Abstract
Background and Purpose: Radiotherapy (RT) is vital for the treatment of locally advanced non-small cell lung cancer (NSCLC), yet its delivery is limited by tolerances of adjacent organs. We sought therefore to identify and characterize gene targets whose inhibition may improve RT. Materials and Methods: Whole genome pooled shRNA cytotoxicity screens were performed in A549 and NCI-H460 using a retroviral library of 74,705 sequences. Cells were propagated with or without daily radiation Monday–Friday. Radiosensitization by top differential dropout hits was assessed by clonogenic assays. Apoptosis was assessed using a caspase 3/7 cell-based activity assay and by annexin V-FITC and PI staining. MCL1 expression was assessed by qPCR and Western blotting. Results: USP9X, a deubiquitinase, was a top hit among druggable gene products. WP1130, a small molecule USP9X inhibitor, showed synergistic cytotoxicity with IR. MCL1, an anti-apoptotic protein deubiquitinated by USP9X, decreased with USP9X inhibition and IR. This was accompanied by increases in caspase 3/7 activity and apoptosis. In a panel of NSCLC lines, MCL1 and USP9X protein and gene expression levels were highly correlated. Lines showing high levels of MCL1 expression were the most sensitive to USP9X inhibition. Conclusions: These data support the use of MCL1 expression as a predictive biomarker for USP9X inhibitors in NSCLC therapy.
Keywords: apoptosis, lung cancer, MCL1, radiosensitizing agents, RNAi screen, USP9X
Abbreviations
- DUB
deubiquitinase
- IR
ionizing radiation
- NSCLC
non-small cell lung cancer
- RT
radiotherapy
- shRNA
short hairpin RNA
- siRNA
small interfering RNA (siRNA)
Introduction
Each year in the United States, approximately 50,000 patients are diagnosed with locally advanced non-small cell lung cancer (NSCLC).1 Due to infiltration of regional structures such as mediastinal lymph nodes, surgical resection alone rarely eliminates the disease. However, unlike distant metastatic disease, locally advanced disease can be eradicated with a combination of radiation therapy (RT), chemotherapy and/or surgery.2 Sensitivity of adjacent organs including the lungs, spinal cord and esophagus however constrains the amount of radiation that can be safely given.3,4 Radiosensitizers are therefore desired to decrease the doses needed to achieve therapeutic effects. Systemic agents such as platinum-based chemotherapy are typically used to radiosensitize tumors, but conventional cytotoxic agents also have dose-limiting toxicities and show modest improvements in patient outcomes.2
Solid malignancies, e.g. NSCLC, typically require high doses of IR for disease control. This is in contrast to many hematologic malignancies. Low-grade follicular lymphomas for example show complete response rates of 50–60% with only 4 Gy in 2 fractions.5 This is in contrast with Stage III NSCLC, with progression-free survival rates of 5–25% at 5 y with doses of 60 Gy or more in 2 Gy fractions.2 This can be understood at least in part because unlike B cells, which undergo TP53-mediated apoptosis in response to low dose IR, NSCLC cells undergo limited apoptosis.6 Agents that increase apoptosis following IR exposure may therefore radiosensitize NSCLC.
To identify the best potential targets for therapy, we performed whole genome pooled shRNA screens in 2 NSCLC cell lines, A549 and NCI-H460. Cells were treated with or without IR 5 d per week, to mimic clinical treatment. In an effort to find radiosensitizers with attenuated effects in unirradiated cells, genes for which shRNA knockdown increased cytotoxicity only in irradiated cells were scored. Gene targets for which inhibitors are available were of particular value, to facilitate development of potential therapeutics.
The top gene candidate was USP9X, a deubiquitinase (DUB). DUBs are key regulators of diverse protein targets including oncogene and tumor suppressor gene products.7 Their principal function is the removal of polyubiquitin chains that target proteins to the proteasome, resulting in prolonged protein expression.8 This has been shown for MCL1, which is an anti-apoptotic protein with a short half-life due to proteasomal degradation. Knockdown of USP9X has been shown to enhance MCL1 turnover9 and radiation-induced decrease of MCL1, sensitizing Jurkat cells to radiation.10 WP1130, a small molecule USP9X inhibitor, promotes accumulation of polyubiquitinated proteins into aggresomes and upregulates apoptosis.11 This inhibitor illustrates an emerging class of therapeutics directed against DUBs for the treatment of cancer.12
Here, we demonstrate 2 findings. First, USP9X inhibition increases radiation-induced apoptosis in NSCLC cells, showing its potential to improve treatment of a solid malignancy. Second, the effects of USP9X inhibition correlate with the expression of MCL1, an anti-apoptotic protein known to be rescued by USP9X. MCL1 expression may therefore serve as a predictive biomarker for USP9X-directed NSCLC therapy.
Results
USP9X is a top hit of a whole genome shRNA screen for NSCLC radiosensitizing gene targets
To identify gene targets whose inhibition may augment the therapeutic index of RT in NSCLC, we performed whole genome pooled shRNA screens with and without IR in 2 cell lines, A549 and NCI-H460. This pair of lines was selected because they harbor similar genetic features including wild-type TP53 and mutant KRAS and STK11 (LKB1).13 It was felt that this similarity would increase the likelihood of finding reproducible hits. KRAS and LKB1 mutant tumors are particularly aggressive and resistant to currently available therapies.14 Also, being TP53 wild-type, the lines were expected to be less vulnerable to genomic instability over the course of the 12 population doublings of the screen.15
The whole genome Hannon-Elledge pooled retroviral shRNA library contains 74,705 distinct shRNA sequences and targets nearly 18,000 known genes.16 After transduction and antibiotic selection, cells were propagated with or without treatment with 1 Gy daily Monday-Friday. The IR schedule was selected to mimic clinical treatment, while a daily dose was selected that showed cytotoxicity yet maintained a sufficient number of cells for repeated culturing. After 2–3 weeks of treatment corresponding to a total of 12 population doublings, cells were harvested and the relative representation of each shRNA sequence before and after treatment was determined using custom Agilent microarrays. Radiosensitizing gene targets were defined as those for which shRNAs exhibited greater-than-threshold cytotoxicity only in the presence of IR. 172 genes met the criteria of having at least one extra shRNA sequence whose abundance decreased reproducibly in both NSCLC lines by at least 2-fold only in the presence of IR (Table S1).
