Abstract
Gastric cancer is one of the most common gastrointestinal malignancies and is associated with poor prognosis. Exploring alterations in the proteomic landscape of gastric cancer is likely to provide potential biomarkers for early detection and molecules for targeted therapeutic intervention. Using iTRAQ-based quantitative proteomic analysis, we identified 22 proteins that were overexpressed and 17 proteins that were downregulated in gastric tumor tissues as compared to the adjacent normal tissue. Calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) was found to be 7-fold overexpressed in gastric tumor tissues. Immunohistochemical labeling of tumor tissue microarrays for validation of CAMKK2 overexpression revealed that it was indeed overexpressed in 94% (92 of 98) of gastric cancer cases. Silencing of CAMKK2 using siRNA significantly reduced cell proliferation, colony formation and invasion of gastric cancer cells. Our results demonstrate that CAMKK2 signals in gastric cancer through AMPK activation and suggest that CAMKK2 could be a novel therapeutic target in gastric cancer.
Keywords: invasion, in vitro labeling, mass spectrometry, proliferation, targeted therapy
Abbreviations
- iTRAQ
Isobaric tags for relative and absolute quantitation
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- SCX
Strong cation exchange chromatography
- PSM
Peptide spectrum match
- siRNA
short interfering RNA
Introduction
Gastric cancer is one of the most common causes of cancer related mortality in the world.1 Adenocarcinoma accounts for 95% of gastric malignancies.2 Gastric cancer is associated with poor prognosis3 and patients with gastric cancer often present symptoms that are non-specific including heartburn, early satiety, mild abdominal pain and nausea.4 In recent years, kinases have become one of the most intensively studied group of proteins as drug targets. To date, the FDA has approved 11 kinase inhibitors for cancer therapy.5 Imatinib, a small molecule inhibitor against BCR-ABL tyrosine kinase revolutionized the treatment of patients with chronic myelogenous leukemia.6,7 Other targeted therapies include erlotinib and gefitinib, targeting EGFR in non small cell lung cancer8 and trastuzumab in HER2 positive breast cancer.9 Targeted therapy in gastric cancer is limited and till date trastuzumab is the only approved targeted agent for HER2positive gastric cancer.10 However, unlike the poor prognosis associated with HER2 positivity in breast cancer, the prognostic value of HER2 in gastric cancer is controversial.11 Apart from HER2, potential therapeutic targets such as MET12 and FGFR213 are pending clinical validation in gastric cancer.
Mass spectrometry-based proteomic analysis has facilitated the discovery of dysregulated proteins in multiple cancer types14-20 and has emerged as an attractive approach to identify candidate biomarkers for diagnosis/prognosis and for identification of potential therapeutic targets. We have previously carried out quantitative proteomic studies leading to the identification of novel biomarkers in esophageal squamous cell carcinoma19 and hepatocellular carcinoma.15 Similar approaches have also been used by us and other groups to identify potential biomarkers in gastric cancer using secretome, plasma and cell based models.21-25 In this study, we used high-resolution mass spectrometry coupled with iTRAQ-based quantitative approach for proteomic analysis of gastric cancer to identify proteins which are differentially expressed. Of the 39 differentially expressed proteins identified in this study, calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) was identified as an overexpressed protein and an attractive target for further exploration due to the well established connection between calcium signaling and cancer pathogenesis.26,27 Alterations in Ca2+ signaling are reported to contribute to multiple aspects of tumor progression such as proliferation, migration, invasion and metastasis.28 Calmodulin (CaM) is one of the key proteins that triggers various signaling events in response to an increase in intracellular levels of Ca2+ ions. Upon binding with Ca2+, CaM activates downstream serine/threonine protein kinases such as calcium/calmodulin-dependent protein kinase kinases (CAMKK) which include CAMKK1 (α) and CAMKK2 (β). These further activate calmodulin-dependent kinases (CaMK) including CAMKI, CAMKII and CAMKIV.29,30 CAMKK2 is also known to regulate 5′adenosine mono phosphate activated protein kinase (AMPK), which is involved in energy homeostasis, cell cycle regulation, cytoskeletal reorganization and autophagy.31,32 CAMKK2 has recently been shown to be an androgen responsive gene and silencing its expression repressed tumor growth in prostate cancer.33,34 However, the role of CAMKK2 in gastric cancer remains unexplored. In this study, we have assessed the potential of CAMKK2 as a novel therapeutic target in gastric cancer.
Results
Identification of proteins with altered expression in gastric adenocarcinoma through quantitative proteomics
Primary gastric adenocarcinoma tissue and corresponding adjacent non-neoplastic tissue from 5 patients was subjected to quantitative proteomic analysis to identify differentially expressed proteins in gastric tumors. Details of the patients and tumors are provided in Table S1. The extracted proteins were digested with trypsin, labeled with iTRAQ reagents, fractionated by SCX chromatography and analyzed using mass spectrometry. The overall experimental workflow used in this study is illustrated in Figure 1. A total of 666 proteins were identified and their fold-change values were calculated based on reporter ion intensities. Proteins identified with a fold change of ≥2 were reported to be overexpressed while those with a fold change of ≤0.5 were considered to be downregulated. With these selection criteria, 22 proteins were found to be overexpressed and 17 proteins were downregulated in gastric adenocarcinoma tissue. Of the 22 overexpressed proteins, 13 were identified for the first time in association with gastric adenocarcinoma. These include CAMKK2, Filamin-A (FLNA) and Prolargin (PRELP) which were overexpressed 7, 2.6 and 2-fold, respectively. A complete list of differentially expressed proteins in gastric adenocarcinoma is provided in Table S2.
Figure 1.
