequently, at stage 2 a significant increase in ROBO3 methylation was seen. Methylation levels of all 4 genes progressively increased with passaging. Their rather early onset of methylation is in line with previous results on cervical biopsies, showing methylation in a substantial subset of precursor lesions (CIN3) and increasing frequencies and levels of methylation in cervical carcinomas.13,15,18,33
Methylation of CYGB was identified as a successive event detectable from stage 3 onwards. To the best of our knowledge, this is the first report showing that HPV-mediated transformation is associated with CYGB methylation. In support of this, our preliminary data on cervical biopsies also revealed frequent CYGB methylation in cervical carcinomas and precursor lesions (unpublished results). CYGB methylation has been described in lung, oral, breast, ovarian and head and neck cancer.28,34-37
A significant increase in DNA methylation levels of FAM19A4, MAL, PHACTR3, and SFRP2 was seen upon progression from stage 3 to stage 4. CADM1 methylation levels showed a slight increase with passaging, and were particularly high in cancer cell lines. Methylation of these genes has previously also been described in a (major) subset of CIN3 lesions and cervical carcinomas, in line with the cell lines representing a premalignant phenotype.13,16,17,38 An earlier onset of MAL methylation compared to CADM1 methylation has been described previously.16,17,39
A number of genes remained unmethylated, such as APC, mir124–1, mir124–3, and RASSF1A. Given the fact that these genes are methylated in cervical carcinomas,15,40-44 methylation-mediated silencing of these genes may occur at later stages of transformation and/or may be cell type dependent. In fact, methylation of mir124–1 and mir124–3 was highly frequent in SCC and only occasionally detected in CIN3 lesions.15 Cell type dependence may particularly account for APC and RASSF1A methylation, which appeared more often in adenocarcinomas than in squamous cell carcinomas.14,40-42,45 Our in vitro models are based on squamous epithelial cells.
Comparison of cells that grew continuously (HPV16-, 18-, 31-, and 33-transduced HFKs) and cells that underwent a crisis period (HPV45-, 66- and, 70-transduced HFKs), revealed a nearly significant (P = 0.059) increase in methylation levels in cells that just escaped from crisis at stage 2. This difference was mainly attributable to significantly increased hTERT methylation. This suggests that methylation of hTERT is particularly advantageous for immortalization of cells harboring less oncogenic HPV types. Methylation of hTERT has previously been described to result in gene activation and increased hTERT expression is associated with immortalization.18,21-23
This phenomenon could be confirmed in the present study, in which upregulated hTERT mRNA expression, as previously demonstrated in all HPV-immortalized cell lines,8 was associated with increased hTERT methylation when compared to hTERT mRNA negative primary cells. However, the CpGs around the transcription start site (region S2) remained largely unmethylated (Fig. 3), which was also described by Renaud et al.25 and Jiang et al.46. This finding suggests that the core promoter needs to remain unmethylated to allow hTERT activation. Methylation of other regions of the hTERT CpG island has often been associated with elevated hTERT expression.18,21,22,25 Two of those regions within the gene contain CTCF binding sites, of which methylation is known to hinder CTCF binding.25 It has therefore been suggested that hTERT activation is due to hypermethylation of the CTCF binding sites located at S2 (CpG 2–7) and S4 (CpG 54–56).25 Except for a few CpGs in HPV45- and HPV66-immortalized cells, frequent methylation within the CTCF binding sites was not observed, in contrast to the flanking CpGs, which is in line with our previous findings.18
It is therefore unlikely that in the studied cell lines hTERT expression is preceded by methylation-mediated inhibition of CTCF binding to established binding sites. Recently, a CTCF-regulated enhancer element 4.5 kb upstream of hTERT, as well as a number of novel candidate CTCF binding sites and non-CTCF repressive elements in the proximal exonic region, have been identified, each of which may be affected by DNA methylation.47,48 Moreover, accumulating evidence indicates that the HPV-encoded E6 protein plays a prominent role in hTERT gene activation by binding to c-Myc or E6AP.29,31,32,49 More recently, E6 was shown to inhibit the binding of MAZ, a novel hTERT repressor, which correspondingly increased SP1 binding and gene activation.50 The E6 proteins of different HPV-types have different capacities to transactivate hTERT. The E6 proteins of HPV16, 18, 31, 33, 35, 51, 52, and 58 showed highest hTERT promoter activation capacity, whereas E6 of HPV66 and 70 displayed lower activity.51
In the present study, least methylation was detected in HPV16-immortalized cells, suggesting that, among the hrHPV types tested, methylation-mediated hTERT activation may be less essential in case of HPV16E6E7 expression. Except for HPV16, no difference in frequency of hTERT methylation between the strongest (HPV18, 31, and 33) and weakest hTERT activators (HPV66 and 70), as described by Van Doorslaer et al.,51 was seen. Therefore, next to differences in E6 activation capacities, site-dependent methylation patterns or other mechanisms may contribute to hTERT gene activation. Although controversial, HPV16 E6-mediated hTERT activation has previously been suggested to rely (in part) on the proximal E-box.32 Except for methylation of a few CpGs in the distal E-box in HPV18- and 45-immortalized cells, the E-boxes were unmethylated in most immortalized cells.
