Abstract
Amphetamine and methamphetamine addiction is described by specific behavioral alterations, suggesting long-lasting changes in gene and protein expression within specific brain subregions involved in the reward circuitry. Given the persistence of the addiction phenotype at both behavioral and transcriptional levels, several studies have been conducted to elucidate the epigenetic landscape associated with persistent effects of drug use on the mammalian brain. This review discusses recent advances in our comprehension of epigenetic mechanisms underlying amphetamine- or methamphetamine-induced behavioral, transcriptional, and synaptic plasticity. Accumulating evidence demonstrated that drug exposure induces major epigenetic modifications—histone acetylation and methylation, DNA methylation—in a very complex manner. In rare instances, however, the regulation of a specific target gene can be correlated to both epigenetic alterations and behavioral abnormalities. Work is now needed to clarify and validate an epigenetic model of addiction to amphetamines. Investigations that include genome-wide approaches will accelerate the speed of discovery in the field of addiction.
Keywords: amphetamine, DNA methylation, histone acetylation, histone methylation, methamphetamine
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPH
amphetamine
- AP1
activator protein 1
- ATF2
activating transcription factor 2
- BASP1
brain abundant signal protein 1
- BDNF
brain derived neurotrophic factor
- CCR2
C‒C chemokine receptor 2
- CoREST
restrictive element 1 silencing transcription factor corepressor
- Cp60
compound 60
- CPP
conditioned place preference
- ChIP
chromatin immunoprecipitation
- CREB
cAMP response element binding protein
- DNMT
DNA methyltransferase
- FOS
Finkel–Biskis–Jinkins murine osteosarcoma viral oncogene
- GABA
γ-aminobutyric acid
- GLUA1
glutamate receptor subunit A1
- GLUA2
glutamate receptor subunit A2
- GLUN1
glutamate receptor subunit N1
- H2Bac
pan-acetylation of histone 2B
- H3
histone 3
- H3K4
lysine 4 of histone 3
- H3K4me3
trimethylation of histone 3 at lysine 4
- H3K9
lysine 9 of histone 3
- H3K9Ac
acetylation of histone 3 at lysine 9
- H3K9me3
trimethylation of histone 3 at lysine 9
- H3K14Ac
acetylation of histone 3 at lysine 14
- H3K18
lysine 18 of histone 3
- H4
histone 4
- H4Ac
pan-acetylation of histone 4
- H4K5
lysine 5 of histone 4
- H4K8
lysine 8 of histone 4
- H4K12Ac
acetylation of histone 4 at lysine 12
- H4K16
lysine 16 of histone 4
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
histone demethylase
- HMT
histone methyltransferase
- IP
intra-peritoneal
- JUN
jun proto-oncogene
- KDM
lysine demethylase
- KLF10
Kruppel-like factor 10
- KMT
lysine methyltransferase
- MeCP2
methyl-CpG binding protein 2
- METH
methamphetamine
- NaB
sodium butyrate
- NAc
nucleus accumbens
- NMDA
N-methyl-D-aspartate
- OfC
orbitofrontal cortex
- PfC
prefrontal cortex
- REST
restrictive element 1 silencing transcription factor
- RNAi
RNA interference
- Ser241
serine 241
- Sin3A
SIN3 transcription regulator family member A
- siRNA
silencing RNA
- TSS
transcription start site
- VPA
valproic acid
- WT1
Wilms tumor protein 1
Introduction
The abuse of amphetamine (AMPH) and its clinically devastating derivative methamphetamine (METH) is widespread. Repeated exposure to these substances can lead to addiction. Addiction is a neuropsychiatric disorder thought to result from neural adaptations at the molecular, cellular, and tissular levels following repeated drug exposure. Current research on addiction to psychostimulants, mainly cocaine and substituted amphetamines, is trying to elucidate the mechanisms underlying such adaptive changes because they might explain the transition from recreational drug use to addicted behaviors. Drug-induced alterations in gene and subsequent protein expression appear necessary to account for drastic dysregulations of physiological brain processes.