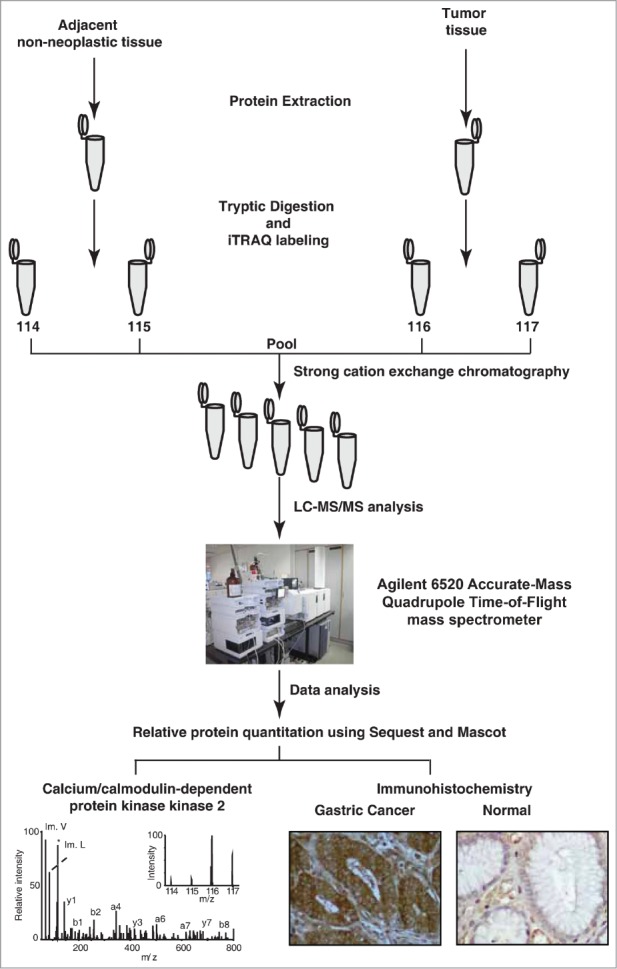
Experimental workflow employed to analyze the gastric cancer tissue proteome. Equal concentrations of protein from both tumor and non-neoplastic tissue were extracted, digested and labeled with iTRAQ reagents. The labeled samples were pooled and fractionated using strong cation exchange chromatography. The sample fractions were analyzed using a quadrupole time-of-flight mass spectrometer. The MS/MS data was searched against Refseq45 protein database using Sequest and Mascot search algorithms. The overexpressed protein CAMKK2 was validated using immunohistochemistry.
Representative MS/MS spectra of a subset of the novel proteins are depicted in Figure 2. In addition, we also identified proteins such as transgelin, biglycan and heat shock 27 kDa protein, which have been reported by other research groups to be overexpressed in gastric cancer.35-37 MS/MS spectra of a subset of known differentially expressed molecules are provided in Figure S1. A partial list of novel and previously reported differentially expressed proteins identified in this study are provided in Table 1.
Figure 2.
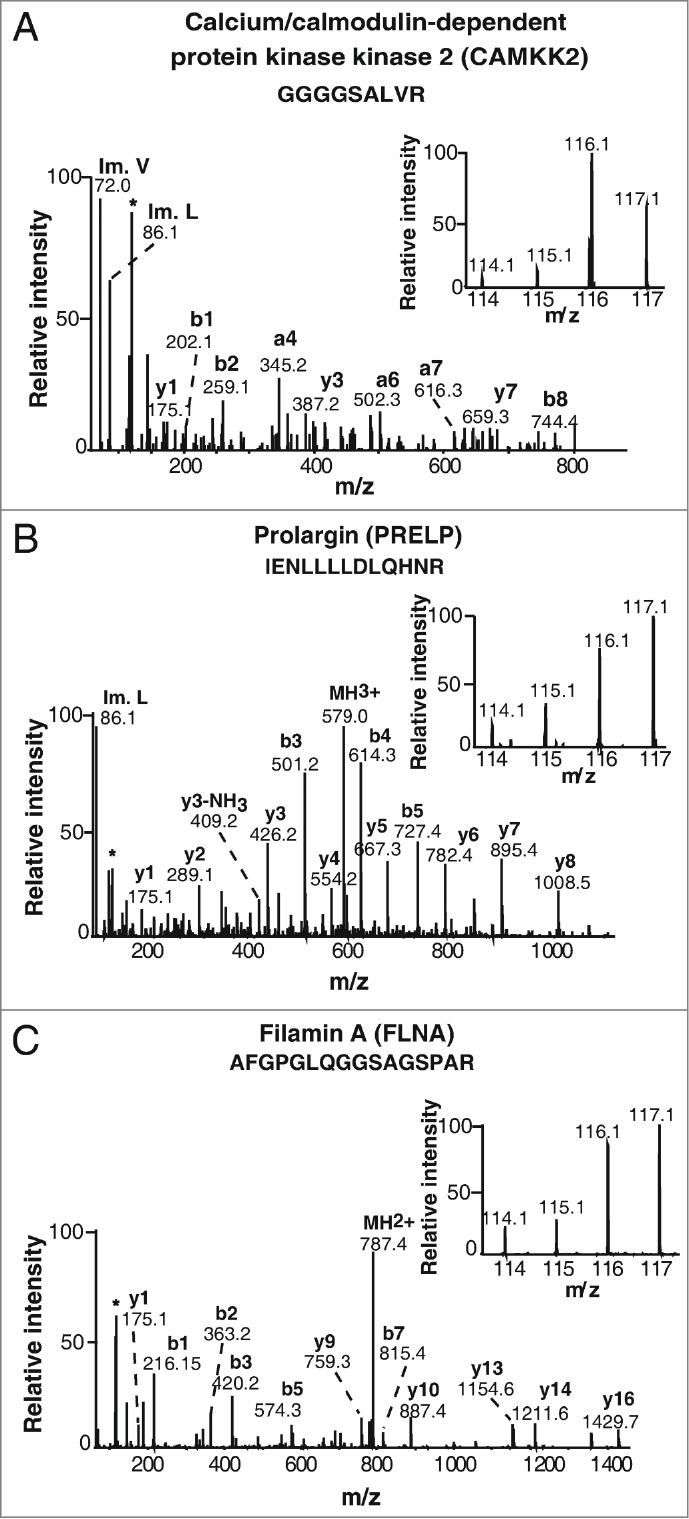
Representative MS/MS spectra of novel overexpressed proteins in gastric adenocarcinoma tissues. (A) Calcium/calmodulin-dependent protein kinase kinase 2 (7-fold). (B) Prolargin (2.6-fold) (C) Filamin-A isoform 1 (2-fold).
Table 1.