In conclusion, passaging of HPV16-, 18-, 31-, 33-, 45-, 66-, and 70-transduced human keratinocytes is correlated with a progressive increase in DNA methylation of selected host cell genes. The timing of the methylation events differed between genes. hTERT, mir124–2, and PRDM14, were the first genes that became methylated, even prior to immortalization (stage 1). Following immortalization, ROBO3 methylation (stage 2) preceded CYGB methylation (stage 3) followed by CADM1, FAM19A4, MAL, PHACTR3 and SFRP2 methylation. Early onset of selected epigenetic host cell alterations during HPV-induced HFK immortalization was mostly independent on the viral oncogenic capacity. The onset of hTERT methylation was inversely related to the immortalization capacity of the HPV types tested, though diminished with passaging. More detailed analysis of the hTERT regulatory sequences indicated that reactivation of hTERT expression in immortal cells is most likely not due to severe methylation of known repressor sites.
Materials and Methods
Cells and cell lines
HFKs were isolated from foreskins of independent donors as described before.52 Cell cultures containing low-risk HPV11, hrHPV16, 18, 31, 33, 45, and probable/possible hrHPV66 and 70 were established by retroviral transduction of HFKs with the E6E7 open reading frames of the respective HPV-type as described before.8 Untransduced HFKs and empty vector (LZRS) transduced HFKs served as controls. HFKs of donor I were transduced with all HPV types. Donor II was transduced with all HPV types except HPV33 and donor III was transduced with HPV16, 33, 45, and 70. Transductants were grown in defined keratinocyte serum-free medium (SFM) (Life Technologies 17005–075) containing 5 ng/ml EGF and 50 ng/ml bovine pituitary extract, 100 U/mL natrium-penicillin G (Astellas Pharma B.V. 117837/ 315932), 100 μg/mL streptomycin (Life Technologies 11860038), and 2 mmol/L l-glutamine (Life Technologies 25030024) and 80μg/ml geneticin (Sigma-Aldrich G8168) selection. The cervical cancer cell lines SiHa (HPV16) and CaSki (HPV16) and the lung cancer cell line A549 (HPV-negative) were obtained from the American Type Culture Collection (Manassas, VA USA). Culture conditions were described previously.52,53 All cells were grown at 37°C and 5% CO2.
DNA isolation and bisulfite modification
Genomic DNA was isolated from cell pellets by proteinase K digestion followed by UltraPureTM Phenol:Chloroform:Isoamyl Alcohol (Life Technology 15593049) extraction as described previously.54
Sodium bisulfite modification was performed on 1μg DNA using the EZ DNA MethylationTM Kit (Zymo Research D5002) according to the manufacturer's protocol, which induces chemical conversion of unmethylated cytosines into uracils while leaving methylated cytosines unchanged.
Quantitative methylation specific PCR (qMSP)
DNA methylation of the promoter regions of 14 host cell genes (i.e., APC, CADM1, CYGB, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, RASSF1A, ROBO3, and SFRP2) was determined by qMSP. For hTERT, 2 different regions were analyzed, located in the promoter (M1) and proximal exonic region (M2).18 miR124–1, -2, and -3 are located at different chromosomal regions and regulated by distinct promoter regions, but all encode the same mature miRNA.15
qMSPs of single and multiple targets (Table 2) were performed in a 12-μl reaction volume containing 50 ng of bisulfite treated DNA. For single target qMSP QuantiTect Probe PCR Kit (Qiagen 204345) was used and for multiplex qMSP Quantitect Multiplex PCR Kit (Qiagen 204545).55 qMSPs were run on the ABI 7500 and/or ABI 7900 Fast Real-Time PCR System (Applied Biosystems).