All psychostimulants are known to transiently facilitate neurotransmission mediated by monoamines including dopamine, norepinephrine, and serotonin in specific brain circuits, notably the reward pathways.1 Reward pathways extend from the ventral tegmental area to the nucleus accumbens (NAc), the dorsal striatum, the hippocampus, and the prefrontal cortex (PfC) through dopaminergic projections.2 Because cocaine and amphetamines increase the amount of dopamine in the synaptic cleft through diverse mechanisms, it is possible that distinct molecular adaptations might occur in response to different psychostimulants. This sentence highlights the need for comprehensive studies of these drugs of abuse. Here, we focus our attention on data specific for AMPH and its highly addictive analog, METH.
AMPH and METH activate neurotransmitter release by reversing monoamine transport, whereas cocaine acts as a monoamine reuptake inhibitor.1 AMPH and METH display very similar molecular effects, notwithstanding some pharmacodynamic variations; for example, at the same dose, METH is described as a more potent stimulant.3 Nevertheless, because a transient excessive monoamine neurotransmission alone cannot account for drug-triggered long-lasting transcriptional and behavioral alterations, there is a need for a multi-systematic yet integrative elucidation of addiction and its consequences.4 Such integration will have to go beyond the dopamine hypothesis of addiction.
These long-lasting aspects of addiction indicate that epigenetic modifications might be key modulators of drug-induced gene expression, because epigenetic alterations can register and maintain durable structural chromatin adaptations.5 This realization has led researchers to try to elucidate the epigenetic landscape associated with psychostimulant addiction. To date, studies have predominantly focused on cocaine exposure,6 with less focus on METH addiction. This review summarizes the evidence linking AMPH or METH exposure to drug-induced epigenetic changes. Those include histone acetylation and methylation, as well as DNA methylation. We will discuss the results examining the effects of these drugs on the expression of histone acetyltransferases (HATs), deacetylases (HDACs), methyltransferases (HMTs/KMTs), demethylases (HDMs/KDMs), and DNA methyltransferases (DNMTs).
Histone acetylation
Histone acetylation remains by far the most studied chromatin modification in animal models of addiction.6 Extended literature has described the role of histone acetylation in cocaine-induced behaviors.7 Similar mechanisms may be involved in METH addiction.
Role of HDAC inhibitors in AMPH and METH addiction: behavioral evidence
A role for histone acetylation in METH addiction was first suggested by behavioral approaches. For instance, METH-induced conditioned place preference (CPP) is associated with increased H3 but not H4 acetylation in the limbic forebrain.8 Models of METH addiction in rodents have demonstrated that METH triggers enhanced locomotor activity, accompanied by behavioral sensitization.9 To assess how histone acetylation might interfere with both phenomena, rodents were treated with either AMPH or METH in conjunction with HDAC inhibitors, usually valproic acid (VPA) and sodium butyrate (NaB). Table 1 lists the experimental details of selected HDAC inhibitors treatments. Studies using either intraperitoneal 10,11 or micro-injections in the amygdala,12 the PfC, 12 and the striatum 12,13 have reported that non-specific HDAC inhibitors can attenuate drug-induced locomotor activity. More interestingly, Cp60, a HDAC1/2-specific inhibitor, is sufficient to prevent AMPH-induced locomotor activity after a single injection,14 suggesting a central role for HDAC1/2 in this behavioral response.
Table 1.
Behavioral and biochemical consequences of HDACs inhibitors and AMPH/METH administration.