The top 10 candidate genes for initial characterization were additionally selected based on 1) availability of small molecule inhibitors and 2) prior evidence of a prognostic role in NSCLC or other cancers. These ten hits were confirmed in a secondary screen performed in A549 and NCI-H460 using pooled siRNAs for each gene (Fig. 1). Out of these 10 hits, 4 (NUAK1, PIN1, USP9X and WEE1) reproducibly showed statistically significantly increased cytotoxicity in the presence of IR in both cell lines. USP9X, a deubiquitinase, emerged as a top hit showing maximum toxicity of multiple shRNAs in the presence of IR.
Figure 1.
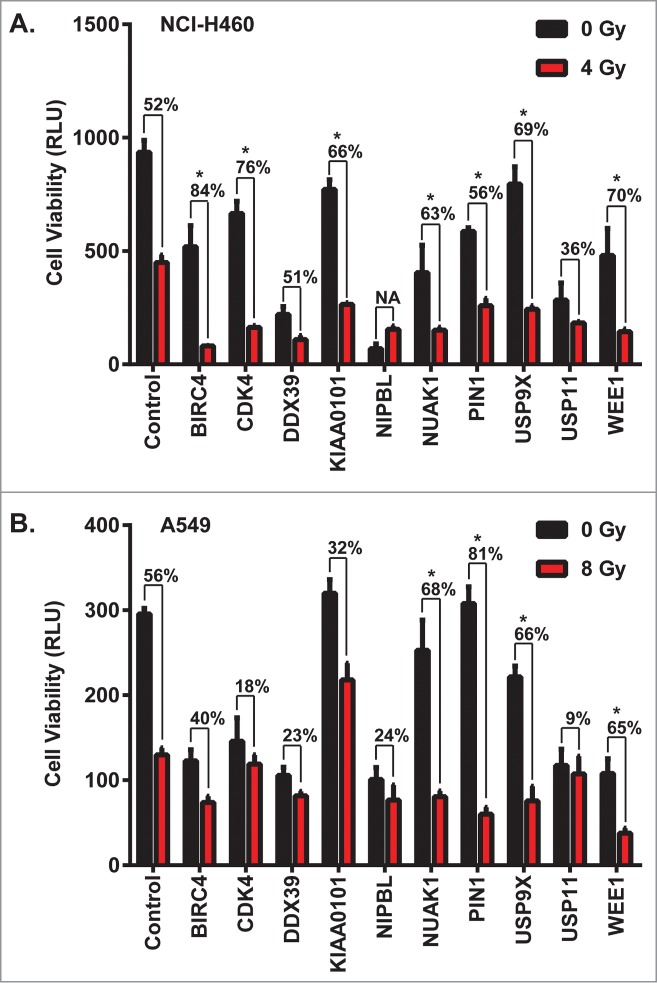
Confirmation of multiple hits from a whole genome shRNA screen ± daily radiation to identify NSCLC radiosensitizers. Ten genes were selected among radiosensitizing hits based on their scores and the availability of small molecule inhibitors targeting their gene products. (A) NCI-H460 and (B) A549 cells were transfected with pools of 4 siRNAs against each gene, and then 24 hours later were treated with a dose of ionizing radiation estimated to yield 50% viability in control cells transfected with a non-silencing siRNA. 24 hours after irradiation, luminescent cell viability assays were performed in triplicate wells. Percentiles above each pair of bars indicate the percentage of cells that were killed following IR and siRNA knockdown compared to siRNA knockdown alone. Percentiles greater than those of non-silencing siRNA indicate synergy, marked by asterisks. Error bars represent standard deviation.
USP9X inhibition radiosensitizes NSCLC cells
To further validate the radiosensitizing properties of USP9X inhibition in NSCLC, we inhibited USP9X expression via independent siRNAs and also used the small molecule deubiquitinase inhibitor WP1130.11 We confirmed robust USP9X knockdown by 3 of 4 tested siRNAs (Fig. 2A). The three confirmed siRNAs were then used to confirm radiosensitization in a cell viability assay combining knockdown ± 4–8 Gy IR (Fig. 2B). Each of the siRNAs led to less than 50% decreases in cell viability when administered alone, but to synergistic decreases in cell viability in combination with IR. Radiosensitization by USP9X knockdown was also observed in clonogenic assays (Fig. 2C). Confirmation with multiple siRNAs strengthened the likelihood that the effect is not off-target.17 A rescue experiment with a non-targetable form of USP9X might further validate the results. However, we opted to assess pharmacologic inhibition of USP9X as a complementary avenue for target validation. WP1130 also yielded synergistic cytotoxicity in combination with IR in clonogenic assays, with dose enhancement factors of around 1.2 or greater (Fig. 2D). Use of both independent siRNAs and a small molecule inhibitor served to confirm NSCLC radiosensitization by USP9X inhibition.
Figure 2.