Table 2.
Primer and probe sequences.
| Gene | Forward primer | Reverse primer | Probe | Length (bp) |
|---|---|---|---|---|
| Single target qMSP | ||||
| hTERT M1 | GAGTAGCGTAGGCGATTTAGGGCGT | GTCCAACAACGCGAAACCGAA | CGCACAACCTCTACAACACTCGAACCACCAACTC | 75 |
| hTERT M2 | TAGATTTTCGGGTTCGTTCG | TCTATACCCGCGAATCCACT | CGACCTAACCCCGACAACGCAACTA | 132 |
| MAL | TTAGGTTATTGGGTTTCGCG | GTACTAACGTCGACCTTAAAACGA | TCCGCGCAAACCTCTCGCTAAC | 86 |
| mir124–1 | CGGCGGGGAGGATGTT | ATAAAAAACGACGCGTATACGTACG | CGGCGTTTTTTATTTTT | 94 |
| mir124–3 | ACGCGGCGAAGACGTTT | CGAACGACGAACGTCGAAA | AAAATCCTCGCCCGAAAAACGCGA | 95 |
| ROBO3 | AGGAGGAGGGTACGAAGAGGTATC | AAAACCCGTAAACTAAAAACCGTAAAC | CCGCTCTCCTACCGATACGCCTAAATACGAT | 120 |
| ACTB | TGGTGATGGAGGAGGTTTAGTAAGT | AACCAATAAAACCTACTCCTCCCTTAAA | ACCACCACCCAACACACAATAACAAACACA | |
| Multiplex qMSP | ||||
| APC | GAACCAAAACGCTCCCCAT | TTATATGTCGGTTACGTGCGTTTATAT | CCCGTCGAAAACCCGCCCGATTA | 74 |
| CADM1 | CGTATGTTATTAGTATTTTATTAGT-TGTTCGTTC | CGCTCGACAACACTACACTCG | ACCTACCTCAAACTAACGACGTTAACTACCTCCGA | 106 |
| CYGB | CGAGGTCGATCGTTAGTTCGTTC | CCAACGACTAACTCGAAAACGCG | CGGCGGTCGTCGTGGATTTAG | 117 |
| FAM19A4 | AGTCGGGCGGTTCGGTTA | CCAAAACGACGCGCAACTA | CCCAACTAACGCGCTAA | 106 |
| mir124–2 | GGGTAATTAATTTGGATTTACG-TCGTTAT | CGTAAAAATATAAACGATACGTAT-ACCTACGT | TTTACAACACACGCCTAAA | 138 |
| MYOD1 | CCAACTCCAAATCCCCTCTCTAT | TGATTAATTTAGATTGGGTTTAGAG-AAGGA | TCCCTTCCTATTCCTAAATCCAACCTAAATACCTCC | 162 |
| PHACTR3 | GGTTATTTTGCGAGCGGTTTC | CGAATACTCTAATTCCACGCGACT | AACCGCGTCGAAAAACGAAAACGACTAC | 114 |
| PRDM14 | TTACGTGTTATTGTCGGGGATTC | ATATCTATTCCTAATACCTAAAAA-CGAAACG | AAACGCCTTAAACGCTAAAAAACTTCGCCTC | 88 |
| RASSF1A | GCGTTGAAGTCGGGGTTC | CCCGTACTTCGCTAACTTTAAACG | ACAAACGCGAACCGAACGAAACCA | 75 |
| SFRP2 | GAGTAGCGTAGGCGATTTAGGGCGT | TCCCGAACCCGCTCTCTT | CGCTAAATACGACTCGAAACCCCGAA | 69 |
| Bisulfite sequencing | ||||
| hTERT S1 | GTTTTTAGGGTTTTTATATTATGG | AAACTAAAAAATAAAAAAACAAAAC | 292 | |
| hTERT S2 | GTTTTGTTTTTTTATTTTTTAGTTT | AACCCTAAAACCCCAAA | 305 | |
| hTERT S3 | TTGGGGTTTTAGGGTTG | ACCAACTCCTTCAAACAAAA | 248 | |
| hTERT S4 | GTAGGTGTTTTGTTTGAAGGA | AACTAAAAACCACCAACACAA | 281 | |
CpGs are indicated in bold.