A partial list of overexpressed proteins identified in gastric adenocarcinoma
| Novel proteins found to be overexpressed in gastric adenocarcinoma | ||||
|---|---|---|---|---|
| Gene symbol | Protein | Function | Fold-change | |
| CAMKK2 | Calcium/calmodulin-dependent protein kinase kinase 2 | Plays a role in calcium/calmodulin-dependent signaling cascade | 7.0 | |
| CNN2 | Calponin-2 | Functions in the structural organization of actin filaments | 2.7 | |
| PRELP | Prolargin | Anchors basement membrane to underlying connective tissue | 2.6 | |
| FLNA | Filamin-A | Involved in cytoskeletal remodeling | 2.0 | |
| Overexpressed proteins that were previously reported in gastric adenocarcinoma | ||||
| Gene symbol | Protein | Function | Fold-change | Citation |
| TAGLN | Transgelin | Involved in actin cross linking | 3.4 | Huang et al, 200835 |
| BGN | Biglycan | Involved in reassembly of collagen filaments and muscle generation | 2.4 | Wang et al, 201137 |
| HSPB1 | Heat shock protein β-1 | Involved in stress resistance and organization of actin filaments | 2.3 | Ren et al, 200636; Ryu et al, 200353 |
| S100A8 | Protein S100-A8 | Involved in cell cycle progression and differentiation | 2.1 | Yong et al, 200743 |
| S100A6 | Protein S100-A6 | Involved in cell cycle progression and differentiation | 2.0 | Wang et al, 201042 |
Immunohistochemical validation of CAMKK2 overexpression in gastric tumor tissues
Tissue microarray-based immunohistochemical validation was carried out using 98 cases of gastric adenocarcinoma and 115 samples of non-neoplastic gastric epithelial tissue. Staining intensity was scored as negative (0), weak (1+), moderate (2+) or strong (3+). About 94% (92 of 98) of gastric adenocarcinoma cases showed moderate to strong staining while 80% (92 of 115) of the non-neoplastic gastric epithelia showed negative to weak staining. A Chi-square test confirmed that the overexpression of CAMKK2 in tumor tissues was statistically significant (p-value=1.9e-26). The results of the immunohistochemical validation are provided as Table 2. CAMKK2 was observed to be predominantly localized in the cytoplasm. Representative staining patterns for CAMKK2 in gastric adenocarcinoma tissues and non-neoplastic mucosa are provided in Figure 3.
Table 2.
Summary of the immunohistochemical validation for CAMKK2 in gastric cancer and normal tissues
| Staining Intensity | Tumor cases | Normal cases |
|---|---|---|
| Strong | 47 | 3 |
| Moderate | 45 | 20 |
| Weak | 5 | 30 |
| Negative | 1 | 62 |
| P-value of significant difference between tumor and normal groups (Chi-square test) | 1.85e-26 | |
Figure 3.
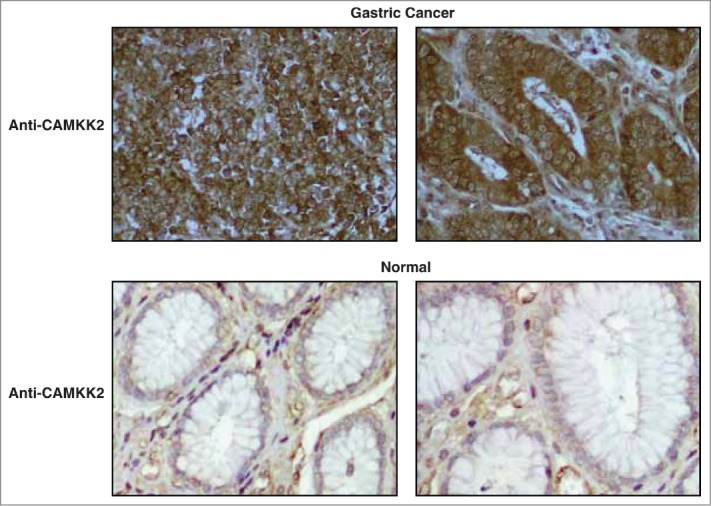
Validation of CAMKK2 by immunohistochemistry. (A) Representative sections from gastric cancer cases stained with anti-CAMKK2 antibody. (B) Representative sections from normal gastric mucosa stained with anti-CAMKK2 antibody.
Silencing of CAMKK2 decreases cellular proliferation in gastric cancer cells
Overexpression of CAMKK2 has been previously reported in prostate cancer where it has been shown to regulate cell proliferation.33,34 Given that we observed CAMKK2 to be abundantly expressed in the gastric cancer, we sought to study the functional significance of its overexpression in gastric cancer. We assessed the expression of CAMKK2 in a panel of gastric cancer cell lines (AGS, NCI-N87, KATO III, and SNU-16) and found detectable expression of the protein in all cell lines (Fig. 4A). Cancer cells often have higher proliferative capacity than their normal counterparts. To determine whether CAMKK2 activity had any effect on cell proliferation, we silenced the expression of CAMKK2 in gastric cancer cell lines using specific siRNAs. Knockdown of CAMKK2 expression led to a decrease in proliferation of AGS, KATO-III, NCI-N87 and SNU-16 cell lines (Fig. 4B–E). As our results indicate that CAMKK2 plays a potential role in proliferation of gastric cancer cells, we next studied the colony forming ability of the gastric cells with suppression of endogenous CAMKK2. In agreement with the cell proliferation data, we observed that siRNA-mediated silencing of CAMKK2 resulted in a significant decrease in the colony formation ability of the gastric cancer cell line AGS (Fig. 5A and B).
Figure 4.
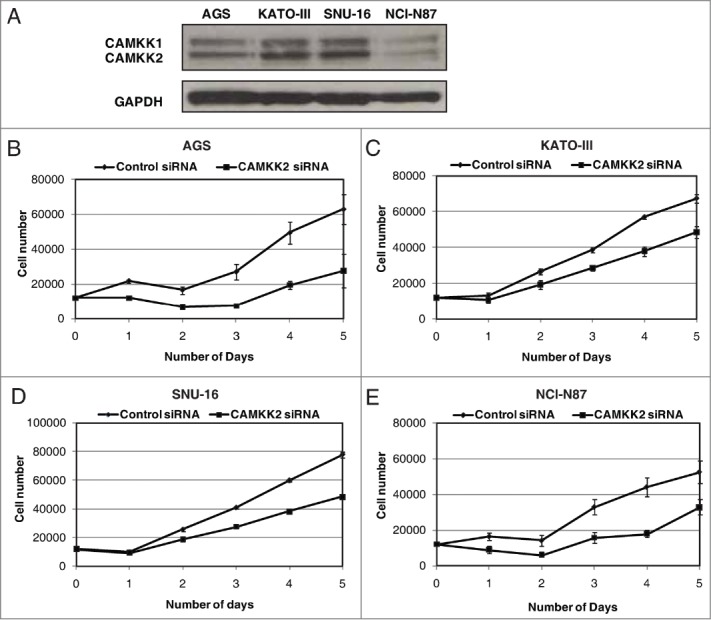
Silencing of CAMKK2 decreases cellular proliferation in gastric cancer cells. (A) Western blotting was done using anti-CAMKK2 antibody to study the relative expression level of CAMKK2 in a panel of gastric cell lines. GAPDH was used as a loading control. Gastric cancer cell lines were transfected with CAMKK2 siRNA and cell proliferation was assessed for (B) AGS, (C) KATO-III, (D) SNU-16 and (E) NCI-N87cell lines.
Figure 5.