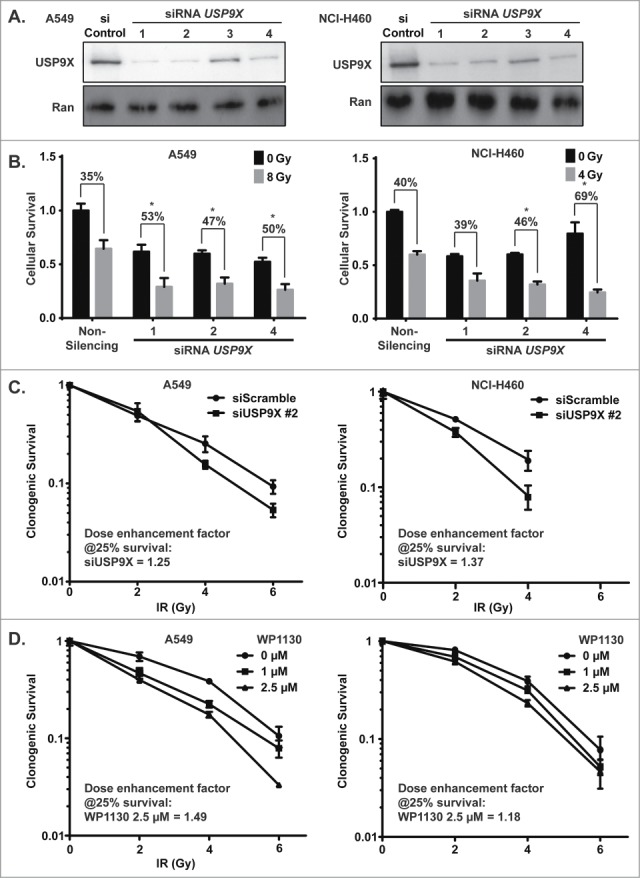
Radiosensitization of NSCLC cells by USP9X inhibition. (A) A549 (left) and NCI-H460 (right) cells were transfected with 4 independent siRNAs against USP9X. Western blotting was performed to assess the extent of knockdown compared to control siRNA transfection. (B) A549 (left) and NCI-H460 (right) cells were transfected with individual siRNAs against USP9X, and then 24 hours later were treated with a dose of ionizing radiation estimated to yield 50% viability in control cells transfected with a non-silencing siRNA. 24 hours after irradiation, luminescent cell viability assays were performed in triplicate wells. Percentiles above each pair of bars indicate the percentage of cells that were killed following IR and siRNA knockdown compared to siRNA knockdown alone. Percentiles greater than those of non-silencing siRNA indicate synergy, marked by asterisks. Error bars represent standard deviation. (C) A549 (left) and NCI-H460 (right) cells were transfected with USP9X siRNA #2, and then 48 hours later received 0–6 Gy IR. Clonogenic assays were performed to assess effects on proliferation. Error bars represent standard deviation. Dose enhancement factors were calculated based on extrapolation of proportional effects on clonogenic survival. (D) A549 (left) and NCI-H460 (right) cells were treated with WP1130, a small molecule USP9X inhibitor, and then 24 hours later received 0–6 Gy IR. Clonogenic assays were performed to assess effects on proliferation. Error bars represent standard deviation. Dose enhancement factors were calculated based on extrapolation of proportional effects on clonogenic survival.
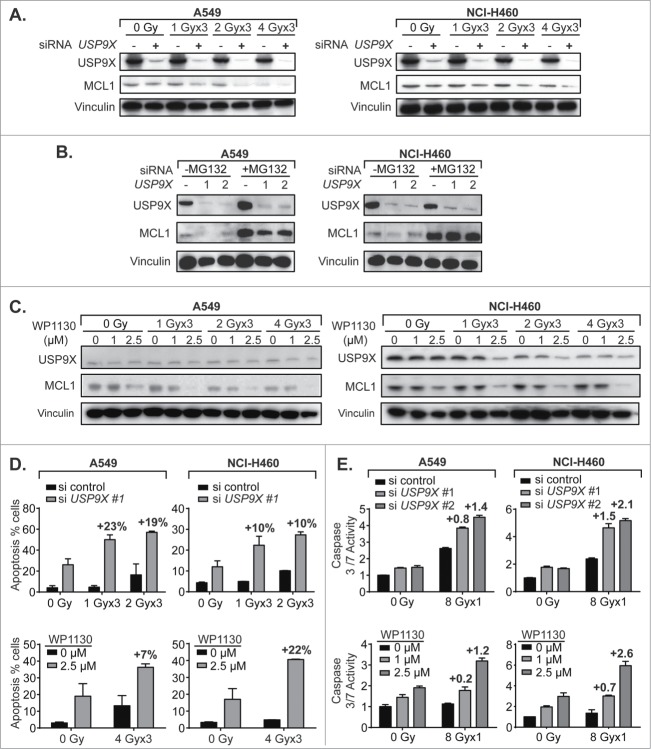
USP9X inhibition decreases MCL1 levels and potentiates apoptosis in NSCLC cells
It has previously been demonstrated that USP9X stabilizes MCL1 by eliminating Lys 48-linked polyubiquitin chains that target the latter for proteasomal degradation.9 MCL1 belongs to the pro-survival BCL2 family of anti-apoptotic genes, and is notable for its rapid turnover.8 We assessed the effect of USP9X siRNA knockdown on MCL1 protein levels in irradiated NSCLC cells, and showed IR dose-dependent decreases in MCL1 expression that were further decreased by USP9X knockdown (Fig. 3A). This decrease was largely reversed by proteasome inhibition with MG132, which strongly increased MCL1 expression with or without USP9X knockdown (Fig. 3B). This was in keeping with the prior observation that MCL1 is degraded by the proteasome, and so whether MCL1 is polyubiquitinated is inconsequential if proteasomal degradation is blocked. The small molecule USP9X inhibitor WP1130 decreased MCL1 expression in a dose-dependent manner, also in combination with IR (Fig. 3C). Although IR alone appeared to have an effect on USP9X and MCL1 expression, the effect was substantially increased with WP1130. Taken together, these data confirmed that USP9X inhibition either by siRNA knockdown or a small molecule inhibitor reduces protein expression of the anti-apoptotic protein MCL1, as seen previously in hematologic malignancies.
Figure 3.
USP9X inhibition increases MCL1 proteasomal degradation and apoptosis in NSCLC cells. (A) Western blot of A549 (left) and NCI-H460 (right) cells showing combined effect of USP9X siRNA knockdown and fractionated IR over 3 consecutive days, on MCL1 levels. (B) Western blot of A549 (left) and NCI-H460 (right) cells showing effect of the proteasome inhibitor MG132 on abundance of MCL1 with or without USP9X siRNA knockdown. (C) Western blot of A549 (left) and NCI-H460 (right) cells showing combined effect of treatment with WP1130, a small molecule USP9X inhibitor, and fractionated IR over 3 consecutive days, on MCL1 levels. (D) Apoptosis in A549 (left) and NCI-H460 (right) cells measured as a percentage of cells staining positive on flow cytometry for annexin V-FITC following fractionated IR over 3 consecutive days with or without USP9X siRNA knockdown (top) or WP1130 treatment (bottom). Percentage points above each pair of bars show the effect due to synergy, by subtracting the effect of siRNA or drug alone as well as the effect of radiation alone. Error bars represent standard deviation. (E) Caspase 3/7 activity in A549 (left) and NCI-H460 (right) cells measured by Caspase-Glo 3/7 luminescent assay (Promega) following single fraction IR with or without USP9X siRNA knockdown (top) or WP1130 treatment (bottom). Values above each pair of bars show the effect due to synergy, by subtracting the effect of siRNA or drug alone as well as the effect of radiation alone. Error bars represent standard deviation.