Methylation levels were normalized to the housekeeping gene MYOD1 (in case of APC, CYGB, and RASSF1A) or ACTB (in case of all other qMSP targets). DNA isolated from cancer cell lines SiHa and A549 served as reference and positive control in the individual qMSPs and were set to 100. SiHa DNA was used as a reference for qMSPs of CADM1, FAM19A4, hTERT, mir124–1, mir124–2, mir124–3, MAL, PHACTR3, PRDM14, ROBO3, and SFRP2; A549 DNA for APC, CYGB, and RASSF1A qMSPs. Methylation levels were determined using the 2-ΔΔCT method,56 resulting in a quantification of DNA methylation in the HPV-transduced cells relative to the positive controls SiHa or A549. All samples had an ACTB Ct-value <32, indicating sufficient DNA quality.
Bisulfite sequencing
For hTERT methylation analysis by bisulfite sequencing methylation independent PCR (MIP) was performed using 4 primer sets (Table 2) spanning the hTERT promoter and first exon from the coding sequence from −476 to +600 bp relative to the ATG. MIP, cloning and sequencing was performed as described before.18 Shortly, purified MIP-products were cloned in pGEM-T using pGEM® -T Easy Vector System (Promega A1360) or into the Zero pCR-Blunt II-TOPO vector (Life Technologies K282020). Approximately ten cloned PCR-fragments of every region and cell line were sequenced using the BigDye Terminator v1.1 cycle sequencing kit on an ABI Prism 3100 sequencer (Applied Biosystems). Sequences were analyzed using Chromas Lite version 2.01.
Statistical analysis
Statistical analysis was performed using SPSS (version 20). The average methylation level of all investigated genes as well as methylation levels per gene were compared between stages and between continuously growing cell lines and cell lineages that encountered a crisis period using the non-parametric Mann-Whitney U test. A two-sided P-value <0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
RDM Steenbergen, PJF Snijders, and CJLM Meijer have relationships with Self-screen B.V., The Netherlands.
Acknowledgments
We thank Leonie Voorwerk, Gaby S Steba, Sabrina Boer, Wina Verlaat, Suzanne Snellenberg, Leontien Bosch, Sylvia Duin, and Lise De Strooper for their excellent technical assistance.
Funding
Stichting HUMAVAC, Dutch Cancer Society VU2010-4668, European Research Council (ERC advanced 2012-AdG, proposal 322986 Mass-care).
References
- 1. IARC. Biological agents . Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum 2012; 100:1-441; PMID:23189750 [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855 [DOI] [PubMed] [Google Scholar]
- 3. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014; 14:395-405; PMID:24854082 [DOI] [PubMed] [Google Scholar]
- 4. Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 2010; 10:550-60; PMID:20592731 [DOI] [PubMed] [Google Scholar]
- 5. Pim D, Banks L. Interaction of viral oncoproteins with cellular target molecules: infection with high-risk vs low-risk human papillomaviruses. APMIS 2010; 118:471-93; PMID:20553529 [DOI] [PubMed] [Google Scholar]
- 6. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11:726-34; PMID:21941284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hiller T, Poppelreuther S, Stubenrauch F, Iftner T. Comparative analysis of 19 genital human papillomavirus types with regard to p53 degradation, immortalization, phylogeny, and epidemiologic risk classification. Cancer Epidemiol, Biomarkers Prev 2006; 15:1262-7; PMID:16835321 [DOI] [PubMed] [Google Scholar]
- 8. Schutze DM, Snijders PJ, Bosch L, Kramer D, Meijer CJ, Steenbergen RD. Differential In Vitro Immortalization Capacity of Eleven (Probably) High-risk Human Papillomavirus Types. J Virol 2013; 88:1714-24; PMID:24257607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lace MJ, Anson JR, Klingelhutz AJ, Lee JH, Bossler AD, Haugen TH, Turek LP. Human papillomavirus (HPV) type 18 induces extended growth in primary human cervical, tonsillar, or foreskin keratinocytes more effectively than other high-risk mucosal HPVs. J Virol 2009; 83:11784-94; PMID:19740985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol 2009; 112:293-9; PMID:19054549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szalmas A, Konya J. Epigenetic alterations in cervical carcinogenesis. Semin Cancer Biol 2009; 19:144-52; PMID:19429477 [DOI] [PubMed] [Google Scholar]