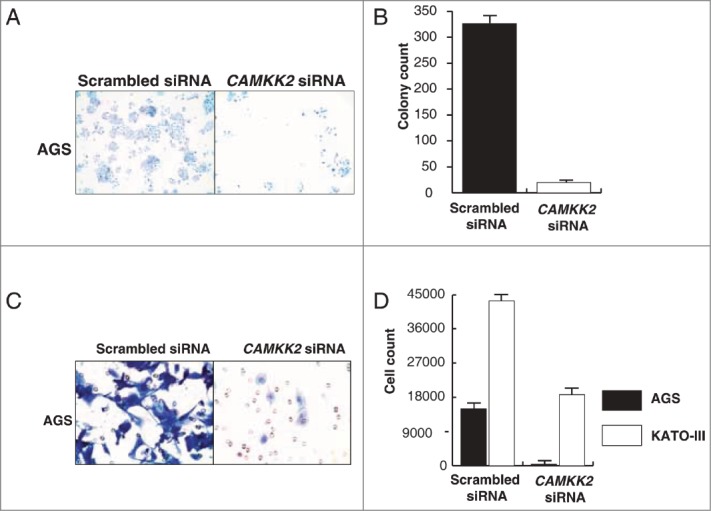
Silencing of CAMKK2 decreases the colony formation ability and invasive property of gastric cancer cell lines. Gastric cancer cell lines were transfected with CAMKK2 siRNA and assessed for either colony formation or invasion ability. (A) Colony formation ability of AGS was decreased post transfection with CAMKK2 siRNA. (B) A graphical representation of the same. (C) Silencing of CAMKK2 led to a decrease in the invasive property of AGS. (D) A graphical representation of the invasive property of AGS and KATO III (KATO III being a suspension line, invasion ability was studied by counting the cells in the upper chamber and the lower chamber of the membrane coated with Matrigel).
Silencing of CAMKK2 decreases the invasive property of gastric cancer cells
Having observed that inhibition of CAMKK2 leads to a decrease in both proliferation and colony forming ability of gastric cell lines, we tested whether CAMKK2 plays a role in gastric cancer metastasis. Gastric cancer cell lines AGS, NCI-N87, KATO-III and SNU-16 were used as in vitro models in which the endogenous expression of CAMKK2 was silenced using CAMKK2 specific siRNA. After 72 h, there was a significant decrease in the invasive ability of the AGS and KATO-III cell lines (Fig. 5C and D). NCI-N87 and SNU-16 cell lines were found to be non-invasive (data not shown). These results suggest that inhibition/silencing of CAMKK2 remarkably decreases the ability of cells to invade the extracellular matrix.
Silencing of CAMKK2 decreases AMPK activity in gastric cancer cells
Since CAMKK2 is known to phosphorylate AMPK on Thr 172 in response to altered calcium levels,38 we studied the effect of CAMKK2 silencing on AMPK phosphorylation in a panel of gastric cancer cells. Silencing of CAMKK2 using siRNA decreased p-AMPKα levels without affecting the total AMPKα levels in the gastric cancer cell lines. As AMPK is reported to phosphorylate FoxO3a on Ser 413,39 we examined the effect of CAMKK2-AMPK pathway on FoxO3a phosphorylation. FoxO3a activity and expression were unaffected by silencing of CAMKK2 (Fig. 6). These results indicate that although CAMKK2 is required for AMPKα phosphorylation, FoxO3a phosphorylation is unaffected in gastric cancer.
Figure 6.
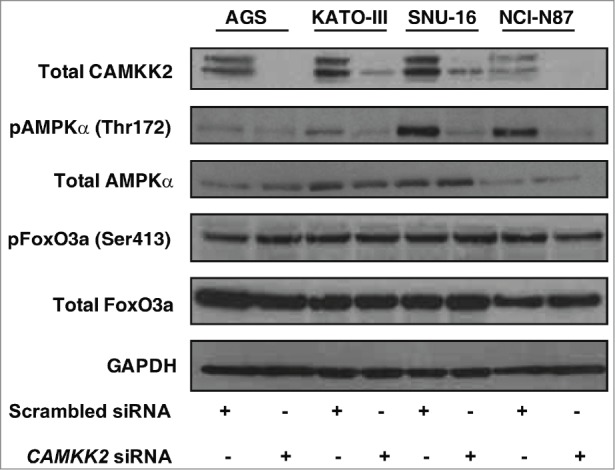
Silencing of CAMKK2 decreases AMPK activity in gastric cancer cells Gastric cancer cell line. AGS, KATO-III SNU-16 and NCI-N87 cell lines were transfected with either CAMKK2 or scrambled siRNA. Expression of Total CAMKK2, p-AMPKα (Thr172), Total AMPKα, p-FoxO3a (Ser413) and Total FoxO3a were probed by Western blot. GAPDH was used as control.
Discussion
Mass spectrometry has become a method of choice to identify differentially expressed proteins facilitating the discovery of potential biomarkers and drug targets. CAMKK2, a serine threonine kinase of Ca(2+)/calmodulin-dependent protein kinase subfamily, was found to be 7-fold overexpressed in gastric tumor tissue, making it an attractive candidate for further verification. Apart from CAMKK2, we also identified several proteins including Filamin A and Prolargin which were overexpressed more than 2-fold. Filamin-A is an actin binding protein involved in cytoskeleton remodeling. It promotes mitosis by binding to cdc25C40 and has been shown to promote cell motility and migration in a variety of cancers such as lung cancer, melanoma, prostate cancer and breast cancer.41 Prolargin or proline/arginine-rich end leucine-rich repeat protein is present in the extracellular matrix where it binds to collagen. There are no reports of differential expression of prolargin in any cancer. These proteins can be pursued for their potential as biomarkers using larger patient cohorts in future studies. In the current study, we focused on the potential of CAMKK2 both as a potential diagnostic marker and a therapeutic target in gastric cancer. Tissue microarray-based immunohistochemical validation confirmed the overexpression of CAMKK2 in a larger cohort, confirming its potential as a diagnostic marker.
Apart from the discovery of biomarkers for early diagnosis, a critical need is the development of pharmacological inhibitors for use as therapeutic agents in cancer including gastric cancer. Previous studies in prostate cancer have demonstrated the potential of CAMKK2 as a therapeutic target. Pathway analysis on our dataset using Ingenuity pathway analysis tool, led to the identification of 6 other proteins with CAMKK2 (ACTA2, CALR, MYH11, TPM1, TPM2, TPM4) which belong to the calcium signaling pathway that were differentially expressed in our study. Additionally, we identified 2 proteins of the S100 family of the calcium-binding proteins overexpressed in gastric tumor tissue (S100A6 - 2-fold and S100A8 - 2.1-fold). Both of these proteins have been reported to be overexpressed in gastric cancer.42,43 These results are indicative of the possible role of aberrant Ca2+ signaling in gastric cancer progression.
