Abstract
Light is both a source of energy and a critically important environmental signal for plant development. Through decades of research, 2 groups of photomorphogenic repressors have been identified. The first group is CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS), which were first identified by genetic screening and then by purification of protein complexes. Another group is the Phytochrome-Interacting Factors (PIFs), which were identified by yeast 2-hybrid screens using phyB as bait. How so many factors work together to repress photomorphogenesis has long been an interesting question. Previously, we demonstrated that CULLIN4 (CUL4) works as a core factor connecting the COP1-SPA complexes, the COP9 signalosome (CSN), and the COP10-DDB1-DET1 (CDD) complex. Recently, we showed that DET1 represses photomorphogenesis through positively regulating the abundance of PIF proteins in the dark. Dr. Huq and his colleagues reported that PIFs may enhance the function of COP1-SPA complexes to promote the degradation of HY5, and thus they synergistically repress photomorphogenesis in the dark. Though much work still needs to be done, these recent breakthroughs shed light on the regulatory relationships among these multiple photomorphogenic repressors.
Keywords: COP/DET/FUS, light signaling, photomorphogenic repressor, PIFs
Multiple Photomorphogenic Repressors in Plants
Seedlings grown in the light display photomorphogenic development with short hypocotyls and open and expanded cotyledons. In contrast, seedlings grown in the dark exhibit skotomorphogenic development with long hypocotyls, closed and unexpanded cotyledons, and apical hooks.1 COP/DET/FUS is a group of pleiotropic genes identified by genetic screens as photomorphogenic repressors since mutants displayed photomorphogenic phenotypes in the dark.2 This group of proteins functions as part of 3 biochemical entities: the COP1-SPA complexes, the COP9 signalosome (CSN), and the CDD complex.
COP1 was the first of these proteins to be characterized molecularly,3 and it was shown to interact with SUPPRESSOR OF PHYTOCHROME A (SPA) proteins to function as Ring-finger type E3 ligases.4,5 In the dark, the COP1-SPA complexes ubiquitinate and degrade ELONGATED HYPOCOTYL 5 (HY5), HY5 HOMOLOG (HYH), LONG HYPOCOTYL IN FAR RED (HFR1) and others via the 26S proteasome to repress photomorphogenesis.6-8 Upon light exposure, the function of COP1 is largely inhibited, at least partially through nuclear export, leading to stabilization of the targeted proteins.9,10
The CSN was initially identified as a repressor of light-dependent development in plants.11 It was later found to be a nuclear-enriched complex consisting of 8 distinct subunits.12 Interestingly, CSN is conserved between plants and mammals, and shows remarkable similarity to the lid subcomplex of the 19S regulatory particle.13 The CSN can interact with CULLIN (CUL)-based E3 ligases to deconjugate Nedd8/Rub from CULLINs and regulate their activities.14,15
The CDD complex is composed of COP10, DET1, and DDB1.16 DET1 is another central repressor of photomorphogenesis identified in 1989.17 It can interact with DAMAGED DNA BINDING PROTEIN1 (DDB1) and bind histone H2B, indicating a role for chromatin remodeling in the regulation of photomorphogensis.18,19 DET1 has also been shown to maintain normal peroxisomal activities in plants to repress photomorphogenesis.20 Recently, DET1 has been found to interact directly with CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) to regulate the circadian clock in vivo.21 COP10 encodes a small ubiquitin E2 variant protein, and both COP10 itself and the CDD complex are known to enhance the activity of several E2s.16,22
In addition to COP/DET/FUS, PIFs function as another group of photomorphogenic repressors in the dark, identified by yeast 2-hybrid screen using phyB as bait.23,24 Although they have different functions in seed germination, hypocotyl elongation, chlorophyll biosynthesis, shade avoidance etc., PIFs (mainly PIF1, PIF3, PIF4 and PIF5) function redundantly to repress photomorphogenesis in the dark.25-30 It is now clear that PIFs accumulate in darkness but are rapidly phosphorylated and degraded upon light exposure.31-33
CUL4 Connects the COP Complexes
All the cop/det/fus mutants show similar photomorphogenic phenotypes in the dark, but how these COP complexes work together was unclear for a long period. COP/DET/FUS proteins are required for the proper nuclear localization of COP1 in the dark.9,12 Gel filtration analysis showed that lack of the CSN reduced the stability of the COP10 complex, and affected its integrity.22 Then our further studies showed that CUL4 might interact with the CDD complex to form an E3 ligase RBX1-CUL4-CDD, and the CSN complex could regulate its activity through derubylation of CUL4.34 Another study showed that both COP1 and SPA proteins have DWD motifs, and could interact with CUL4-DDB1 to form large E3 ligases.35 Interestingly, upon UV exposure, the COP1-SPA complexes may disassociate from CUL4-DDB1 and interact with the UV-B receptor UVR8 to form a UVR8-COP1-SPA complex and stabilize HY5 proteins in vivo.36 These results indicate that the activity of COP complexes can be connected by the function of CUL4.