- 12. Duenas-Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer 2005; 4:38; PMID:16248899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steenbergen RD, Ongenaert M, Snellenberg S, Trooskens G, van der Meide WF, Pandey D, Bloushtain-Qimron N, Polyak K, Meijer CJ, Snijders PJ, et al. . Methylation-Specific Digital Karyotyping of HPV16E6E7 expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J Pathol 2013; 231:53-62; PMID:23674368 [DOI] [PubMed] [Google Scholar]
- 14. Henken FE, Wilting SM, Overmeer RM, van Rietschoten JG, Nygren AO, Errami A, Schouten JP, Meijer CJ, Snijders PJ, Steenbergen RD. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer 2007; 97:1457-64; PMID:17971771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 2010; 9:167; PMID:20579385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Overmeer RM, Henken FE, Snijders PJ, Claassen-Kramer D, Berkhof J, Helmerhorst TJ, Heideman DA, Wilting SM, Murakami Y, Ito A, et al. . Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol 2008; 215:388-97; PMID:18498117; http://dx.doi.org/ 10.1002/path.2367 [DOI] [PubMed] [Google Scholar]
- 17. Overmeer RM, Henken FE, Bierkens M, Wilting SM, Timmerman I, Meijer CJ, Snijders PJ, Steenbergen RD. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol 2009; 219:327-36; PMID:19662663; http://dx.doi.org/ 10.1002/path.2598 [DOI] [PubMed] [Google Scholar]
- 18. de Wilde J, Kooter JM, Overmeer RM, Claassen-Kramer D, Meijer CJ, Snijders PJ, Steenbergen RD. hTERT promoter activity and CpG methylation in HPV-induced carcinogenesis. BMC Cancer 2010; 10:271; PMID:20534141; http://dx.doi.org/ 10.1186/1471-2407-10-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung MT, Lai HC, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Liu HS, Chu DW, Lin YW. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol 2009; 112:646-53; PMID:19095296; http://dx.doi.org/ 10.1016/j.ygyno.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 20. Snellenberg S, Cillessen SA, van Criekinge W, Bosch L, Meijer CJ, Snijders PJ, Steenbergen RD. Methylation-mediated repression of PRDM14 contributes to apoptosis evasion in HPV-positive cancers. Carcinogenesis 2014; 35:2611-8; PMID:25233927; http://dx.doi.org/ 10.1093/carcin/bgu197 [DOI] [PubMed] [Google Scholar]
- 21. Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Exp Cell Res 2003; 289:326-34; PMID:14499633; http://dx.doi.org/ 10.1016/S0014-4827(03)00281-7 [DOI] [PubMed] [Google Scholar]
- 22. Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer 2002; 101:335-41; PMID:12209957; http://dx.doi.org/ 10.1002/ijc.10593 [DOI] [PubMed] [Google Scholar]
- 23. Kumari A, Srinivasan R, Vasishta RK, Wig JD. Positive regulation of human telomerase reverse transcriptase gene expression and telomerase activity by DNA methylation in pancreatic cancer. Ann Surg Oncol 2009; 16:1051-9; PMID:19194757; http://dx.doi.org/ 10.1245/s10434-009-0333-8 [DOI] [PubMed] [Google Scholar]
- 24. Wang Z, Xu J, Geng X, Zhang W. Analysis of DNA methylation status of the promoter of human telomerase reverse transcriptase in gastric carcinogenesis. Arch Med Res 2010; 41:1-6; PMID:20430247; http://dx.doi.org/ 10.1016/j.arcmed.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 25. Renaud S, Loukinov D, Abdullaev Z, Guilleret I, Bosman FT, Lobanenkov V, Benhattar J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res 2007; 35:1245-56; PMID:17267411; http://dx.doi.org/ 10.1093/nar/gkl1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steenbergen RD, Kramer D, Meijer CJ, Walboomers JM, Trott DA, Cuthbert AP, Newbold RF, Overkamp WJ, Zdzienicka MZ, Snijders PJ. Telomerase suppression by chromosome 6 in a human papillomavirus type 16-immortalized keratinocyte cell line and in a cervical cancer cell line. J Natl Cancer Inst 2001; 93:865-72; PMID:11390536; http://dx.doi.org/ 10.1093/jnci/93.11.865 [DOI] [PubMed] [Google Scholar]
