Abstract
Plant cellulose biosynthesis is a complex process involving cellulose-synthase complexes (CSCs) and various auxiliary factors essential for proper orientation and crystallinity of cellulose microfibrils in the apoplast. Among them is KORRIGAN1 (KOR1), a type-II membrane protein with multiple N-glycans within its C-terminal cellulase domain. N-glycosylation of the cellulase domain was important for KOR1 targeting to and retention within the trans-Golgi network (TGN), and prevented accumulation of KOR1 at tonoplasts. The degree of successful TGN localization of KOR1 agreed well with in vivo-complementation efficacy of the rsw2–1 mutant, suggesting non-catalytic functions in the TGN. A dynamic interaction network involving microtubules, CSCs, KOR1, and currently unidentified glycoprotein component(s) likely determines stress-triggered re-organization of cellulose biosynthesis and resumption of cell-wall growth under stress.
Keywords: Arabidopsis; complex N-glycans; cell wall; trans-Golgi network; β1,; 4-endoglucanase
Abbreviations
- CSC
cellulose-synthase complex
- TGN
trans-Golgi network
- GHF9
glycosyl-hydrolase family 9
- ER
endoplasmic reticulum
- PM
plasma membrane
Arabidopsis thaliana KORRIGAN1/RADIALLY SWOLLEN 2 (AtKOR1/RSW2) is a type-II membrane protein consisting of an N-terminal cytoplasmic domain, a transmembrane domain, and a C-terminal luminal/extracellular cellulase domain. KOR1 is central for maintaining cellulose biosynthesis in both primary and secondary cell walls,1,2 because kor1 loss-of-function mutations resulted in various corresponding cell-wall defects and decreased cellulose contents. The KOR1 C-terminal domain resembles the catalytic domain of β1,4-endoglucanases in glycosyl-hydrolase family 9 (GHF9) and is thought to cleave cellulose polymers in vivo. Catalytic function of KOR1 is important, since the jia1 allele of KOR1 with amino-acid substitution (A577V) in the GHF9 active site-signature motif cannot fully support plant growth.3
GHF9 endoglucanases are widespread in both Prokaryota and Eukaryota including all plant cellulases. The KOR1 sequence contains 8 N-glycosylation motifs (N1-N8), of which 5 are highly conserved among plant GHF9 proteins,4 suggesting that N-glycosylation of KOR1 may be essential for function. Our genetic interaction analyses detected strong synergistic effects between KOR1 partial loss-of-function allele rsw2–1 and other mutations defective in either N-glycan attachment (stt3a-2) in the endoplasmic reticulum (ER) or complex N-glycan maturation in the Golgi apparatus (cgl1–3).5 In contrast, prokaryotic GHF9 proteins entirely lack N-glycosylation.6,7 Early studies used soluble recombinant plant orthologs of KOR1 (BnCel16 and PttCel9A) expressed in Pichia pastoris to test the role of N-glycans for protein function.7,8 Enzymatic deglycosylation caused an activity loss of over 90% in BnCel16 and up to 80% in PttCel9A, leading to the conclusion that N-glycans are essential for catalytic function. Another study with recombinant KOR1 expressed in insect cells showed that removal of single N-glycans affects enzymatic activity in a position-specific manner. Decrease of KOR1 activity by single mutations was approximately 20–30%.9
While above studies indicated that N-glycans influence the enzymatic function of KOR1, interpretation of specific activity data of purified KOR1 variants, as well as of enzymatically deglycosylated KOR1, disregard potential differences in N-glycosylation and folding efficacy within host cells that can influence the stability of individual KOR1 variants. In addition, biological significance of the enzymatic activity losses observed in vitro remained unclear. To determine the in-vivo function of KOR1 N-glycans, a systematic mutagenesis of all 8 N-glycosylation sites was conducted in our recent study.4 Functional complementation using GFP-KOR1 variants, whose expression was driven by native KOR1 promoter and terminator sequences, were analyzed in KOR1-deficient mutant host rsw2–1, and the N-glycosylation state of KOR1 and the root-growth phenotype of the transformants were scored. From these analyses several conclusions could be drawn. First, all KOR1 N-glycosylation sites are used in Arabidopsis thaliana, consistent with the data obtained for KOR1 in a heterologous