The molecular mechanisms regulating the enzymatic activity of CAMKK2 are not clearly understood. CAMKI, CAMKIV and AMPK have been reported as downstream targets of CAMKK2.44 It has been shown that CAMKK2 activation is required for it to bind to AMPK, although not reported as a requirement for its association with the other substrates.45 CAMKK2 has been reported to stimulate AMPK in prostate cancer.34 We studied the effect of CAMKK2 knockdown by siRNA in gastric cancer cell lines. We observed CAMKK2 knockdown led to decreased phosphorylation of AMPKα without affecting FoxO3a activity. The exact mechanism of this signaling network in gastric cancer is unclear from our current data and is beyond the scope of this manuscript. Our results reveal that silencing CAMKK2 significantly decreased gastric cancer cell viability. Cancer cells have the increased ability to invade local tissues as well as metastasize to distant organs and often negatively influence the treatment outcome. We report that CAMKK2 plays a significant role in the invasive property and colony formation ability of the gastric cancer cells. It is clear from our findings that CAMKK2 plays a crucial role in gastric cancer cell proliferation and can serve as a potential therapeutic target in gastric cancer. A detailed understanding of the function of CAMKK2 in gastric cancer is much needed and requires in vivo and in vitro models. Our work provides a scaffold for future studies to systematically investigate the role of CAMKK2 in gastric cancer.
Materials and Methods
Tissue samples
Tumor and adjacent non-neoplastic tissue samples were procured from operable cases of gastric cancer at Kidwai Memorial Institute of Oncology, Bangalore, India. Fresh tissue samples were collected from patients during surgery with informed consent after obtaining Institutional Review Board approval. Patients undergoing or with prior radio or chemotherapy were excluded from this study. Tumor and adjacent non-neoplastic gastric epithelial regions of the resected tissue specimens were determined by an experienced pathologist. The tissues were stored at −80°C until use.
Sample preparation
Five pairs of tumor and adjacent non-neoplastic tissue specimens were used for 4-plex iTRAQ labeling experiment. Sample preparation and iTRAQ labeling was carried out as described earlier.19 Briefly, 100 mg of tissue was homogenized in 0.5% SDS using a hand-held disruptor and sonicated. The protein concentrations of tissue lysates were estimated using Lowry's method46 and the samples were normalized based on protein concentration. Equal amounts of protein were pooled from the tumor and adjacent non-neoplastic tissue lysates to obtain 2 pools each with a final concentration of 160 μg. The samples were then reduced using tris (2-carboxyethyl) phosphine (TCEP) at 60°C for 1 hr. Subsequently, alkylation was carried out using methyl methanethiosulfonate (MMTS) at room temperature for 10 min. The samples were digested using sequencing grade modified trypsin (Promega, catalog #V5111) (1:20) at 37°C overnight. The tryptic digests were then vacuum dried and stored at −80°C until further analysis.
iTRAQ labeling and SCX fractionation
The tryptic peptides were reconstituted in 25 μl dissolution buffer and the non-neoplastic and tumor samples were split in to 2 fractions each serving as technical replicates. The peptide digests were labeled with one of the 4 iTRAQ reagents at room temperature for an hour. Peptides from non-neoplastic tissues were labeled with iTRAQ reagents containing 114 and 115 reporter ions, while peptides from tumor tissues were labeled with iTRAQ reagents containing 116 and 117 reporter ions. The labeled samples were then combined, diluted 4-fold and the pH was adjusted to 2.7 with 10 mM KH2PO4, 20% acetonitrile solution. The pooled sample was then subjected to strong cation exchange chromatography using a PolySULFOETHYL A column (200 × 2.1 mm, 5 μm, 200 Å, PolyLC Inc., catalog#202SE0502). Peptides were loaded onto the column at a flow rate of 200 μl/min and eluted using a 30 min gradient from 8–50% of 10 mM KH2PO4, 350 mM KCl, 20% acetonitrile solution. The fractions collected were dried, reconstituted in 0.1% TFA, desalted using C18 Stage Tips (3M Empore high-performance extraction disks) and stored at −20°C until further analysis.
Mass spectrometry and protein quantitation
LC-MS/MS analysis was carried out on a 6520 Accurate Mass Quadrupole Time-of-Flight (QTOF) mass spectrometer (Agilent Technologies, Santa Clara, California, USA) interfaced with an HPLC chip cube system (Agilent Technologies, catalog # G4240-62001). Samples were injected onto the enrichment column using Agilent 1200 series capillary liquid chromatography system fitted with an autosampler. For the enrichment step, an injection flush volume of 4 μl was used. The peptides were eluted using a gradient of 3-40% of 90% acetonitrile containing 0.1% formic acid at a flow rate of 400 nl/min for 30 min. Data-dependent acquisition was carried out using Mass Hunter software. A precursor survey scan was carried out for 1 second (from 350–1,800 m/z) followed by 3 MS/MS scans at a rate of 3 spectra per second (50–2,000 m/z). Three most intense ions with a charge state preference, in the order +2, +3, +4, were used for fragmentation. The precursor ions selected to be fragmented were excluded for next 30 sec. MS/MS data (.d files) were processed to generate mascot generic format (.mgf) files using Mascot distiller (Matrix Science Inc., version 2.4.0). The data were then searched against human RefSeq Build 45 protein sequence database (containing 34,349 sequences with common contaminants) using Mascot (version 2.2) and Sequest search algorithms through Proteome Discoverer (Thermo Scientific, Bremen, Germany, version 1.3.0.339). The search criteria included oxidation of methionine as dynamic modification and methylthio modification of cysteine, iTRAQ 4-plex modification at peptide N-terminus and Lysine (K) as static modifications. For both the searches, precursor mass tolerance of 0.1 Da, fragment mass tolerance of 0.1 Da and one missed cleavage were allowed. The data were searched against a decoy database to calculate the false discovery rate (FDR) at the peptide level. For protein identification, a cutoff of 1% FDR at the peptide spectrum matches (PSMs) level was employed. Relative protein level between non-neoplastic and tumor was quantitated using Reporter Ions Quantifier node of Proteome Discoverer. Network generation and canonical pathway analysis was carried out using Ingenuity Pathway Analysis software. A data set containing differentially expressed genes with their corresponding identifiers and expression values was entered as input into the application. Every gene was mapped to its corresponding entry in Ingenuity's Knowledge Base using the accession numbers. This list of network eligible molecules was overlaid onto a global molecular network developed from information present in Ingenuity's Knowledge Base.
