Abstract
Arabinogalactan proteins are abundant cell surface proteoglycans in plants and are implicated to act as developmental markers during plant growth. We previously reported that AtGALT31A, AtGALT29A, and AtGLCAT14A-C, which are involved in the biosynthesis of arabinogalactan proteins, localize not only to the Golgi cisternae but also to smaller compartments, which may be a part of the unconventional protein secretory pathway in plants. In Poulsen et al.,1 we have demonstrated increased targeting of AtGALT29A to small compartments when Y144 is substituted with another amino acid, and we implicated a role for Y144 in the subcellular targeting of AtGALT29A. In this paper, we are presenting another aspect of Y144 substitution in AtGALT29A; namely, Y144A construct demonstrated a 2.5-fold increase while Y144E construct demonstrated a 2-fold decrease in the galactosyltransferase activity of AtGALT29A. Therefore, the electrostatic status of Y144, which is regulated by an unknown kinase/phosphatase system, may regulate AtGALT29A enzyme activity. Moreover, we have identified additional proteins, apyrase 3 (APY3; At1g14240) and UDP-glucuronate epimerases 1 and 6 (GAE1, At4g30440; GAE6, At3g23820), from Arabidopsis thaliana that co-localize with AtGALT31A in the small compartments when expressed transiently in Nicotiana benthamiana. These proteins may play roles in nucleotide sugar metabolism in the small compartments together with arabinogalactan glycosyltransferases.
Keywords: arabinogalactan proteins, exocyst-positive organelle, glycosyltransferases, protein O-glycosylation, plant proteoglycan, plant cell wall, unconventional protein secretion
Cell surface arabinogalactan proteins (AGPs) change structure spatially and developmentally in a tightly regulated manner and are thus considered developmental markers.2 However, little is known about the molecular mechanisms that control the synthesis and degradation of AGPs. AGPs are synthesized as post-translational modifications of proteins, which begin in the ER and continue in the Golgi apparatus.3 We previously reported that at least some of the AGP glycosyltransferases (GTs) localize to an unknown subcellular compartment of ∼0.5 μm in diameter in addition to the Golgi cisternae of ∼1.0 μm in diameter when these proteins are expressed transiently in N. benthamiana and stably in an Arabidopsis thaliana mutant.1 AtGALT31A was particularly highly localized to the small compartments and did not localize with the trans-Golgi network that was defined by SYP61, early endosomes stained by FM4-64, and prevacuolar compartments induced by Wortmannin treatment.1 Instead, AtGALT31A-defined small compartments were partially co-localized with EXO70E2, a marker for the exocyst-positive organelle (EXPO4), which was recently described to mediate the unconventional protein secretory (UPS) pathway5 in plants. N-glycosylation-processing enzymes were rarely found in the AtGALT31A-defined small compartments, indicating that the AtGALT31A-defined small compartments are mostly dedicated to AGP biosynthesis (protein O-glycosylation) and not the N-glycosylation of proteins.1
We previously presented that the site-directed mutagenesis of a phosphorylation site of AtGALT29A (Y144) that was identified by proteomics increased the localization frequency of the protein to the small compartments from 25% to 60–70% compared to the unmodified AtGALT29A when expressed in N. benthamiana.1 The result indicated the importance of Y144 in the subcellular targeting of AtGALT29A; however, this targeting is most likely not regulated by the electrostatic status of Y144 as a result of phosphorylation since both Y144A and Y144E constructs altered the localization in the same way. In this paper, we present the effect of Y144 mutagenesis on the enzyme activity of AtGALT29A. The recombinant AtGALT29A Y144A expressed in N. benthamiana showed 2.5-fold increased galactosyltransferase (GalT) activity, while the recombinant AtGALT29A Y144E showed 2-fold decreased GalT activity compared to wild type AtGALT29A (Fig. 1A). The results indicate that the charge status of Y144 likely affects the AtGALT29A GalT activity, which may be regulated by an unknown kinase/phosphatase system. The predicted 3D structure of AtGALT29A using rat α-2,6-sialyltransferase6 as a template indicates that the Y144 site is located in the globular catalytic domain, far (35 Å) from the catalytic site, and on the surface of the globular domain (Fig. 1B). Surface charge status often affects protein conformation; thus, the regulatory mechanisms of AtGALT29A enzyme activity by the electrostatic status of Y144 might be due to conformational changes in the globular catalytic domain. Thus, change of the subcellular targeting of AtGALT29A by substitution of Y144 is not likely mediated by Y144 phosphorylation, while AtGALT29A enzyme activity seems to be regulated by Y144 phosphorylation status. We previously demonstrated that AtGALT29A and AtGALT31A form a heterodimer, and the enzyme complex exhibits increased GalT activity.7An interaction with AtGALT31A also occurs with AtGALT29A Y144A and Y144E.1 Thus, how the 2 factors that enhance enzyme activity work with each other, namely whether the electrostatic status of Y144 and heterodimer formation with AtGALT31A act additively, synergistically, or in an antagonistic manner, in the regulation of AtGALT29A activity is an interesting question.