plant system (Nicotiana benthamiana).9 Second, no single N-glycosylation site of KOR1 is essential. Indeed, KOR1 variants lacking up to 6 N-glycosylation sites supported root growth similar to wild-type KOR1. Third, in-vivo efficacies of singly N-glycosylated KOR1 variants vary depending on position of the attached N-glycan. Variants containing only the 1st (N108), 2nd (N133), or 8th (N567) N-glycan did not support root growth effectively. These N-glycosylation sites lie close to the N- or C-terminus of the catalytic domain and show less evolutional conservation than the other sites. Among highly conserved (close to 100%) N-glycosylation sites, the 4th (N324) and 7th (N425) position conferred highest rsw2–1 complementation efficacy. These two sites are flanking the active site cleft of KOR1 and may support proper direction of cellulose substrates into the catalytic site. Fourth, even non-glycosylated KOR1 can partially complement rsw2–1, suggesting that KOR1 without N-glycans retains some activity, or KOR1 may have non-catalytic functions as suggested previously.3,4
In wild-type plants, intact GFP-KOR1 localized predominately in the trans-Golgi network (TGN), the plasma membrane (PM), and at low levels also in tonoplasts.4 Cycling of KOR1 between the PM and heterogeneous intracellular compartments has been reported.10 During cell division, KOR1 becomes enriched at the cell plate, which is promoted by a polarized targeting signal (LL and YXXΦ) in the cytoplasmic domain.11 Among these locations, growth-supporting capacity of the KOR1 N-glycosylation variants correlated well with their TGN/PM profile. Loss of up to 6 N-glycans rarely affected localization of GFP-KOR1, despite decrease of GFP labels at the PM. Functionally compromised GFP-KOR1 variants with single or no N-glycans, as well as the G429R mutation (fully N-glycosylated RSW2–1), on the other hand, increased GFP labeling at tonoplasts. This suggests that retention in the TGN is integral to KOR1's in-vivo function. Liebminger et al.9 reported that the RSW2–1 protein was retained in the ER and likely degraded during ER-quality control, based on low KOR1 abundance in rsw2–1 and biochemical analysis of the RSW2–1 protein in N. benthamiana. However, in our study, using Arabidopsis as host, GFP-RSW2–1 was able to exit the ER but accumulated at the tonoplast. Mutations in STT3a (encoding an oligosaccharyltransferase subunit) markedly decreased N-glycosylation of KOR1 in the ER.4 This was particularly evident for less conserved sites, i.e., N-glycosylation at N1 or N2 were undetectable, and N8 showed approximately 50% occupation. In the salt-sensitve stt3a-2 mutant, GFP-KOR1 predominantly accumulated at the tonoplast with some retention in the TGN, which was much more severe than expected from the moderate underglycosylation of KOR1 at 3 to 4 sites, suggesting that additional (glyco)proteins might function in retaining KOR1 within the TGN.4
The precise role of KOR1 in cellulose biosynthesis remains unclear. The proposed function of KOR1 orthologs includes cleavage of sitosterol glycoside primers at the PM,12 relieving tension of cellulose microfibrils in the apoplast,13 and modulation of their crystallinity.14 Recent data, on the other hand, predict that KOR1 also fulfills functions independent of its catalytic properties, possibly in aligning cellulose microfibrils without cleaving, similar to the recently proposed function of the Arabidopsis COBRA protein at the PM.15 Although earlier studies failed to detect interactions between KOR1 and the cellulose-synthase complex (CSC),1 two new studies show that KOR1 interacts with CesA proteins. Lei et al.3 found that the central cytoplasmic CesA domains can interact with KOR1 in vitro. Vain et al.16 used BiFC and yeast 2-hybrid assays to demonstrate that the transmembrane region of KOR1 (amino acids 70–94), rather than the cytoplasmic domain, is required for KOR1-CSC (CesA1/CesA3/CesA6) interaction as well as in-vivo dimerization of KOR1. Our observations, however, do not support self-dimerization of KOR1 in vivo, because GFP-KOR1 localization was similar in wild-type and in rsw2–1, plus GFP-RSW2–1 localization was not influenced by endogenous KOR1 in wild-type host cells.4 Therefore, the mode of KOR1-CSC interaction has yet to be fully understood.