- 27. van der Meide WF, Snellenberg S, Meijer CJ, Baalbergen A, Helmerhorst TJ, van der Sluis WB, Snijders PJ, Steenbergen RD. Promoter methylation analysis of WNT/β-catenin signaling pathway regulators to detect adenocarcinoma or its precursor lesion of the cervix. Gynecol Oncol 2011; 123:116-22; PMID:21726894; http://dx.doi.org/ 10.1016/j.ygyno.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 28. Hubers AJ, Heideman DA, Herder GJ, Burgers SA, Sterk PJ, Kunst PW, Smit HJ, Postmus PE, Witte BI, Duin S, et al. . Prolonged sampling of spontaneous sputum improves sensitivity of hypermethylation analysis for lung cancer. J Clin Pathol 2012; 65:541-5; PMID:22461647; http://dx.doi.org/ 10.1136/jclinpath-2012-200712 [DOI] [PubMed] [Google Scholar]
- 29. Gewin L, Galloway DA. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J Virol 2001; 75:7198-201; PMID:11435602; http://dx.doi.org/ 10.1128/JVI.75.15.7198-7201.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis 2003; 24:1167-76; PMID:12807729; http://dx.doi.org/ 10.1093/carcin/bgg085 [DOI] [PubMed] [Google Scholar]
- 31. Oh ST, Kyo S, Laimins LA. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J Virol 2001; 75:5559-66; PMID:11356963; http://dx.doi.org/ 10.1128/JVI.75.12.5559-5566.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Veldman T, Liu X, Yuan H, Schlegel R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci U S A 2003; 100:8211-6; PMID:12821782; http://dx.doi.org/ 10.1073/pnas.1435900100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narayan G, Goparaju C, Arias-Pulido H, Kaufmann AM, Schneider A, Durst M, Mansukhani M, Pothuri B, Murty VV. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol Cancer 2006; 5:16; PMID:16700909; http://dx.doi.org/ 10.1186/1476-4598-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer 2006; 94:561-8; PMID:16449996; http://dx.doi.org/ 10.1038/sj.bjc.6602972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shaw RJ, Omar MM, Rokadiya S, Kogera FA, Lowe D, Hall GL, Woolgar JA, Homer J, Liloglou T, Field JK, et al. . Cytoglobin is upregulated by tumour hypoxia and silenced by promoter hypermethylation in head and neck cancer. Br J Cancer 2009; 101:139-44; PMID:19568272; http://dx.doi.org/ 10.1038/sj.bjc.6605121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shivapurkar N, Stastny V, Okumura N, Girard L, Xie Y, Prinsen C, Thunnissen FB, Wistuba II, Czerniak B, Frenkel E, et al. . Cytoglobin, the newest member of the globin family, functions as a tumor suppressor gene. Cancer Res 2008; 68:7448-56; PMID:18794132; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wojnarowicz PM, Provencher DM, Mes-Masson AM, Tonin PN. Chromosome 17q25 genes, RHBDF2 and CYGB, in ovarian cancer. Int J Oncol 2012; 40:1865-80; PMID:22344671 [DOI] [PubMed] [Google Scholar]
- 38. Chung MT, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Chu TY, Lai HC, Lin YW. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol 2009; 112:301-6; PMID:19038436; http://dx.doi.org/ 10.1016/j.ygyno.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 39. Overmeer RM, Louwers JA, Meijer CJ, van Kemenade FJ, Hesselink AT, Daalmeijer NF, Wilting SM, Heideman DA, Verheijen RH, Zaal A, et al. . Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int J Cancer 2011; 129:2218-25; PMID:21190187; http://dx.doi.org/ 10.1002/ijc.25890 [DOI] [PubMed] [Google Scholar]
- 40. Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res 2001; 7:1982-6; PMID:11448914 [PubMed] [Google Scholar]
- 41. Kang S, Kim JW, Kang GH, Lee S, Park NH, Song YS, Park SY, Kang SB, Lee HP. Comparison of DNA hypermethylation patterns in different types of uterine cancer: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. Int J Cancer 2006; 118:2168-71; PMID:16331610; http://dx.doi.org/ 10.1002/ijc.21609 [DOI] [PubMed] [Google Scholar]