Figure 1.
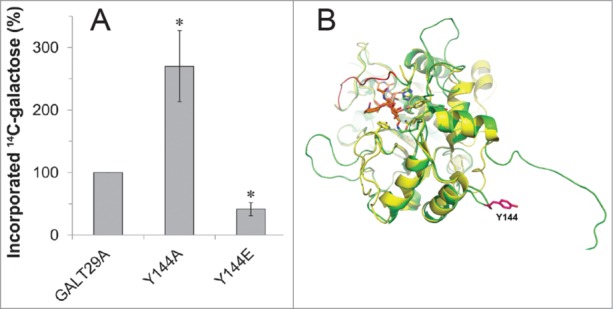
(A) Galactosyltransferase activity of AtGALT29A and site-directed mutants of AtGALT29A Y144A and Y144E. Affinity-purified recombinant proteins expressed in N. benthamiana leaves were incubated with UDP-14[C]-Gal in the presence of a mixture of AGP acceptors (SP32) as described previously.7 Relative activity (%) is compared to the activity level of AtGALT29A, which represents 100% activity. The error bar indicates standard deviation (n = 3). * indicates significant differences compared to wild type AtGALT29A (p < 0.05, Student's t-test). (B) Predicted 3D structure of AtGALT29A (green) using the crystal structure elucidated from rat α-2,6-sialyltransferase6 (2WNB, yellow). Y144 is located on the surface of a globular catalytic domain that is 35 Å from the predicted catalytic site (His319 of 2WNB); the substrate analog (orange) is indicated.
In this paper, we report proteins in addition to those analyzed previously that localize in the small compartments when expressed in N. benthamiana.1 Under the same experimental condition as used for proteins analyzed previously, a nucleoside phosphatase family protein (NTPDase; apyrase; APY3), which is encoded by At1g14240, and 2 UDP-glucuronic acid 4-epimerases (GAE1 and 6 encoded by At4g30440 and At3g23820, respectively) from Arabidopsis thaliana frequently co-localized with AtGALT31A in the small compartments (Fig. 2). In A. thaliana, 2 NTPDases (AtAPY1 or AtAPY2) have been characterized as functional NTPDases.8 Both AtAPY1 and 2 are integral membrane proteins that localize to the Golgi membrane9,10 and likely play a role in the metabolism of nucleotide diphosphate (NDP)-sugar, especially uridine diphosphate (UDP)-sugar, which is the substrate for the GTs reactions that occur in the Golgi lumen. Namely, the UDP produced after the GTs reaction is hydrolyzed to uridine monophosphate (UMP) by NTPDase, and UMP is transported to the cytosol by NDP-sugar transporters (antiporter) on the Golgi membrane. In the cytosol, UDP is used to generate new UDP-sugars that are transported by NDP-sugar transporters to the Golgi lumen where the UDP-sugars are then used for the GTs reactions. APY3 has not been biochemically characterized but might be a NTPDase dedicated to UDP metabolism in the small compartments. GAE1 and 6 have a topology similar to type II membrane proteins, and the recombinant proteins catalyze the epimerization of UDP-α-D-glucuronic acid (UDP-GlcA) to UDP-α-D-galacturonic acid (UDP-GalA).11,12 UDP-GalA is a major precursor for the synthesis of pectin in plants, and pectin biosynthesis occurs in the Golgi apparatus;13 however, our data indicate that GAE1 and 6 at least partially co-localize with AtGALT31A in the small compartments when expressed in N. benthamiana. A form of AGP that is covalently linked to pectin has recently been reported in APAP1 from A. thaliana,14, thus GAE1 and 6 might provide UDP-GalA for the synthesis of a APAP1-type structure together with AGP GTs in the small compartments. .