The next step was to observe whether this correlated with increased apoptosis. Apoptosis was observed in a minority of A549 or NCI-H460 cells treated with IR, despite wild-type TP53. However, USP9X inhibition with either siRNAs or WP1130 increased by at least 2- to 3-fold the percentage of cells undergoing apoptosis measured by annexin V FITC staining (Fig. 3D). For further confirmation, intracellular effector caspase 3 and 7 activity levels were measured and found to be increased 3- to 4-fold in irradiated cells treated with USP9X siRNAs or WP1130 (Fig. 3E). Thus, it appeared that USP9X may radiosensitize NSCLC cells by augmenting the apoptotic cell death pathway.
Sensitivity of NSCLC cells to USP9X inhibition correlates with MCL1 expression
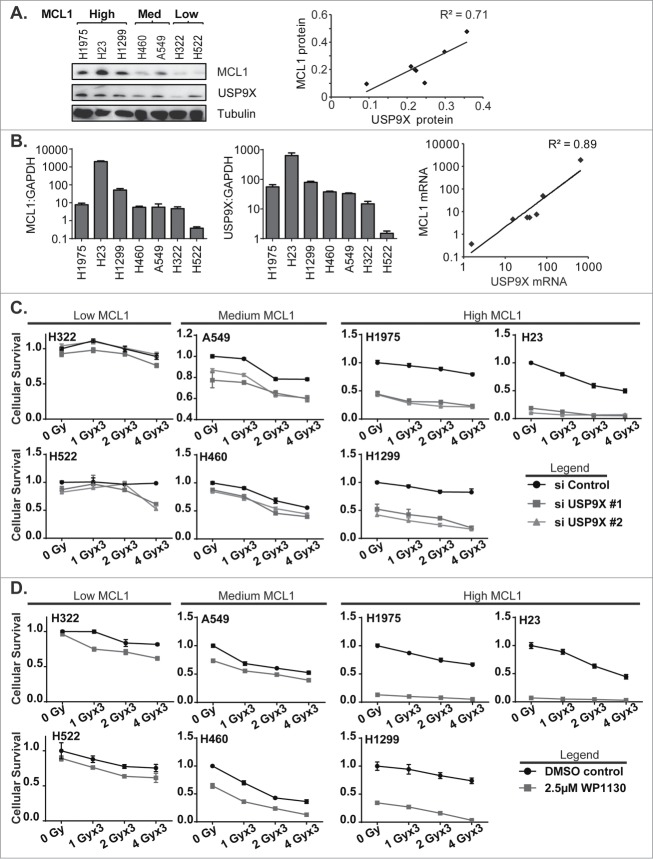
To confirm the effects of USP9X inhibition on MCL1 levels and associated increases in apoptotic cell death, we broadened the panel of NSCLC cell lines tested to include NCI-H23, NCI-H322, NCI-H522, NCI-H1299 and NCI-H1975, in addition to A549 and NCI-H460. We observed a wide variation in MCL1 protein expression among the 7 lines tested, and grouped the cell lines accordingly (Fig. 4A). USP9X and MCL1 protein levels correlated fairly well, with a correlation coefficient of 0.71. Likewise, we observed correlations in USP9X and MCL1 mRNA expression levels among the 7 NSCLC lines (Fig. 4B). On an exponential fit, given variations in expression levels over several orders of magnitude, the correlation coefficient was 0.89.
Figure 4.
(See previous page). Correlation of MCL1 and USP9X levels and sensitivity to USP9X inhibition in NSCLC cells. (A) Western blot showing MCL1 and USP9X levels in 7 NSCLC cell lines grouped in tertiles by MCL1 protein expression levels (left). Correlation of USP9X and MCL1 protein expression levels by densitometry of the Western blot, with linear regression (right). (B) Quantitative PCR for MCL1 and USP9X mRNA expression levels normalized to GAPDH (left). Correlation of USP9X and MCL1 mRNA levels by qPCR, with power regression (left). (C) Luminescent cell viability assays in the 7 NSCLC cell lines grouped in tertiles by MCL protein expression levels, following fractionated radiation over 3 consecutive days with or without USP9X siRNA knockdown. Error bars reflect standard deviation. (D) As C, but with WP1130 treatment in lieu of USP9X siRNA knockdown.
We hypothesized that USP9X inhibition would exert cytotoxic effects in an MCL1-dependent manner. NSCLC cells were therefore grouped by MCL1 protein expression levels, e.g., low, medium and high, and tested prospectively. As predicted, NSCLC cell lines showing low or medium levels of MCL1 showed relatively low to moderate levels of radiosensitization following USP9X siRNA knockdown, compared to lines showing high levels of MCL1 (Fig. 4C). This was also recapitulated using the USP9X inhibitor WP1130, which showed a radiosensitizing effect in the USP9X lines expressing low to medium levels of MCL1, but was nearly sufficient on its own to kill NSCLC cells expressing high MCL1 (Fig. 4D). In the latter context, USP9X inhibitors may show effectiveness as systemic therapies rather than strictly as radiosensitizers.