Immunohistochemical labeling
Immunohistochemical analysis for CAMKK2 was carried out on custom tissue microarrays generated by Dr. Juan Carlos Roa. Anti-CAMKK2 rabbit polyclonal antibody was procured from Sigma (catalog# HPA017389). Paraffin embedded sections of normal esophagus were used as controls for the analysis. These sections were obtained from the Kidwai Memorial Institute of Oncology, Bangalore. The immunohistochemical staining was carried out as previously described.47 Briefly, formalin fixed paraffin-embedded tissue sections were deparaffinized followed by heat-induced antigen retrieval in citrate buffer containing 0.05% Tween20, pH6.0 for 10 min. Endogenous peroxidases were quenched using a blocking solution consisting of hydrogen peroxide. The tissue sections were incubated with anti-CAMKK2 rabbit polyclonal antibody (1:100) at 4°C overnight. Normal esophagus sections used as positive control were stained with the primary antibody while those used as negative control were treated with antibody diluent. The tissue sections were subsequently washed with PBS buffer and treated with goat anti-rabbit secondary antibody conjugated with horse radish peroxidase. Excess secondary antibody was washed off using PBS followed by addition of 3,3′-diaminobenzidine peroxidase substrate (Vector Laboratories Inc., catalog# SK-4100). Once the signal developed, the reaction was stopped by rinsing the tissue sections with water. The slides were counter-stained with hematoxylin, mounted and observed under the microscope. The immunohistochemical labeling was assessed by an experienced pathologist (RVK) and staining intensity was scored as negative (0), weak (1+), moderate (2+) and strong (3+). A Chi-square test was carried out to determine the significance of the immunohistochemistry results.
Cell culture
Gastric cancer cell lines, AGS, KATO-III, NCI-N87, and SNU-16 were obtained from ATCC (Manassas, VA). All cell lines were grown in RPMI medium (Gibco, Grand Island, NY) supplemented with 10% FBS and were maintained at 37°C in humidified atmosphere with 5% CO2.
Western blot analysis
Gastric cancer cells were cultured in their specified media. Whole cell extracts were prepared using RIPA buffer (10 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton-X-100, 0.1% SDS) containing protease inhibitors (Roche, Indianapolis, IN). Rabbit polyclonal anti-CAMKK2 antibody was obtained from Sigma (St. Louis, MO; catalog # HPA017389). Total AMPKα (#2532), Phospho-AMPKα (Thr172) (#2535), FoxO3a (#2497), Phospho-FoxO3a (Ser413) (#8174) and GAPDH (#2118) antibodies were obtained from Cell Signaling Technology (Cell Signaling Technology, Beverly, MA). Western blot analysis was performed as previously described48 using 30 μg protein lysate.
siRNA transfection
ON-TARGETplus SMARTpool control siRNA and CAMKK2 siRNA were obtained from Dharmacon (Lafayette, CO). The adherent cell lines AGS and NCI-N87 were transfected with RNAiMAX (Invitrogen, Grand Island, NY) following manufacturer's instructions. Transfection was carried out as previously described.49 Cells were subjected to immunoblot analysis, invasion assay and viability assay 48 hrs post-transfection, unless otherwise stated.
siRNA transfection of suspension cell lines SNU-16 and KATO-III were carried out using nucleofection technology (Amaxa, Cologne, Germany) as previously described.50 Briefly, 1.5 × 106 cells were collected and suspended in 100 μl of Nucleofector solution V (AmaxaTM cell line optimization NuclefectorTM kit, Catalog number- VCO-1001N). The cells were transfected by electroporation using previously optimized electric pulse T20 of Nucleofector II TM (Amaxa Biosystems, Nattermannallee 1, 50829, Koeln Germany) according to the manufacturer's instructions.
Cell proliferation
AGS, KATO-III, NCI-N87 and SNU16 cells were seeded in 12-well plates (1.2 × 104 cells/well) and transfected with CAMKK2 or scrambled siRNA. Cells were counted every 24 hours post-transfection for 120 hours (5 days). All experiments were performed in triplicate and standard deviation was calculated.
Colony formation assays
Gastric cancer cell lines were transfected with CAMKK2 or scrambled siRNA. After 48h post transfection, 1 × 105 cells were seeded into 100-mm Petri dishes with complete media. The resulting colonies were fixed with methanol, and stained with Giemsa (Sigma, St. Louis, MO). The number of colonies per dish was counted. All experiments were performed in triplicate and standard deviation was calculated.
Invasion assays
Invasion assays were performed in a transwell system (BD Biosciences, San Jose, CA) with Matrigel-coated filters, and cellular invasion was evaluated after 72 hours as described.51 Briefly, invasive property of the cells was assayed in the membrane invasion culture system using PET membrane (8-μm pore size).The upper compartment of a transwell coated with Matrigel (BD BioCoat Matrigel Invasion Chamber). Cells were harvested using trypsin-EDTA buffer and washed with serum free media. The cells were seeded at density of 2.0 × 104 cells per 500 μl of serum free media on the Matrigel-coated PET membrane in the upper compartment. The lower compartment was filled with complete growth media and the plates were incubated at 37°C for 72 hours. Post incubation time, the upper surface of the membrane was wiped with a cotton-tip applicator to remove non migratory cells. Cells that migrated to the lower surface of membrane were fixed and stained by methylene blue. Each measurement was performed in duplicate and the experiments were repeated 3 times.
Availability of proteomic data
The data generated in the course of this study has been deposited in publicly available databases to make it accessible to the scientific community. The mass spectrometry data files were submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE public data repository.52 The data can be accessed using dataset identifier PXD000585. The immunohistochemistry data and lists of proteins and peptides identified in this study were also submitted to Human Proteinpedia18 (http://www.humanproteinpedia.org). The protein and peptide lists can be visualized using the link http://www.humanproteinpedia.org/data_display?exp_id=00697. The immunohistochemistry data for CAMKK2 can be visualized with the link http://www.humanproteinpedia.org/Experimental_details?exp_id=TE-245973 for adjacent non-neoplastic tissue and http://www.humanproteinpedia.org/Experimental_details?can_id=105421 for tumor tissue.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We also thank Dr SK Shankar and Dr Anita Mahadevan of National Institute of Mental Health and Neurological Sciences (NIMHANS), for providing access to the microscopy imaging facility. We also thank Agilent Technologies for access to instrumentation.