| Reference | Drug | HDACs inhibitor(s) | Tested behavior | Administration and testing schedule | Effect of HDACs inhibitors | Global biochemical alterations |
|---|---|---|---|---|---|---|
| Frey et al., 2006 | AMPH IP | VPA IP, 2x/d | Locomotor hyperactivity | Prevented | / | |
| Reversed | ||||||
| Arent et al., 2011 | METH IP | NaB / VPA Micro-injections | Locomotor hyperactivity | Attenuated | / | |
| Schroeder et al., 2013 | AMPH IP | Cp60 SAHA IP | Locomotor hyperactivity | Cp60: prevented SAHA: no effect | PfC, ventral striatum, hippocampus: ↑H2BAc, ↑H3K9Ac, ↑H4K12Ac | |
| Kalda et al., 2007 | AMPH IP | NaB / VPA IP | Acquisition of behavioral sensitization | Enhanced | Striatum: ↑H4K12Ac with additive effects | |
| Maintenance of behavioral sensitization | Attenuated | / | ||||
| Coccurello et al., 2007 | METH IP | VPA IP | Acquisition of behavioral sensitization | Attenuated | / | |
| Harkness et al., 2013 | METH IP | BA IP | Acquisition of behavioral sensitization | Enhanced | NaB reverts METH-induced H3K14Ac | |
| Maintenance of behavioral sensitization | Enhanced | / |
Administration schedules are pictured with the following symbols: a black square corresponds to one daily drug injection, a white square to one daily HDACs inhibitor injection, and the red arrows indicate each behavioral test. Abbreviations: IP, intra-peritoneal injection. NaB, sodium butyrate; VPA, valproic acid.
Surprisingly, treatments with HDAC inhibitors showed apparently discrepant consequences both on the acquisition and the maintenance of METH- or AMPH-induced locomotor behavioral sensitization15-19 with animals exhibiting distinct behavioral responses to distinct experimental regimens. Non-specific pharmacological approaches have also resulted in divergent conclusions in the case of cocaine-injected animals.7 Assessing the role of each specific HDAC by viral overexpression or genetic knockout may be essential to a better understanding of drug-induced epigenetic adaptations. In particular, class I and class II HDACs seem to play distinct roles, while NaB and VPA have been identified in vitro as potential inhibitors of class I but not class II HDACs.20 In addition, VPA, but not NaB, has been reported as a regulator of GABAergic signaling, which in turn modulates the activity of dopamine neurons,21 thus complicating the use of non-specific pharmacological agents.
HDACs inhibitors can cause additive increasing effects on METH- or AMPH-induced histone acetylation in the striatum, notably on H4 acetylation.15,17 In contrast, NaB exhibits weaker additive effects compared to VPA, or even some opposite effects.18 These discrepant results may be explained by different dosing regimens, diverse behavioral testing paradigms, or multiple biochemical targets in the brain. Importantly, METH or AMPH use may increase global acetylation in the striatum.22 HDACs inhibitors could potentiate these drug effects, while having variable consequences on drug-elicited behavioral responses.
Regulation of HDACs expression, histone acetylation, and transcriptional response
Acetylation of H3 and H4 appears to play a central role in drug-induced transcriptional responses. Specifically, a single METH injection was reported to induce global time-dependent increases in acetylated H4K5 and H4K8, but a global time-dependent decrease in H3K9, H3K18, and H4K16 acetylation in the NAc.23 This study also correlated patterns of histone acetylation with a METH-induced decrease in HDAC1, but an increase in HDAC2 and ATF2 protein levels. Thus, H4K5 and H4K8 hyperacetylation could have resulted from both a prior METH-induced increase in ATF2 expression and a decrease in HDAC1 expression, since RNAi-mediated knockdown against HDAC1 was shown to increase H4K5 acetylation.24 In contrast, increased HDAC2 expression, which also accompanies HDAC1 decrease after RNAi treatment, putatively as a compensatory mechanism,24 may account for H3K9, H3K18, and H4K16 hypoacetylation. This study suggests that METH differentially modulates the expression of HDAC1, HDAC2, and ATF2 in the NAc, with the resulting pattern of histone acetylation differentially regulating the expression of many genes.23 The time course expression of class I HDAC1 and HDAC2, as well as class II HDAC4 and HDAC5, revealed surprising results in the PfC,25 suggesting a unique role for the PfC or NAc in addiction. In the PfC, HDAC1 mRNA level appears reduced after acute METH treatment, similar to the decreases observed in the NAc.23 HDAC1 expression is, however, not affected after chronic treatment or withdrawal. On the other hand, HDAC2 expression in the PfC was decreased after both acute and chronic METH injections.25 HDAC4 and HDAC5 were decreased only after withdrawal, while global HDAC activity is increased.25 These complex results highlight a potential shift from the involvement of class I to class II HDACs during withdrawal.25 Similarly, models of cocaine addiction have hypothesized opposite roles for class I and class II HDACs, as evidenced by behavioral studies. For example, class I HDAC1/2 are thought to enhance cocaine effects,26 whereas overexpression of class II HDAC5 in the NAc is reported to inhibit cocaine-induced CPP.27