Cooperation of COP/DET/FUS and PIFs
COP/DET/FUS and PIFs are 2 groups of photomorphogenic repressors identified using different strategies. During the past decade, the underlying mechanisms regulating interactions between these 2 groups of factors were largely unknown, except that the abundance of PIF3 proteins was reduced in cop1 mutants.31
We recently found that DET1 could physically interact with 4 PIF (PIF1, PIF3, PIF4 and PIF5) proteins and positively regulate their levels. The PIF3 levels in the mutants of DET1, COP10 and CUL4 were all lower than in wild type plants, and the protein levels correlated well with their photomorphogenic phenotypes, which indicated that this positive regulation might be accomplished by the CUL4-CDD complex. We then demonstrated that DET1 could stabilize the 4 PIF (PIF1, PIF3, PIF4 and PIF5) proteins in the dark, while DET1 only positively regulated the PIF3 transcription level. Genetic analysis showed that pif single mutations enhanced while PIF overexpression partially suppressed the phenotypes of det1-1. Most of the genes regulated by both light and DET1, were also regulated by PIFs. Taken together, we proposed that DET1 repressed photomorphogenesis partially by stabilizing PIFs in the dark.37
At the same time, Dr. Huq and his colleagues reported another functional relationship between COP1-SPA complexes and PIFs. They found that PIFs could promote COP1-SPA-mediated HY5 degradation in the dark. The pif1 single mutant could promote photomorphogenesis synergistically with cop1 and spa123 mutants. The HY5 protein levels in pif single and higher order mutants indicated that PIFs redundantly promoted HY5 protein degradation. Further genetic analysis showed that hy5 mutants could partially suppress the photomorphogenic phenotypes of both cop1-6 pif1 and pifq mutants in the dark. Biochemical analyses showed that PIF1 could form complexes with COP1, HY5, and SPA1 and that PIF1 could enhance the substrate recruitment and autoubiquitylation and transubiquitylation activities of COP1. Together, PIFs can enhance the ligase activity of COP1-SPA and synergistically repress photomorphogenesis in the dark.38
Known and Unknown
Based on the current knowledge, we propose a model of how COP/DET/FUS and PIFs work together to repress photomorphogenesis in the dark (Fig. 1). However, there are still many questions waiting to be further investigated. For example, what's the regulatory mechanism that controls the COP1-SPA complexes to act as E3 ligases themselves or as adaptors of CUL4-based E3 ligases? How does DET1 stabilize PIFs in the dark? Do PIFs also promote the E3 ligase activity of COP1-SPA toward other targets of COP1? Why do the functions of COP1- and DET1-containing E3 ligases rely on each other during photomorphogenesis? These photomorphogenic repressors play important roles not only in light signal transduction, but also in many other processes regulating plant development. Further investigation of these photomorphogenic repressors will advance our views of regulatory mechanisms underlying plant development and their responses to environmental cues.
Figure 1.