Funding
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics, Bangalore. T.S. Keshava Prasad and Akhilesh Pandey are supported by DBT Program Support on Neuroproteomics and infrastructure for proteomic data analysis (BT/01/COE/08/05) to IOB. This work was supported by NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036), an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520) and a contract (HHSN268201000032C) from the National Heart, Lung and Blood Institute. Dr. Harsha Gowda is a Wellcome Trust/DBT India Alliance Early Career Fellow. YS, NS, SR and SSM are recipients of Senior Research Fellowship from University Grants Commission (UGC), Government of India. SMP and JS are recipients of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. RR is a recipient of research associateship from Department of Biotechnology, Government of India.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: Cancer J Clin 2011; 61:69-90; PMID:21296855 [DOI] [PubMed] [Google Scholar]
- 2. Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg 2005; 241:27-39; PMID:15621988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roth AD. Curative treatment of gastric cancer: towards a multidisciplinary approach? Crit Rev Oncol Hematol 2003; 46:59-100; PMID:12672518; http://dx.doi.org/ 10.1016/S1040-8428(02)00160-9 [DOI] [PubMed] [Google Scholar]
- 4. Krejs GJ. Gastric cancer: epidemiology and risk factors. Digest Dis 2010; 28:600-3; PMID:21088409; http://dx.doi.org/ 10.1159/000320277 [DOI] [PubMed] [Google Scholar]
- 5. Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009; 9:28-39; PMID:19104514; http://dx.doi.org/ 10.1038/nrc2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 1996; 2:561-6; PMID:8616716; http://dx.doi.org/ 10.1038/nm0596-561 [DOI] [PubMed] [Google Scholar]
- 7. Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med 2001; 344:1038-42; PMID:11287973; http://dx.doi.org/ 10.1056/NEJM200104053441402 [DOI] [PubMed] [Google Scholar]
- 8. Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005; 353:123-32; PMID:16014882; http://dx.doi.org/ 10.1056/NEJMoa050753 [DOI] [PubMed] [Google Scholar]
- 9. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344:783-92; PMID:11248153; http://dx.doi.org/ 10.1056/NEJM200103153441101 [DOI] [PubMed] [Google Scholar]
- 10. Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010; 376:687-97; PMID:20728210; http://dx.doi.org/ 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 11. Sheng WQ, Huang D, Ying JM, Lu N, Wu HM, Liu YH, Liu JP, Bu H, Zhou XY, Du X. HER2 status in gastric cancers: a retrospective analysis from four Chinese representative clinical centers and assessment of its prognostic significance. Ann Oncol 2013; 24:2360-4; PMID:23788757; http://dx.doi.org/ 10.1093/annonc/mdt232 [DOI] [PubMed] [Google Scholar]
- 12. Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A 2006; 103:2316-21; PMID:16461907; http://dx.doi.org/ 10.1073/pnas.0508776103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 2012; 61:673-84; PMID:22315472; http://dx.doi.org/ 10.1136/gutjnl-2011-301839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sutton CW, Rustogi N, Gurkan C, Scally A, Loizidou MA, Hadjisavvas A, Kyriacou K. Quantitative proteomic profiling of matched normal and tumor breast tissues. J Proteome Res 2010; 9:3891-902; PMID:20560667; http://dx.doi.org/ 10.1021/pr100113a [DOI] [PubMed] [Google Scholar]
- 15. Chaerkady R, Harsha HC, Nalli A, Gucek M, Vivekanandan P, Akhtar J, Cole RN, Simmers J, Schulick RD, Singh S, et al. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J Proteome Res 2008; 7:4289-98; PMID:18715028; http://dx.doi.org/ 10.1021/pr800197z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen JS, Chen KT, Fan CW, Han CL, Chen YJ, Yu JS, Chang YS, Chien CW, Wu CP, Hung RP. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and iTRAQ labeling mass spectrometry. FEBS J 2010; 277:3028-38; PMID:20546304; http://dx.doi.org/ 10.1111/j.1742-4658.2010.07712.x [DOI] [PubMed] [Google Scholar]
- 17. Gagné J-P, Éthier C, Gagné P, Mercier G, Bonicalzi M-È, Mes-Masson A-M, Droit A, Winstall E, Isabelle M, Poirier GG. Comparative proteome analysis of human epithelial ovarian cancer. Proteome Sci 2007; 5:16; PMID:17892554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, et al. Human Proteinpedia enables sharing of human protein data. Nat Biotechnol 2008; 26:164-7; PMID:18259167; http://dx.doi.org/ 10.1038/nbt0208-164 [DOI] [PubMed] [Google Scholar]
- 19. Pawar H, Kashyap MK, Sahasrabuddhe NA, Renuse S, Harsha HC, Kumar P, Sharma J, Kandasamy K, Marimuthu A, Nair B, et al. Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery. Cancer Biol Ther 2011; 12:510-22; PMID:21743296; http://dx.doi.org/ 10.4161/cbt.12.6.16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Jiang J, Arellano M, Zhang L, Yan X, Wong DT, Hu S. Quantification of serum proteins of metastatic oral cancer patients using LC-MS/MS and iTRAQ labeling. Open Proteomics J 2008; 1:72; PMID:20485468; http://dx.doi.org/ 10.2174/1875039700801010072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chong PK, Lee H, Loh MC, Choong LY, Lin Q, So JB, Lim KH, Soo RA, Yong WP, Chan SP, et al. Upregulation of plasma C9 protein in gastric cancer patients. Proteomics 2010; 10:3210-21; PMID:20707004; http://dx.doi.org/ 10.1002/pmic.201000127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng L, Su T, Leng A, Zhang X, Xu M, Yan L, Gu H, Zhang G. Upregulation of soluble resistance-related calcium-binding protein (sorcin) in gastric cancer. Med Oncol 2010; 27:1102-8; PMID:19885748; http://dx.doi.org/ 10.1007/s12032-009-9342-5 [DOI] [PubMed] [Google Scholar]
- 23. Loei H, Tan HT, Lim TK, Lim KH, So JB-Y, Yeoh KG, Chung MC. Mining the gastric cancer secretome: identification of GRN as a potential diagnostic marker for early gastric cancer. J Proteome Res 2012; 11:1759-72; PMID:22204653; http://dx.doi.org/ 10.1021/pr201014h [DOI] [PubMed] [Google Scholar]
- 24. Marimuthu A, Subbannayya Y, Sahasrabuddhe NA, Balakrishnan L, Syed N, Sekhar NR, Katte TV, Pinto SM, Srikanth SM, Kumar P, et al. SILAC-based quantitative proteomic analysis of gastric cancer secretome. Proteomics Clin Appl 2013; 7:355-66; PMID:23161554; http://dx.doi.org/ 10.1002/prca.201200069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y, Lim SK, Choong LY, Lee H, Chen Y, Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG, et al. Cathepsin S mediates gastric cancer cell migration and invasion via a putative network of metastasis-associated proteins. J Proteome Res 2010; 9:4767-78; PMID:20812763; http://dx.doi.org/ 10.1021/pr100492x [DOI] [PubMed] [Google Scholar]