Figure 2.
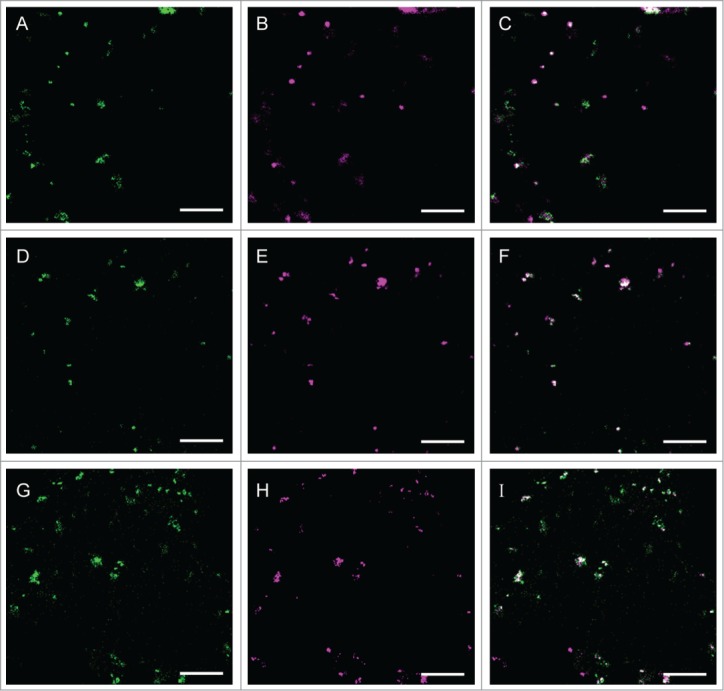
Co-localization of APY3, GAE1, and GAE6 with AtGALT31A in the small compartments. AtGALT31A-mCer3 (A-I, green) was co-expressed with APY3-YFP (A-C, magenta), GAE1-YFP (D-F, magenta), and GAE6-YFP (G-I, magenta) in N. benthamiana leaves. AtGALT31A-mCer3 frequently co-localized with APY3-YFP, GAE1-YFP, and GAE6-YFP in the small compartments, which are approximately 0.5 μm in diameter. Scale bars = 10 μm.
Collectively, we present evidence that Y144 phosphorylation may regulate AtGALT29A enzyme activity in addition to Y144's involvement in determining the subcellular targeting of AtGALT29A, as reported previously.7 Furthermore, we identified that A. thaliana APY3, GAE1, and GAE6 localize to the small compartments together with AtGALT31A when expressed transiently in N. benthamiana. Previously we have observed the dual localization of AtGALT31A in the Golgi and in the small compartments in both expression systems using N. benthamiana and A. thaliana mutant as hosts1. Also, we rarely observed localization of N-glycan processing enzymes in the small compartments when expressed in N. benthamiana,1 which is consistent with the localization of the native enzymes detected by antibodies. Therefore, we consider the dual localization observed for APY3, GAE1 and GAE6 when expressed in N. benthamiana is not likely an artifact of heterologous and transient expression using N. benthamiana as a host. However, we have previously observed a difference for the subcellular distribution for AtGALT31A expressed in N. benthamiana and A. thaliana.1 We currently do not know the factors that caused the difference, but it might be due to different tissues, plant species, and/or expression methods of the proteins. Similar phenomenon might be true for APY3, GAE1 and GAE6, which should be investigated in future.