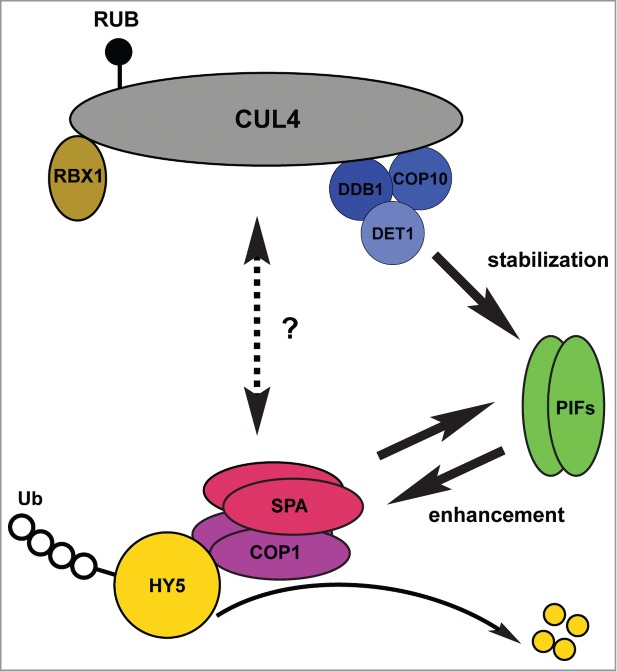
Multiple factors work together to repress photomorphogenesis in the dark. The CDD complex interacts with CUL4 to form the RBX1-CUL4-CDD E3 ligase in the dark. The RBX1-CUL4-CDD complex positively regulates PIF abundance primarily by stabilizing PIF proteins. On the other hand, PIFs promote the degradation of HY5 by enhancing the interactions between COP1-SPA complexes and HY5 and enhancing the ubiquitylation activities of COP1. Thus, these 2 newly- identified regulatory processes explain why the functions of COP1-SPA complexes rely on the CUL4-CDD complex to a certain extent. COP1 also positively regulates PIF3 abundance, although the mechanism is unclear. CSN regulates the activity of CUL4-based E3 ligases by deconjugating Nedd8/Rub from CULLINs (CSN is not shown in the model). The complete mechanism(s) by which these multiple photomorphogenesis factors work in concert need further investigation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Our work was supported by grants from the National Natural Science Foundation of China (31271294, 31330048, U1031001), the Ministry of Science and Technology of China (2011CB100101, 2012CB910900, 2012AA10A304), the Ministry of Agriculture of China (2011-G2B), Peking-Tsinghua Center for Life Sciences, and State Key Laboratory of Protein and Plant Gene Research.
References
- 1.Von Arnim A, Deng XW. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol 1996; 47:215-43; PMID:15012288; http://dx.doi.org/ 10.1146/annurev.arplant.47.1.215 [DOI] [PubMed] [Google Scholar]
- 2.Sullivan JA, Shirasu K, Deng XW. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet 2003; 4: 948-58; PMID:14631355; http://dx.doi.org/ 10.1038/nrg1228 [DOI] [PubMed] [Google Scholar]
- 3.Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 1992; 71:791-801; PMID:1423630; http://dx.doi.org/ 10.1016/0092-8674(92)90555-Q [DOI] [PubMed] [Google Scholar]
- 4.Hoecker U, Quail PH. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 2001; 276:38173-8; PMID:11461903 [DOI] [PubMed] [Google Scholar]
- 5.Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHO GENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 2008; 20:2307-23; PMID:18812498; http://dx.doi.org/ 10.1105/tpc.107.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000; 405:462-6; PMID:10839542; http://dx.doi.org/ 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- 7.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 2002; 16:1247-59; PMID:12023303; http://dx.doi.org/ 10.1101/gad.969702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang IC, Yang JY, Seo HS, Chua NH. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 2005; 19:593-602; PMID:15741320; http://dx.doi.org/ 10.1101/gad.1247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Arnim AG, Osterlund MT, Kwok SF, Deng XW. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 1997; 114:779-88; PMID:9232869; http://dx.doi.org/ 10.1104/pp.114.3.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacin M, Legris M, Casal JJ. Rapid decline in nuclear constitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol 2014; 164:1134-8; PMID:24434030; http://dx.doi.org/ 10.1104/pp.113.234245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei N, Chamovitz DA, Deng XW.. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 1994; 78:117-24; PMID:8033203 [DOI] [PubMed] [Google Scholar]
- 12.Chamovitz D. A., Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW. The COP9 complex a novelmultisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 1996; 86:115-21; PMID:8689678; http://dx.doi.org/ 10.1016/S0092-8674(00)80082-3 [DOI] [PubMed] [Google Scholar]
- 13.Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol 1998; 8:919-22; PMID:9707402; http://dx.doi.org/ 10.1016/S0960-9822(07)00372-7 [DOI] [PubMed] [Google Scholar]
- 14.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ.. Promotion ofNEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001; 292:1382-5; PMID:11337588; http://dx.doi.org/ 10.1126/science.1059780 [DOI] [PubMed] [Google Scholar]