USP9X inhibition increases apoptosis in NSCLC cells in an MCL1-dependent manner
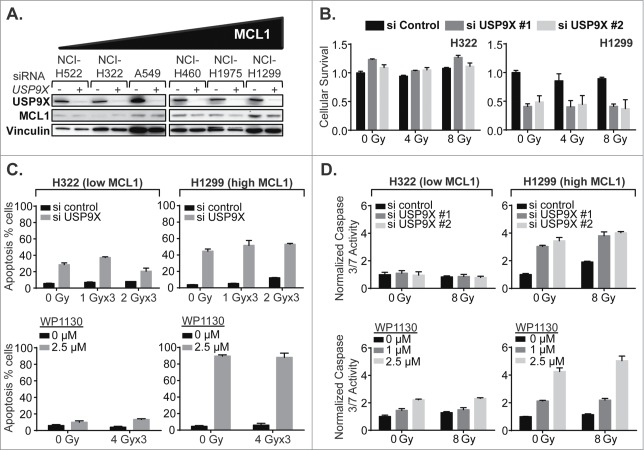
Next, the effect of USP9X inhibition on MCL1 protein expression levels was assessed in the panel of NSCLC lines showing varying levels of MCL1 expression. The greatest absolute effects were observed in lines showing high MCL1, as expected since these lines had the greatest quantities of protein to potentially lose (Fig. 5A). We then selected one NSCLC cell line showing low MCL1 expression, NCI-H322, and one cell line showing high MCL1, NCI-H1299, and performed the following experiments prospectively. We confirmed that USP9X siRNA knockdown radiosensitized only the NSCLC line showing high MCL1 expression, observed as decreases in cell viability with IR and knockdown in NCI-H1299 but not NCI-H322 (Fig. 5B). Some apoptosis as measured by annexin V-FITC staining was observed following USP9X siRNA knockdown in NCI-H322, although this was greater in NCI-H1299, the high MCL1 line (Fig. 4C). WP1130 only increased apoptosis in the high MCL1 line (Fig. 5C). More conclusively, caspase 3 and 7 activity were not significantly increased by USP9X siRNA inhibition in the low MCL1 line but were substantially increased in the high MCL1 line, and WP1130 increased caspase 3 and 7 activity much more substantially in the NSCLC line expressing high MCL1 (Fig. 5D). We therefore largely confirmed our hypothesis that USP9X inhibition increases apoptosis in NSCLC cells more substantially in cells expressing high versus low MCL1.
Figure 5.
USP9X inhibition impacts apoptotic cell death in NSCLC cells expressing high but not low MCL1. (A) Western blot showing effect of USP9X siRNA knockdown on MCL1 expression in cell lines arranged by basal MCL1 expression. (B) Luminescent cell viability assays in a low MCL1 expressing line, NCI-H322 (left), compared to a high MCL1 expressing line, NCI-H1299 (right), treated with IR with or without USP9X siRNA knockdown. Error bars represent standard deviation. (C) Apoptosis in a low MCL1 expressing line, NCI-H322 (left), compared to a high MCL1 expressing line, NCI-H1299 (right), measured as a percentage of cells staining positive on flow cytometry for annexin V-FITC following fractionated IR over 3 consecutive days with or without USP9X siRNA knockdown (top) or WP1130 treatment (bottom). Error bars represent standard deviation. (D) Caspase 3/7 activity in NCI-H322 (left) and NCI-H1299 (right) cells measured by Caspase-Glo 3/7 luminescent assay (Promega) following single fraction IR with or without USP9X siRNA knockdown (top) or WP1130 treatment (bottom). Error bars represent standard deviation.
Discussion
The main aim of the study was to identify and characterize a gene target for NSCLC radiosensitization identified by whole genome shRNA screening. The main advantage of this approach is that it offered agnostic discovery of novel gene targets whose silencing reproducibly impacts viability of NSCLC cells treated with ionizing radiation. The cells were irradiated 5 d per week to approximate fractionated radiotherapy, with the hope this would ultimately yield treatments that would synergize with standard treatment for locally advanced NSCLC.
We selected USP9X as a top druggable hit, and demonstrated that inhibition of this deubiquitinase by either siRNAs or a small molecule inhibitor, WP1130, augments IR-induced cell death in vitro. In keeping with published findings,13 USP9X inhibition was accompanied by decreases in expression of the anti-apoptotic protein MCL1, and this was associated with increases in apoptosis measured by annexin V-FITC staining and caspase 3/7 activity assays. In a panel of NSCLC cell lines, MCL1 and USP9X protein and mRNA expression levels varied concomitantly. We hypothesized that lines showing high MCL1 levels would be more susceptible to USP9X inhibition, and confirmed this by comparing cytotoxicity in cells grouped by MCL1 protein expression. USP9X inhibition therefore emerges as a promising strategy for NSCLC tumors expressing high MCL1 expression.
Novel strategies that increase apoptosis in response to cytotoxic therapies will likely have substantial value. Apoptosis is not ordinarily the principal mode of cell death in NSCLC in response to IR. High doses, e.g. irradiation of A549 or NCI-H460 cells with single 20 Gy fractions, yield rates of apoptosis of only 5–35%.14 Following USP9X inhibition, however, we observed 2- to 3-fold increases in apoptosis. This may yield NSCLC cell death at lower doses than are required for mitotic catastrophe, which requires the introduction of a sufficient number of chromosomal abnormalities to result in failed mitosis and cytokinesis.
There is already a recognized need for MCL1 inhibitors. MCL1 is overexpressed in multiple tumors and promotes chemoresistance in a number of cancers.18,19 BH3 mimetics have been developed to target the BCL2 family of proteins including BCL2 and BCLXL, including ABT737 and GDC-0199. These inhibitors however showed lack of specificity and toxicity in normal adjacent tumors.20
Interestingly, the levels of USP9X and MCL1 expression correlated well at the levels of protein and mRNA expression. Similar correlation was previously observed for protein expression in follicular lymphoma samples.9 Because USP9X deubiquitinates MCL1 and rescues it from rapid proteasomal turnover, it stands to reason that high USP9X protein levels correlate with high MCL1. However, the correlation between mRNA expression levels suggests concordant transcriptional regulation, possibly through shared transcription factors or microRNA regulation. Further investigation may help elucidate whether the 2 proteins participate in a shared pathway to regulate anti-apoptotic processes.