Nevertheless, at least partially APY3, GAE1 and GAE6 target to the small compartments together with AtGALT31A when expressed in N. benthamiana and therefore these enzymes may function together with AGP GTs in the small compartments. Further characterization of: (i) the proteins residing in the small compartments, e.g., protein kinases and phosphatases; (ii) targeting and retaining mechanism of those proteins in the small compartments; and (iii) biogenesis of the small compartments; are interesting questions to address to understand the function of the small compartments with respect to so-called the unconventional protein secretion in plants. Besides, the obtained knowledge may offer novel tools to make use of the unconventional protein secretion pathway of plants to produce useful recombinant products.15-17
Methods
DNA cloning
DNA constructs of AtGALT31A-mCer37, AtGALT29A-mCer37, AtGALT29A Y144A1 and Y144E1 were described previously. Full-length cDNAs encoding APY3 (At1g14240), GAE1 (At4g30440), and GAE6 (At3g23820) were kindly provided by Dr. Joshua Heazlewood (Joint BioEnergy Institute, Lawrence Berkeley National Laboratory). Genes without stop codons were cloned into the Gateway vector, pDONR223, using following primer pairs: APY3-sense: GGGGACAAGTTTGTACAAA-AAAGCAGGCTTCATGACACCGGAGACGGATG; APY3-antisense: GGGGACCACTTTGTACAAGAAAGCTGGGTG-AAAGCCAAGATATTTTCTAG; GAE1-sense: GGGGACAA-GTTTGTACAAAAAAGCAGGCTTCATGCCTTCAATAGA-AGATG; GAE1-antisense: GGGGACCACTTTGTACAAGA-AAGCTGGGTGATGTACAAGCTTGGCTTTAG; GAE6-sense: GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAT-GCCCCTGTCGGCGAC; and GAE6-antisense: GGGGACC-ACTTTGTACAAGAAAGCTGGGTGAGCGGAATCTTCG-GCGTG.
The inserts were further moved to the expression vectors (pEarleyGate mCer3 and pEarleyGate 101) and transformed into the Agrobacterium tumefaciens strain C58C1 pGV3850 according to the method described previously.18
Expression of recombinant proteins and enzyme assay
Transient expression of the constructs in N. benthamiana leaves, purification of recombinant proteins, and the enzyme assay were performed following the method described previously.7 The relative protein concentrations were estimated by densitometric analysis of the Western blot using ImageJ,19 and the enzyme activities were adjusted to protein concentration.
Co-localization study
Subcellular localization of recombinant proteins was studied in N. benthamiana leaves 2 days after infiltration of Agrobacterium harboring the respective construct using a Leica TCS SP5 inverted confocal laser scanning microscope (www.leica.com) as described previously.7 For detecting the mCer3 and YFP signals, excitation was performed by argon laser at 458 and 514 nm, and emission was detected at 475–505 and 525–600 nm.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Danish Agency for Science, Technology and Innovation: the Strategic Research for Health, Food and Welfare (DSF:09-067059) and Danish Agency for Science, Technology and Innovation: the Technology and Production (FTP:274-09-0113) to NG.