In unchallenged wild-type Arabidopsis seedlings, KOR1 accumulates in the SYP61-positive TGN4,17 and at the PM. At the PM, KOR1 partially colocalizes with CSCs in small punctate structures that travel on linear tracks; and upon inhibition of cellulose biosynthesis by isoxaben, KOR1 and CSCs colocalize in donut-shaped intracellular compartments proposed to represent the Golgi apparatus.3 However, in newly transfected Arabidopsis rsw2–1 protoplasts, we found that GFP-KOR1 accumulates in extensive, SYP61-negative structures on the cell surface after prolonged culture (Fig. 1). Such non-mobile GFP labels at the PM were also observed in tobacco protoplasts (data not shown). Therefore, after exiting from the TGN, KOR1 seems to be faithfully delivered to the PM but remains confined to regularly spaced domains. These findings are consistent with lack of a stable polar axis and isodiametric organization of the cytoskeleton in non-embedded protoplasts. By contrast, CesA-trafficking inhibitor CESTRIN promoted extensive colocalization of both KOR1 and CSCs with TGN marker SYP61 in walled cells of intact tissue.18 Such induced colocalization was observed also by either isoxaben or osmotic stress treatment.19
Figure 1.
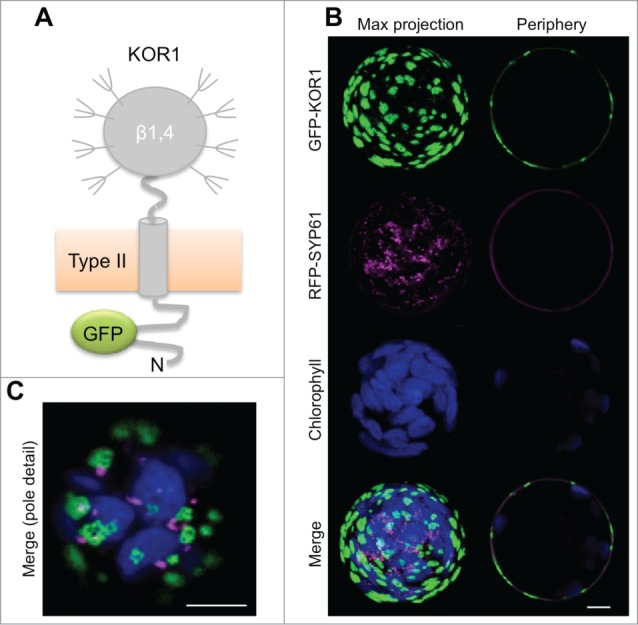
GFP-KOR1 patterns at the surface of KOR1-deficient Arabidopsis rsw2–1 protoplasts upon co-expression of TGN marker RFP-SYP61. (A) Membrane topology of the GFP-KOR1 reporter protein. (B) Maximum (Max) projection of about 30 sections (left) and a central section (right) are shown for single and merged channels. (C) Pole detail of the same cell (Max projection of merged signals). Fluorescent signals were recorded ca. 48 h post-transfection. Green, GFP-KOR1; magenta, TGN marker RFP-SYP61; blue, chlorophyll. EG, endoglucanase domain; N, N-terminus. Bars represent 5 5 μm.