- 26. Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ. Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer 2007; 7:519-30; PMID:17585332; http://dx.doi.org/ 10.1038/nrc2171 [DOI] [PubMed] [Google Scholar]
- 27.Monteith GR, Davis FM, Roberts-Thomson SJ. Calcium channels and pumps in cancer: changes and consequences. J Biol Chem 2012; 287:31666-73. PMID: 22822055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berridge MJ. Calcium signalling and cell proliferation. Bioessays 1995; 17:491-500; PMID:7575490; http://dx.doi.org/ 10.1002/bies.950170605 [DOI] [PubMed] [Google Scholar]
- 29. Racioppi L. CaMKK2: a novel target for shaping the androgen-regulated tumor ecosystem. Trends Mol Med 2013; 19:83-8; PMID:23332598; http://dx.doi.org/ 10.1016/j.molmed.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soderling TR. The Ca-calmodulin-dependent protein kinase cascade. Trends Biochem Sci 1999; 24:232-6; PMID:10366852; http://dx.doi.org/ 10.1016/S0968-0004(99)01383-3 [DOI] [PubMed] [Google Scholar]
- 31. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251-62; PMID:22436748; http://dx.doi.org/ 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 2005; 280:29060-6; PMID:15980064; http://dx.doi.org/ 10.1074/jbc.M503824200 [DOI] [PubMed] [Google Scholar]
- 33. Karacosta LG, Foster BA, Azabdaftari G, Feliciano DM, Edelman AM. A regulatory feedback loop between Ca2+/Calmodulin-dependent protein kinase kinase 2 (CaMKK2) and the androgen receptor in prostate cancer progression. J Biol Chem 2012; 287:24832-43; PMID:22654108; http://dx.doi.org/ 10.1074/jbc.M112.370783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 2011; 30:2719-33; PMID:21602788; http://dx.doi.org/ 10.1038/emboj.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Q, Huang Q, Chen W, Wang L, Lin W, Lin J, Lin X. Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol 2008; 134:1219-27; PMID:18446369; http://dx.doi.org/ 10.1007/s00432-008-0398-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren H, Du N, Liu G, Hu H-T, Tian W, Deng Z-P, Shi J-S. Analysis of variabilities of serum proteomic spectra in patients with gastric cancer before and after operation. World J Gastroenterol 2006; 12:2789; PMID:16718772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang B, Li G-X, Zhang S-G, Wang Q, Wen Y-G, Tang H-M, Zhou C-Z, Xing A-Y, Fan J-W, Yan D-W. Biglycan expression correlates with aggressiveness and poor prognosis of gastric cancer. Exp Biol Med 2011; 236:1247-53; PMID:21998129; http://dx.doi.org/ 10.1258/ebm.2011.011124 [DOI] [PubMed] [Google Scholar]
- 38. Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 2011; 13:1016-23; PMID:21892142; http://dx.doi.org/ 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 2007; 282:30107-19; PMID:17711846; http://dx.doi.org/ 10.1074/jbc.M705325200 [DOI] [PubMed] [Google Scholar]
- 40. Telles E, Gurjar M, Ganti K, Gupta D, Dalal SN. Filamin A stimulates cdc25C function and promotes entry into mitosis. Cell Cycle 2011; 10:776-82; PMID:21325883; http://dx.doi.org/ 10.4161/cc.10.5.14954 [DOI] [PubMed] [Google Scholar]
- 41. Zhong Z, Yeow W-S, Zou C, Wassell R, Wang C, Pestell RG, Quong JN, Quong AA. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res 2010; 70:2105-14; PMID:20179208; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang XH, Zhang LH, Zhong XY, Xing XF, Liu YQ, Niu ZJ, Peng Y, Du H, Zhang GG, Hu Y, et al. S100A6 overexpression is associated with poor prognosis and is epigenetically up-regulated in gastric cancer. Am J Pathol 2010; 177:586-97; PMID:20581057; http://dx.doi.org/ 10.2353/ajpath.2010.091217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yong HY, Moon A. Roles of calcium-binding proteins, S100A8 and S100A9, in invasive phenotype of human gastric cancer cells. Arch Pharm Res 2007; 30:75-81; PMID:17328245; http://dx.doi.org/ 10.1007/BF02977781 [DOI] [PubMed] [Google Scholar]
- 44. Racioppi L, Means AR. Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J Biol Chem 2012; 287:31658-65; PMID:22778263; http://dx.doi.org/ 10.1074/jbc.R112.356485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Green MF, Anderson KA, Means AR. Characterization of the CaMKKbeta-AMPK signaling complex. Cell Signal 2011; 23:2005-12; PMID:21807092; http://dx.doi.org/ 10.1016/j.cellsig.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951; 193:265-75; PMID:14907713 [PubMed] [Google Scholar]
- 47. Marimuthu A, Jacob HKC, Jakharia A, Subbannayya Y, Keerthikumar S, Kashyap MK, Goel R, Balakrishnan L, Dwivedi S, Pathare S, et al. Gene expression profiling of gastric cancer. J Proteomics Bioinform 2011; 4:74-82; http://dx.doi.org/ 10.4172/jpb.1000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chatterjee A, Chang X, Nagpal JK, Chang S, Upadhyay S, Califano J, Trink B, Sidransky D. Targeting human 8-oxoguanine DNA glycosylase to mitochondria protects cells from 2-methoxyestradiol-induced-mitochondria-dependent apoptosis. Oncogene 2008; 27:3710-20; PMID:18246124; http://dx.doi.org/ 10.1038/onc.2008.3 [DOI] [PubMed] [Google Scholar]
- 49. Chang X, Ravi R, Pham V, Bedi A, Chatterjee A, Sidransky D. Adenylate kinase 3 sensitizes cells to cigarette smoke condensate vapor induced cisplatin resistance. PLoS One 2011; 6:e20806; PMID:21698293; http://dx.doi.org/ 10.1371/journal.pone.0020806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Toni F, Racaud-Sultan C, Chicanne G, Mas VM, Cariven C, Mesange F, Salles JP, Demur C, Allouche M, Payrastre B, et al. A crosstalk between the Wnt and the adhesion-dependent signaling pathways governs the chemosensitivity of acute myeloid leukemia. Oncogene 2006; 25:3113-22; PMID:16407823; http://dx.doi.org/ 10.1038/sj.onc.1209346 [DOI] [PubMed] [Google Scholar]
- 51. Ebrahimnejad A, Streichert T, Nollau P, Horst AK, Wagener C, Bamberger AM, Brummer J. CEACAM1 enhances invasion and migration of melanocytic and melanoma cells. Am J Pathol 2004; 165:1781-7; PMID:15509546; http://dx.doi.org/ 10.1016/S0002-9440(10)63433-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 2013; 41:D1063-9; PMID:23203882; http://dx.doi.org/ 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci 2003; 18:505-9; PMID:12923326; http://dx.doi.org/ 10.3346/jkms.2003.18.4.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