Loss of USP9X appears to have context-dependent effects. In pancreatic ductal adenocarcinoma (PDAC), it enhances transformation, protects cancer cells from anoikis and accelerates tumorigenesis in Kras mutant mice.21 This is in contrast to the antineoplastic effects of USP9X inhibition that have been observed in other contexts. USP9X knockdown reduced anchorage-dependent growth and WP1130 induced significant cytotoxicity in 5 PDAC cell lines, which were felt to represent the malignancy at a more advanced stage.22 In a related vein, USP9X downregulation by pemetrexed appears to reduce MCL1 and increase apoptosis of NSCLC cells treated with this cytotoxic agent.23
USP9X has additional targets beyond MCL1, including SMAD4, a critical component of TGFβ signaling.24 SMAD4 monoubiquitination destabilizes its interaction with SMAD2, resulting in TGFβ pathway inhibition. USP9X inhibition would therefore be anticipated to also result in TGFβ pathway inhibition, which may be pro-proliferative. Also, USP9X deubiquitinates and stabilizes SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1), which is required for cellular motility; depletion of USP9X significantly impairs migration of MDA-MB-231 breast cancer cells.25 USP9X inhibition may therefore have pleiotropic effects, which makes the elucidation of biomarkers that can predict prevailing responses all the more essential.
Of note, several known DSB repair genes, including ATM, DNA-PK (PRKDC), BRCA1 and MRE11A, did not score as hits in our screen based on the scoring criteria. ShRNAs that showed inconsistent results between replicates were not included. Then, only those genes with at least one shRNA sequence that dropped out in the presence of IR but not without radiation scored as hits. There was a BRCA1 shRNA that reproducibly dropped out of the pool of cells treated with radiation, but it also decreased partially without radiation, so was not considered a radiosensitizing gene hit. None of the ATM, DNA-PK or MRE11A shRNAs scored as hits with or without radiation. Likewise, MCL1 appears to be a false negative in the primary screen. Although it was represented in the shRNA pool, its shRNAs did not vary consistently with or without IR. The extent of false negatives is a limitation of the screen. This may be a property of genome-wide RNAi screens in general.26 Like other whole genome pooled shRNA screens, our screen revealed new potential targets, yet additional targets are likely to be picked up by alternate means.
Several key experiments remain. Perhaps most importantly, the efficacy of USP9X inhibitors including WP1130 should be tested in vivo, preferably in genetically-engineered mouse models of NSCLC that recapitulate common driver oncogenes including mutant KRAS and EGFR and loss of tumor suppressor genes including TP53 and STK11 (LKB1). It is reasonably likely that these tumors will vary in MCL1 expression, and if so, the hypothesis will be that the high MCL1 expressing tumors will be the most responsive to USP9X inhibitor treatment. These data would further support the use of MCL1 protein level as a predictive biomarker in subsequent clinical studies. Also, these experiments would help address the potential normal tissue toxicities of USP9X inhibitors, in particular in irradiated lung and other thoracic organs-at-risk. One may hypothesize that neoplastic cells are more dependent on anti-apoptotic factors including MCL1 when faced with cytotoxic therapies. In vivo studies will be crucial to generate preclinical evidence of the therapeutic index of USP9X inhibitors as radiosensitizers.
Materials and Methods
Reagents
WP1130 (S2243) was purchased from Selleck Chemicals. 200 mM stocks in DMSO were stored at −20°C and prior to use were diluted and stored at 4°C for up to one week. Based on IC50 concentrations, all WP1130 treatments were at a final concentration of 1 μM and 2.5 μM for A549 and NCI-H460, unless stated otherwise. The following antibodies were used at the listed dilutions for Western immunoblots: USP9X (A301–350A, Bethyl Lab, 1:1000), MCL1 (4572S, Cell Signaling Technology, 1:1000), tubulin (sc-32293, Santa Cruz Biotechnology, Santa Cruz, CA, 1:1000), vinculin (sc-25336, Santa Cruz Biotechnology, 1:1000), RAN1 (sc-58467, Santa Cruz Biotechnology, 1:1000), anti-mouse (NA931v, GE Healthcare UK United, 1:3000) and anti-rabbit (NA934v, GE Healthcare UK United, 1:3000) horseradish peroxidase-linked secondary antibodies.
Cell lines
Human non-small cell lung cancer (NSCLC) cell lines A549, NCI-H23, NCI-H322, NCI-H460, NCI-H522, NCI-H1299 and NCI-H1975 were gifted by Dr. Matthew Meyerson; they can be purchased from American Type Culture Collection (ATCC). All cell lines were grown at 37°C in humidified 5% CO2 in Gibco RPMI 1640 (Life Technologies) containing 10% fetal bovine serum (FBS, Sigma-Aldrich) and 1 μg/ml Normocin (InvivoGen).
Whole genome pooled shRNA screen
The screen was performed essentially as previously described [ref]. The Hannon-Elledge whole genome pooled shRNA library consists of 6 viral pools each containing approximately 13,000 different MSCV-PM retroviral shRNA particles targeting human genes. For each pool, 3 replicates of at least 1.3 × 107 cells of A549 or NCI-H460 cells were incubated with an equivalent number of retroviral colony-forming units in media containing 8 μg/ml polybrene (Sigma-Aldrich), for a 1000-fold representation of each shRNA sequence at a multiplicity-of-infection (MOI) of 1. After selection for stable integrants using 1 μg/ml puromycin, cells were passaged for a total of 12 population doublings (PD), at all times maintaining a minimum of 1.3 × 107 cells per replicate. Irradiated cells were treated with ionizing radiation (IR) using a Gammacell 40 Exactor Cs-137 irradiator (Best Theratronics) in 1 Gy daily fractions Monday-Friday. Genomic DNA was extracted from cells harvested both before and after the 12 population doublings. Half-hairpin shRNA-containing sequences were amplified by PCR, purified by agarose gel electrophoresis and labeled with Cy5 (PD 0) or Cy3 (PD 12). Competitive hybridization to custom Agilent microarrays was performed.