NaCl/osmotic stress induces transient depolymerization and repolymerization of microtubules, which is essential for stress acclimation20,21 and triggered by tubulin kinase PHS1.22 Because CSCs associate with cortical microtubules via the CSI1/POM2 protein,23,24 and microtubules are required for both, guiding CSCs at the PM as well as their secretion and internalization,19,25 stress-induced microtubule depolymerization likely results in transient disruption of CSC and KOR1 organization. Perhaps, re-establishing microtubule-association of CSCs and KOR1 are essential steps for recovery from the salt shock and resuming growth under persisting salt stress. Inability of Arabidopsis N-glycosylation and modification pathway mutants (stt3a, cgl1, hgl1, fucTa fucTb xylT) to fully re-establish anisotropic growth at the root tip is likely linked to their defects in re-building a functional KOR1/CSC network in the TGN under persisting stress conditions. Our observation that the cgl1 mutation affects KOR1 function in trans4 supports this hypothesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was in part financially supported by Deutsche Forschungsgemeinschaft (DFG) grants SCHA 541/9 and 11 (to A.v.S.) and by USDA-CSREES “Designing food for health” grants 2008-34402-19195, 2009-34402-19831, and 2010-34402-20875 (to H.K.).
References
- 1.Szyjanowicz PM, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J 2004; 37:730-40; PMID:14871312; http://dx.doi.org/ 10.1111/j.1365-313X.2003.02000.x [DOI] [PubMed] [Google Scholar]
- 2.Lane DR, Wiedemeier A, Peng L, Hofte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al.. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 2001; 126:278-88; PMID:11351091; http://dx.doi.org/ 10.1104/pp.126.1.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei L, Zhang T, Strasser R, Lee CM, Gonneau M, Mach L, Vernhettes S, Kim SH, J Cosgrove D, Li S, et al.. The jiaoyao1 Mutant Is an Allele of korrigan1 That Abolishes Endoglucanase Activity and Affects the Organization of Both Cellulose Microfibrils and Microtubules in Arabidopsis. Plant Cell 2014; 26:2601-16; http://dx.doi.org/ 10.1105/tpc.114.126193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rips S, Bentley N, Jeong IS, Welch JL, von Schaewen A, Koiwa H. Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 2014; 26:3792-808; http://dx.doi.org/ 10.1105/tpc.114.129718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al.. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 2008; 105:5933-8; PMID:18408158; http://dx.doi.org/ 10.1073/pnas.0800237105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung ED, Lao G, Irwin D, Barr BK, Benjamin A, Wilson DB. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl Environ Microbiol 1993; 59:3032-43; PMID:8215374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Master ER, Rudsander UJ, Zhou W, Henriksson H, Divne C, Denman S, Wilson DB, Teeri TT. Recombinant expression and enzymatic characterization of PttCel9A, a KOR homologue from Populus tremula x tremuloides. Biochemistry 2004; 43:10080-9; PMID:15287736; http://dx.doi.org/ 10.1021/bi049453x [DOI] [PubMed] [Google Scholar]
- 8.Molhoj M, Ulvskov P, Dal Degan F. Characterization of a functional soluble form of a Brassica napus membrane-anchored endo-1,4-β-glucanase heterologously expressed in Pichia pastoris. Plant Physiol 2001; 127:674-84; PMID:11598241; http://dx.doi.org/ 10.1104/pp.010269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebminger E, Grass J, Altmann F, Mach L, Strasser R. Characterizing the link between glycosylation state and enzymatic activity of the endo-beta1,4-glucanase KORRIGAN1 from Arabidopsis thaliana. J Biol Chem 2013; 288:22270-80; PMID:23782689; http://dx.doi.org/ 10.1074/jbc.M113.475558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert S, Bichet A, Grandjean O, Kierzkowski D, Satiat-Jeunemaitre B, Pelletier S, Hauser MT, Höfte H, Vernhettes S. An Arabidopsis endo-1,4-β-D-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 2005; 17:3378-89; PMID:16284310; http://dx.doi.org/ 10.1105/tpc.105.036228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH. KORRIGAN , an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. The Plant cell 2000; 12:1137-52; PMID:10899980; http://dx.doi.org/ 10.1105/tpc.12.7.