References
- 1. Poulsen CP, Dilokpimol A, Mouille G, Burow M, Geshi N. Arabinogalactan glycosyltransferases target to a unique subcellular compartment that may function in unconventional secretion in plants. Traffic 2014; 15(11):1219-34; PMID:25074762; http://dx.doi.org/ 10.1111/tra.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol 2007; 58:137-61; PMID:17201686; http://dx.doi.org/ 10.1146/annurev.arplant.58.032806.103801 [DOI] [PubMed] [Google Scholar]
- 3. Knoch E, Dilokpimol A, Geshi N. Arabinogalactan proteins: focus on carbohydrate active enzymes. Front Plant Sci 2014; 5:198; PMID:24966860; http://dx.doi.org/ 10.3389/fpls.2014.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J, Ding Y, Wang J, Hillmer S, Miao Y, Lo SW, Wang X, Robinson DG, Jiang L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010; 22:4009-30; PMID:21193573; http://dx.doi.org/ 10.1105/tpc.110.080697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding Y, Wang JJ, Stierhof Y-D, Robinson DG, Jiang L. Unconventional protein secretion. Trends Plant Sci 2012; 17:606-15; PMID:22784825; http://dx.doi.org/ 10.1016/j.tplants.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 6. Kuhn B, Benz J, Greif M, Engel AM, Sobek H, Rudolph MG. The structure of human α-2,6-sialyltransferase reveals the binding mode of complex glycans. Acta Crystallogr D Biol Crystallogr 2013; 69:1826-38; PMID:23999306; http://dx.doi.org/ 10.1107/S0907444913015412 [DOI] [PubMed] [Google Scholar]
- 7. Dilokpimol A, Poulsen CP, Vereb G, Kaneko S, Schulz A, Geshi N. Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol 2014; 14:90; PMID:24693939; http://dx.doi.org/ 10.1186/1471-2229-14-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steinebrunner I, Jeter C, Song C, Roux SJ. Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem 2000; 38:913-22; http://dx.doi.org/ 10.1016/S0981-9428(00)01209-2 [DOI] [Google Scholar]
- 9. Schiller M, Massalski C, Kurth T, Steinebrunner I. The Arabidopsis apyrase AtAPY1 is localized in the Golgi instead of the extracellular space. BMC Plant Biol 2012; 12:123; PMID:22849572; http://dx.doi.org/ 10.1186/1471-2229-12-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiu T-Y, Christiansen K, Moreno I, Lao J, Loqué D, Orellana A, Heazlewood JL, Clark G, Roux SJ. AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol 2012; 53:1913-25; PMID:23034877; http://dx.doi.org/ 10.1093/pcp/pcs131 [DOI] [PubMed] [Google Scholar]
- 11. Mølhøj M, Verma R, Reiter W-D. The biosynthesis of D-Galacturonate in plants. functional cloning and characterization of a membrane-anchored UDP-D-Glucuronate 4-epimerase from Arabidopsis. Plant Physiol 2004; 135:1221-30; PMID:15247385; http://dx.doi.org/ 10.1104/pp.104.043745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Usadel B, Schlüter U, Mølhøj M, Gipmans M, Verma R, Kossmann J, Reiter W-D, Pauly M. Identification and characterization of a UDP-D-glucuronate 4-epimerase in Arabidopsis. FEBS Lett 2004; 569:327-31; PMID:15225656; http://dx.doi.org/ 10.1016/j.febslet.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 13. Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol 2008; 11:266-77; PMID:18486536; http://dx.doi.org/ 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 14. Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 2013; 25:270-87; PMID:23371948; http://dx.doi.org/ 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Czlapinski JL, Bertozzi CR. Synthetic glycobiology: Exploits in the Golgi compartment. Curr Opin Chem Biol 2006; 10:645-51; PMID:17056297; http://dx.doi.org/ 10.1016/j.cbpa.2006.10.009 [DOI] [PubMed] [Google Scholar]
- 16. Strasser R. Challenges in O-glycan engineering of plants. Front Plant Sci 2012; 3:218; PMID:23015807; http://dx.doi.org/ 10.3389/fpls.2012.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strasser R, Stadlmann J, Schähs M, Stiegler G, Quendler H, Mach L, Glössl J, Weterings K, Pabst M, Steinkellner H. Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol J 2008; 6:392-402; PMID:18346095; http://dx.doi.org/ 10.1111/j.1467-7652.2008.00330.x [DOI] [PubMed] [Google Scholar]
- 18. Knoch E, Dilokpimol A, Tryfona T, Poulsen CP, Xiong G, Harholt J, Petersen BL, Ulvskov P, Hadi MZ, Kotake T, et al. A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J 2013; 76:1016-29; PMID:24128328; http://dx.doi.org/ 10.1111/tpj.12353 [DOI] [PubMed] [Google Scholar]
- 19. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671-5; PMID:22930834; http://dx.doi.org/ 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]