- 15.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 2001; 292:1379-82; PMID:11337587; http://dx.doi.org/ 10.1126/science.1059776 [DOI] [PubMed] [Google Scholar]
- 16.Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, et al.. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 2004; 18:2172-81; PMID:15342494; http://dx.doi.org/ 10.1101/gad.1229504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 1989; 58:991-9; PMID:2776216; http://dx.doi.org/ 10.1016/0092-8674(89)90950-1 [DOI] [PubMed] [Google Scholar]
- 18.Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogensis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosomal context. Curr Biol 2002; 12:1529-34; PMID:12225670; http://dx.doi.org/ 10.1016/S0960-9822(02)01105-3 [DOI] [PubMed] [Google Scholar]
- 19.Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 2002; 12:1462-72; PMID:12225661; http://dx.doi.org/ 10.1016/S0960-9822(02)01106-5 [DOI] [PubMed] [Google Scholar]
- 20.Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 2002; 297:405-9; PMID:12130786; http://dx.doi.org/ 10.1126/science.1073633 [DOI] [PubMed] [Google Scholar]
- 21.Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell 2011; 43:703-12; PMID:21884973; http://dx.doi.org/ 10.1016/j.molcel.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev 2002; 16:554-9; PMID:11877375; http://dx.doi.org/ 10.1101/gad.964602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni M, Tepperman JM, Quail PH.. PIF3, a phytochrome- interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998; 95:657-67; PMID:9845368; http://dx.doi.org/ 10.1016/S0092-8674(00)81636-0 [DOI] [PubMed] [Google Scholar]
- 24.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 1999; 400:781-4; PMID:10466729; http://dx.doi.org/ 10.1038/23500 [DOI] [PubMed] [Google Scholar]
- 25.Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004; 305:1937-41; PMID:15448264; http://dx.doi.org/ 10.1126/science.1099728 [DOI] [PubMed] [Google Scholar]
- 26.Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004; 16:3045-58; PMID:15486102; http://dx.doi.org/ 10.1105/tpc.104.025163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Yi H, Choi G, Shin B, Song PS. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 2003; 15:2399-407; PMID:14508006; http://dx.doi.org/ 10.1105/tpc.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 2008; 53:312-23; PMID:18047474; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- 29.Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A 2009; 106:7660-5; PMID:19380720; http://dx.doi.org/ 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 2008; 18:1815-23; PMID:19062289; http://dx.doi.org/ 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al.. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 2004; 16:1433-45; PMID:15155879; http://dx.doi.org/ 10.1105/tpc.021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 2006; 23:439-46; PMID:16885032; http://dx.doi.org/ 10.1016/j.molcel.2006.06.011 [DOI] [PubMed] [Google Scholar]
- 33.Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014; 344:1160-4; PMID:24904166; http://dx.doi.org/ 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al.. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 2006; 18:1991-2004; PMID:16844902; http://dx.doi.org/ 10.1105/tpc.106.043224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al.. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 2010; 22:108-23; PMID:20061554; http://dx.doi.org/ 10.1105/tpc.109.065490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci U S A 2013; 110:16669-74; PMID:24067658; http://dx.doi.org/ 10.1073/pnas.1316622110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 2014; 26:3630-45; PMID:25248553; http://dx.doi.org/ 10.1105/tpc.114.130666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, Paik I, Zhu L, Bu Q, Huang X, Deng XW, Huq E. PHYTOCHROME INTERACTING FACTOR1 Enhances the E3 Ligase Activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to Synergistically Repress Photomorphogenesis in Arabidopsis. Plant Cell 2014; 26:1992-2006; PMID:24858936; http://dx.doi.org/ 10.1105/tpc.114.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]