Genome-wide analysis using a ChIP-Sequencing approach gives access to precise patterns of histone acetylation, and allows for a better comparison between regulation of histone acetylation and gene expression. Using this approach, Cadet et al.22 have reported that acute METH injection induces H4K5 acetylation around the transcription start sites (TSSs) of genes in the dorsal striatum. This results are consistent with previous global results15,17 and reflect changes in gene expression.23 Similar positive correlation between H4K5 acetylation in the TSS and gene expression was found for chronic METH treatment,22 although chronically regulated genes are different from acutely regulated ones. However, the correlation appeared weaker for chronically treated animals, suggesting that METH-induced novel H4K5 acetylation might be necessary but not sufficient to maintain transcriptional changes in gene expression. Microarray analysis also demonstrated that acute METH mainly causes a global increase in gene expression, whereas chronic METH is linked with a global decrease.22 Global downregulation after chronic METH could correlate with the observation that H4K5 acetylation does not necessarily elicit significant changes in gene expression. Thus, combinatorial epigenetic influences could involve in transcriptional regulation after METH treatment.
Microarray analysis has also identified putative proteins that might interact with HDACs and consequently account for complex regulation of chromatin structure. In a long-access model of METH self-administration, the drug was shown to affect the expression of genes that act in complexes with HDACs as either activators or repressors.28 For example, the study reported increased brain abundant signal protein 1 (basp1) mRNA level after METH self-administration. BASP1 can co-repress WT1 targets in the NAc by recruiting HDAC1.29 In a similar fashion, the protein product of Kruppel-like factor 10 (klf10) gene that was also upregulated by METH self-administration contains a R1 domain that allows interaction with the HDAC co-repressor, Sin3A.30 The striatal upregulation of these 2 transcripts could lead to decreased transcription due to HDAC-mediated histone hypoacetylation.28
Transcriptional regulation of members of the AP1 complex through histone acetylation
Activator Protein-1 (AP1) complexes work as transcription factors that can regulate gene expression in response to numerous physiological and pathological stimuli.31 Some members of these complexes, including JunD and ΔfosB, which is a splicing variant of FosB, appear to be crucial regulators of responses to rewarding stimuli.32 Specifically, drug-induced ΔfosB accumulates in the striatum due to its high stability, leading to the speculation that this accumulation might be responsible, in part, for the transcriptional differences observed after acute and chronic drug use.33 Indeed, acute METH was found to increase FosB/ΔfosB, c-Fos, c-Jun and JunB mRNA levels the striatum, whereas chronic METH appeared to blunt this effect.28,34 Consistently with their higher stability, protein levels of FosB/ΔfosB are reported to be increased after METH self-administration in various brain regions.35
Histone acetylation may also play a role at different steps of these processes. First, H4 acetylation in the striatum appears to be an important regulator of fosB gene expression after chronic AMPH treatment, in a fashion consistent with increased ΔfosB expression.17 Authors also reported that AMPH increases striatal CREB phosphorylation, thus reducing CREB:HDAC1 interaction and recruitment onto the promoter region of ΔfosB.17 Histone acetylation could also explain the blunting of c-fos expression that occurs with chronic drug exposure (see Fig. 1). In fact, ChIP experiments in the striatum have revealed that chronic AMPH treatment increases ΔfosB binding onto c-fos promoter and increases ΔfosB-mediated HDAC1 recruitment.36 HDAC activity could have led to c-fos promoter hypo-acetylation and subsequent reduced c-fos expression. Viral overexpression of either ΔfosB or HDAC1 confirmed those results.36 Therefore, c-fos desensitization after chronic AMPH exposure appears to be mediated by ΔfosB and HDAC1. More studies are thus needed to test whether this mechanism is involved in the blunting of other neuroplasticity genes. These investigations should provide a partial window to the molecular adaptations underlying the transition of recreational use to compulsive abuse.