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng L, Kawagoe Y, Hogan P, Delmer D. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 2002; 295:147-50; PMID:11778054; http://dx.doi.org/ 10.1126/science.1064281 [DOI] [PubMed] [Google Scholar]
- 13.Ueda T. Cellulase in Cellulose Synthase: A Cat among the Pigeons? Plant Physiol 2014; 165:1397-8; PMID:25085884; http://dx.doi.org/ 10.1104/pp.114.245753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi J, Rudsander UJ, Hedenstrom M, Banasiak A, Harholt J, Amelot N, Immerzeel P, Ryden P, Endo S, Ibatullin FM, et al.. KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol 2009; 50:1099-115; PMID:19398462; http://dx.doi.org/ 10.1093/pcp/pcp062 [DOI] [PubMed] [Google Scholar]
- 15.Sorek N, Sorek H, Kijac A, Szemenyei HJ, Bauer S, Hematy K, Wemmer DE, Somerville CR. The Arabidopsis COBRA protein facilitates cellulose crystallization at the plasma membrane. J Biol Chem 2014; 289:34911-20; PMID:25331944; http://dx.doi.org/ 10.1074/jbc.M114.607192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Vain T, Crowell EF, Timpano H, Biot E, Desprez T, Mansoori N, Trindade LM, Pagant S, Robert S, Höfte H, et al.. The Cellulase KORRIGAN Is Part of the Cellulose Synthase Complex. Plant Physiol 2014; 165:1521-32; PMID:24948829; http://dx.doi.org/ 10.1104/pp.114.241216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al.. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 2012; 22:413-24; PMID:21826108; http://dx.doi.org/ 10.1038/cr.2011.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worden N, Wilkop TE, Esteve VE, Jeannotte R, Lathe R, Vernhettes S, Weimer B, Hicks G, Alonso J, Labavitch J, et al.. CESA TRAFFICKING INHIBITOR Inhibits Cellulose Deposition and Interferes with the Trafficking of Cellulose Synthase Complexes and Their Associated Proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol 2015; 167:381-93; PMID:25535279; http://dx.doi.org/ 10.1104/pp.114.249003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 2009; 11:797-806; PMID:19525940; http://dx.doi.org/ 10.1038/ncb1886 [DOI] [PubMed] [Google Scholar]
- 20.Shoji T, Suzuki K, Abe T, Kaneko Y, Shi H, Zhu JK, Rus A, Hasegawa PM, Hashimoto T. Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol 2006; 47:1158-68; PMID:16861712; http://dx.doi.org/ 10.1093/pcp/pcj090 [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Li J, Yuan M. Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol 2007; 48:1534-47; PMID:17906320; http://dx.doi.org/ 10.1093/pcp/pcm123 [DOI] [PubMed] [Google Scholar]
- 22.Fujita S, Pytela J, Hotta T, Kato T, Hamada T, Akamatsu R, Ishida Y, Kutsuna N, Hasezawa S, Nomura Y, et al.. An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr Biol 2013; 23:1969-78; PMID:24120637; http://dx.doi.org/ 10.1016/j.cub.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 23.Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S. POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 2012; 24:163-77; PMID:22294619; http://dx.doi.org/ 10.1105/tpc.111.093575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Lei L, Somerville CR, Gu Y. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci U S A 2012; 109:185-90; PMID:22190487; http://dx.doi.org/ 10.1073/pnas.1118560109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 2009; 21:1141-54; PMID:19376932; http://dx.doi.org/ 10.1105/tpc.108.065334 [DOI] [PMC free article] [PubMed] [Google Scholar]