For scoring, the mean log2 (Cy3/Cy5) ratio was determined for each shRNA sequence using triplicate data. Those shRNAs for which the standard deviation of log2 (Cy3/Cy5) ratios was greater than the absolute mean log2 (Cy3/Cy5) ratio were excluded from further analyses, to reduce false positive hits. For each gene, one point was assigned for each different shRNA sequence targeting that gene for which the mean log2 (Cy3/Cy5) ratio was less than or equal to -1, representing a decrease in representation of the shRNA sequence of at least 2-fold during passaging. One half point was assigned for each shRNA sequence for which the mean log2 (Cy3/Cy5) ratio was less than or equal to −0.5. A negative point was assigned for each shRNA sequence for which the mean log2 (Cy3/Cy5) ratio was greater than or equal to +1, and a negative half point was assigned for each shRNA sequence for which the mean log2 (Cy3/Cy5) ratio was greater than or equal to +0.5, to penalize discordant results among shRNA sequences targeting a particular gene. Finally, genes were ranked based on descending total score, with higher rankings given to genes with equivalent total scores but fewer different shRNA sequences targeting the gene. Those genes that showed the greatest differences in score between unirradiated and irradiated cells were given the highest ranking as radiosensitizers.
Secondary screen for radiosensitizing gene hits and caspase 3/7 assays
96-well white, clear bottom polystyrene tissue culture plates (#3903, Corning) were seeded with NSCLC cells in 100 μl media to achieve 20% confluence. Twenty-four h after seeding, cells were transfected with 2 nM siGenome siRNAs (Dharmacon) or AllStars negative control (Qiagen) using Lipofectamine RNAiMax (Life Technologies). Twenty-four h after transfection, cells were irradiated with single doses of 4 or 8 Gy. Twenty-four h after irradiation, CellTiter-Glo cell viability assays (Promega) were performed. Alternately, 6 hours after irradiation, Caspase 3/7 assays (Promega) were performed. Results were normalized relative to those of the negative control.
USP9X inhibition, irradiation and cell survival assays
Twenty-four h prior to transfection or drug treatment, 1 × 105 NSCLC cells were plated in 6-well tissue culture plates (Corning). For RNAi knockdown, cells were transfected with 20 nM pooled or individual USP9X siRNAs (Dharmacon) or AllStars negative control (Qiagen) using RNAiMax (Life Technologies) according to manufacturer's instructions. For drug inhibition, cells were treated with 1 μM or 2.5 μM of WP1130 or DMSO control, which was maintained on subsequent days. Starting 24 h later, cells were treated with 0–6 Gy. For clonogenic assays, 6 hours after IR, cells were trypsinized, counted, and seeded into 10 cm dishes in triplicate to generate isolated colonies. Once colonies became visible by eye, plates were stained with 1% crystal violet (Sigma-Aldrich) and colonies of at least 50 cells were counted by eye.
Annexin FITC staining
The Annexin FITC V staining kit II (BD PharMingen, San Jose, CA) was used per manufacturer's instructions. Adherent and NSCLC cells treated with WP1130 or USP9X siRNA were collected together. Cells were washed with cold PBS and resuspended 1 × 106/mL in cold binding buffer containing propidium iodide and annexin FITC. Flow cytometry was performed on at least 104 cells on a BD LSR Fortessa flow cytometer (BD Biosciences). FITC and PtdIns fluorescences were passed through 520 nm and 630 nm bandpass filters, respectively. All results were normalized to cells treated with DMSO vehicle control for WP1130 treatment or negative control siRNA for RNAi knockdown.
Western immunoblots
To generate whole cell lysates, 1 ml/107 cells of ice-cold RIPA buffer containing 50 mM Tris, pH 8.0 (Bio-Rad), 150 mM NaCl (Sigma-Aldrich), 1.0% IGEPAL CA-630 (Sigma-Aldrich), 0.5% sodium deoxycholate (Sigma-Aldrich), 0.01% SDS (Bio-Rad), and Complete proteasome inhibitor (11 836 153 001, Roche Diagnostics), was used. Semi-dry transfer was performed using the Trans-Blot SD apparatus (Bio-Rad) onto PVDF membrane (Bio-Rad) per manufacturer's instructions. Western Lightning Plus enhanced chemiluminesence substrate (Perkin-Elmer) was used for visualization on Amersham Hyperfilm (GE Healthcare).
Quantitative RT-PCR
For gene expression analyses, total RNA was extracted from cell lines using the RNeasy kit (Qiagen) following manufacturer's instructions. First strand cDNA synthesis was performed with 0.5 ug total RNA using a mix of random primers and oligo dT primers using the iScript cDNA synthesis kit (Bio-Rad). cDNA was purified using a PCR purification kit (Qiagen) following manufacturer's instructions. Quantitative RT-PCR was performed using a Chromo 4 DNA Engine Thermal Cycler (Bio-Rad), with MCL1 and USP9X intron-spanning primers (Eurofins MWG Operon) and SYBR Green (Bio-Rad) master mix; MCL1: CATTCCTGATGC CACCTTCT (forward) and TCGTAAGGACAAAACGGGAC (reverse), USP9X: TTCTCCAAGTTT-GGCATGGT (forward) and CACCCC TTAGAGATGGAGCA (reverse), GAPDH: TGCACCACCAACTGCTTAGC (forward) and GGCATGGACTGTGGTCATGAG (reverse). Data was normalized using GAPDH, with values reported as relative fold increases over control.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
DK is supported by the American Society for Radiation Oncology (ASTRO) Junior Faculty Career Research Training Award, a Joint Center for Radiation Therapy Foundation Grant, a Dana-Farber/Harvard Cancer Center SPORE Developmental Research Project Award in Lung Cancer Research and the National Cancer Institute of the National Institutes of Health under Award Number K08CA172354.