- 42. Kuzmin I, Liu L, Dammann R, Geil L, Stanbridge EJ, Wilczynski SP, Lerman MI, Pfeifer GP. Inactivation of RAS association domain family 1A gene in cervical carcinomas and the role of human papillomavirus infection. Cancer Res 2003; 63:1888-93; PMID:12702579 [PubMed] [Google Scholar]
- 43. Widschwendter A, Gattringer C, Ivarsson L, Fiegl H, Schneitter A, Ramoni A, Muller HM, Wiedemair A, Jerabek S, Muller-Holzner E, et al. . Analysis of aberrant DNA methylation and human papillomavirus DNA in cervicovaginal specimens to detect invasive cervical cancer and its precursors. Clin Cancer Res 2004; 10:3396-400; PMID:15161694; http://dx.doi.org/ 10.1158/1078-0432.CCR-03-0143 [DOI] [PubMed] [Google Scholar]
- 44. Wisman GB, Nijhuis ER, Hoque MO, Reesink-Peters N, Koning AJ, Volders HH, Buikema HJ, Boezen HM, Hollema H, Schuuring E, et al. . Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer 2006; 119:1908-14; PMID:16736496; http://dx.doi.org/ 10.1002/ijc.22060 [DOI] [PubMed] [Google Scholar]
- 45. Cohen Y, Singer G, Lavie O, Dong SM, Beller U, Sidransky D. The RASSF1A tumor suppressor gene is commonly inactivated in adenocarcinoma of the uterine cervix. Clin Cancer Res 2003; 9:2981-4; PMID:12912945 [PubMed] [Google Scholar]
- 46. Jiang J, Zhao LJ, Zhao C, Zhang G, Zhao Y, Li JR, Li XP, Wei LH. Hypomethylated CpG around the transcription start site enables TERT expression and HPV16 E6 regulates TERT methylation in cervical cancer cells. Gynecol Oncol 2012; 124:534-41; PMID:22108635; http://dx.doi.org/ 10.1016/j.ygyno.2011.11.023 [DOI] [PubMed] [Google Scholar]
- 47. Eldholm V, Haugen A, Zienolddiny S. CTCF mediates the TERT enhancer-promoter interactions in lung cancer cells: identification of a novel enhancer region involved in the regulation of TERT gene. Int J Cancer 2014; 134:2305-13; PMID:24174344; http://dx.doi.org/ 10.1002/ijc.28570 [DOI] [PubMed] [Google Scholar]
- 48. Wong TC, Sokol ES, Schep AN, Punjiya M, Tran DA, Allan D, Drewell RA. Transcriptional repression by the proximal exonic region at the human TERT gene. Gene 2011; 486:65-73; PMID:21787851; http://dx.doi.org/ 10.1016/j.gene.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 49. Veldman T, Horikawa I, Barrett JC, Schlegel R. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol 2001; 75:4467-72; PMID:11287602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu M, Katzenellenbogen RA, Grandori C, Galloway DA. An unbiased in vivo screen reveals multiple transcription factors that control HPV E6-regulated hTERT in keratinocytes. Virology 2013; 446:17-24; PMID:24074563; http://dx.doi.org/ 10.1016/j.virol.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Van Doorslaer K, Burk RD. Association between hTERT activation by HPV E6 proteins and oncogenic risk. Virology 2012; 433:216-9; PMID:22925336; http://dx.doi.org/ 10.1016/j.virol.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steenbergen RD, Walboomers JM, Meijer CJ, van der Raaij-Helmer EM, Parker JN, Chow LT, Broker TR, Snijders PJ. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene 1996; 13:1249-57; PMID:8808699 [PubMed] [Google Scholar]
- 53. Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, Snijders PJ. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst 2004; 96:294-305; PMID:14970278; http://dx.doi.org/ 10.1093/jnci/djh031 [DOI] [PubMed] [Google Scholar]
- 54. van Zeeburg HJ, Snijders PJ, Pals G, Hermsen MA, Rooimans MA, Bagby G, Soulier J, Gluckman E, Wennerberg J, Leemans CR, et al. . Generation and molecular characterization of head and neck squamous cell lines of fanconi anemia patients. Cancer Res 2005; 65:1271-6; PMID:15735012; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-3665 [DOI] [PubMed] [Google Scholar]
- 55. Snellenberg S, De Strooper LM, Hesselink AT, Meijer CJ, Snijders PJ, Heideman DA, Steenbergen RD. Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape. BMC Cancer 2012; 12:551; PMID:23176198; http://dx.doi.org/ 10.1186/1471-2407-12-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101-8; PMID:18546601; http://dx.doi.org/ 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]