References
- 1. Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. . SEER Cancer Statistics Review, 1975-2011, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2011/, based on November 2013 SEER data submission, posted to the SEER web site, April 2014. [Google Scholar]
- 2. Scarpaci A, Mitra P, Jarrar D, Masters GA. Multimodality approach to management of stage III non-small cell lung cancer. Surg Oncol Clin N Am 2013; 22:319-28; PMID:23453337; http://dx.doi.org/ 10.1016/j.soc.2012.12.014 [DOI] [PubMed] [Google Scholar]
- 3. Marks LB, Yorke ED, Jackson A, Ten Haken RK, Constine LS, Eisbruch A, Bentzen SM, Nam J, Deasy JO. Use of normal tissue complication probability models in the clinic. Int J Radiat Oncol Biol Phys 2010; 76:S10-9; PMID:20171502; http://dx.doi.org/ 10.1016/j.ijrobp.2009.07.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Werner-Wasik M, Yu X, Marks LB, Schultheiss TE. Normal-tissue toxicities of thoracic radiation therapy: esophagus, lung, and spinal cord as organs at risk. Hematol Oncol Clin North Am 2004; 18:131-60, x-xi; PMID:15005286; http://dx.doi.org/ 10.1016/S0889-8588(03)00150-3 [DOI] [PubMed] [Google Scholar]
- 5. Ganem G, Cartron G, Girinsky T, Haas RL, Cosset JM, Solal-Celigny P. Localized low-dose radiotherapy for follicular lymphoma: history, clinical results, mechanisms of action, and future outlooks. Int J Radiat Oncol Biol Phys 2010; 78:975-82; PMID:20970029; http://dx.doi.org/ 10.1016/j.ijrobp.2010.06.056 [DOI] [PubMed] [Google Scholar]
- 6. Stuschke M, Sak A, Wurm R, Sinn B, Wolf G, Stuben G, Budach V. Radiation-induced apoptosis in human non-small-cell lung cancer cell lines is secondary to cell-cycle progression beyond the G2-phase checkpoint. Int J Radiat Biol 2002; 78:807-19; PMID:12428922; http://dx.doi.org/ 10.1080/09553000210148903 [DOI] [PubMed] [Google Scholar]
- 7. Lim KH, Baek KH. Deubiquitinating enzymes as therapeutic targets in cancer. Curr Pharm Des 2013; 19:4039-52; PMID:23181570; http://dx.doi.org/ 10.2174/1381612811319220013 [DOI] [PubMed] [Google Scholar]
- 8. Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta 2014; 1843:114-28; PMID:23845989; http://dx.doi.org/ 10.1016/j.bbamcr.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, et al. . Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 2010; 463:103-7; PMID:20023629; http://dx.doi.org/ 10.1038/nature08646 [DOI] [PubMed] [Google Scholar]
- 10. Trivigno D, Essmann F, Huber SM, Rudner J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia 2012; 14:893-904; PMID:23097624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res 2010; 70:9265-76; PMID:21045142; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-1530 [DOI] [PubMed] [Google Scholar]
- 12. Pal A, Young MA, Donato NJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res 2014; 74:4955-66; PMID:25172841; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-1211 [DOI] [PubMed] [Google Scholar]
- 13. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. . The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483:603-7; PMID:22460905; http://dx.doi.org/ 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, et al. . A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012; 483:613-7; PMID:22425996; http://dx.doi.org/ 10.1038/nature10937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem 2012; 113:433-9; PMID:22006292; http://dx.doi.org/ 10.1002/jcb.23400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Zhu S, Kozono D, Ng K, Futalan D, Shen Y, Akers JC, Steed T, Kushwaha D, Schlabach M, et al. . Genome-wide shRNA screen revealed integrated mitogenic signaling between dopamine receptor D2 (DRD2) and epidermal growth factor receptor (EGFR) in glioblastoma. Oncotarget 2014; 5:882-93; PMID:24658464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sigoillot FD, King RW. Vigilance and validation: Keys to success in RNAi screening. ACS Chem Biol 2011; 6:47-60; PMID:21142076; http://dx.doi.org/ 10.1021/cb100358f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S, Wang HG, Zhang X, Bullrich F, Croce CM, et al. . Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with In vitro and In vivo chemoresponses. Blood 1998; 91:3379-89; PMID:9558396 [PubMed] [Google Scholar]
- 19. Lestini BJ, Goldsmith KC, Fluchel MN, Liu X, Chen NL, Goyal B, Pawel BR, Hogarty MD. Mcl1 downregulation sensitizes neuroblastoma to cytotoxic chemotherapy and small molecule Bcl2-family antagonists. Cancer Biol Ther 2009; 8:1587-95; PMID:19556859; http://dx.doi.org/ 10.4161/cbt.8.16.8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Billard C. BH3 mimetics: status of the field and new developments. Mol Cancer Ther 2013; 12:1691-700; PMID:23974697; http://dx.doi.org/ 10.1158/1535-7163.MCT-13-0058 [DOI] [PubMed] [Google Scholar]
- 21. Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grützmann R, Aust D, Rümmele P, et al. . The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012; 486:266-70; PMID:22699621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cox JL, Wilder PJ, Wuebben EL, Ouellette MM, Hollingsworth MA, Rizzino A. Context-dependent function of the deubiquitinating enzyme USP9X in pancreatic ductal adenocarcinoma. Cancer Biol Ther 2014; 15:1042-52; PMID:24841553; http://dx.doi.org/ 10.4161/cbt.29182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan J, Zhong N, Liu G, Chen K, Liu X, Su L, Singhal S. Usp9x- and Noxa-mediated Mcl-1 downregulation contributes to pemetrexed-induced apoptosis in human non-small-cell lung cancer cells. Cell Death Dis 2014; 5:e1316; PMID:24991768; http://dx.doi.org/ 10.1038/cddis.2014.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. . FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009; 136:123-35; PMID:19135894; http://dx.doi.org/ 10.1016/j.cell.2008.10.051 [DOI] [PubMed] [Google Scholar]
- 25. Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, Wilson C, Nathans R, Zhang J, Kirschner MW, et al. . Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem 2013; 288:2976-85; PMID:23184937; http://dx.doi.org/ 10.1074/jbc.M112.430066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hao L, He Q, Wang Z, Craven M, Newton MA, Ahlquist P. Limited agreement of independent RNAi screens for virus-required host genes owes more to false-negative than false-positive factors. PLoS Comp Biol 2013; 9:e1003235; PMID:24068911; http://dx.doi.org/ 10.1371/journal.pcbi.1003235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.