Figure 1.
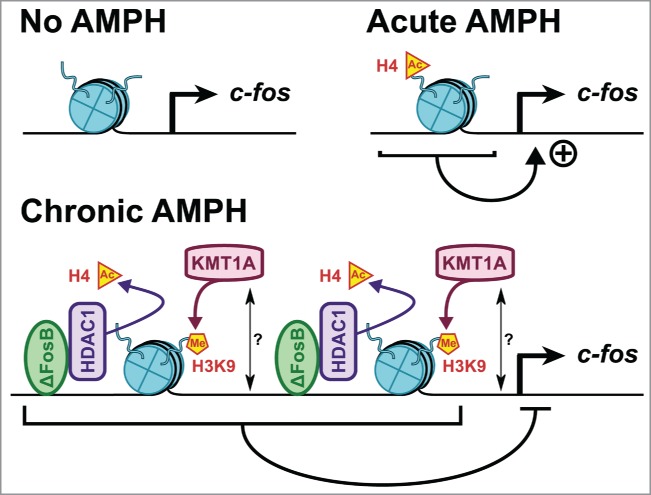
Epigenetic desensitization of c-fos after chronic AMPH treatment. Striatal c-fos expression is induced by an acute drug challenge but blunted after chronic AMPH exposure. This transcriptional desensitization correlates with increased binding of ΔfosB and HDAC1 recruitment onto the c-fos promoter. There is coincident increased KMT1A expression after chronic AMPH. Together, these enzymes reshape surrounding chromatin into a repressive conformation for c-fos transcription by catalyzing H4 deacetylation and H3K9 methylation, resulting in blunted c-fos response to acute drug challenge.36
Histone methylation
Although histone methylation has not been well studied in addiction models,6 this modification may modulate gene expression in a much finer way because histone methylation can promote either activation or inhibition of transcription.37 Trimethylation of histone H3 at lysine 4 (H3K4me3), which is usually associated with active transcription, was found to be increased in the NAc in models of METH-induced behavioral sensitization 38 and CPP.39 This increase in H3K4me3 occurred on the promoter of C-C chemokine receptor 2 (ccr2) gene during behavioral sensitization.38 Interestingly, ccr2 knockout mice showed impaired maintenance of behavioral sensitization to METH.38 H3K4me3 is also linked with the upregulation of several genes in the NAc during METH-associated memory formation, including c-fos.39 Using siRNA-mediated approaches, Aguilar-Valles et al. 39 also reported that KMT2A, an enzyme involved in histone H3 trimethylation at K4, was upregulated with chronic METH injections and was also necessary for METH-associated memory formation and maintenance. Another enzyme, KDM5C, which demethylates H3K4, was also involved in the maintenance of METH CPP.39 Taken together, these studies suggest that chronic METH may specifically alter the abundance of H3K4me3 on the promoter of genes responsible for long-lasting adaptations.
It is important to note that METH self-administration was not associated with increased H3K4me3 abundance on the promoters of c-fos and fosB in the dorsal striatum.40 Contradictory results could be explained by distinct roles of dorsal and ventral striatum or by different neural adaptations to non-contingent or contingent injections of the drug. It is also to be noted that desensitization of the c-fos gene after chronic AMPH treatment (see Fig. 1) is correlated with increased expression of KMT1A and repressive H3K9me3 on its promoter.36 Crosstalks between permissive (H3K4me3) and repressive (H3K9me3, hypoacetylated histones) marks might also explain time-dependent variations of epigenetic modifications on various drug-related genes. An interesting candidate for such interactions is KLF10, which is upregulated in the striatum after METH self-administration.28 KLF10 interacts with KDM5B, a histone demethylase (HDM) for H3K4me3,41 in addition to its interaction with HDAC:Sin3a complex.30 METH-induced increase in HDM activity might serve as a compensatory response to METH-induced H3K4me3,38,39 thus resulting in a lack of changes in H3K4me3 on the promoter of several genes even in the presence of increased H3K4me3 protein levels.40 It is also likely that genome-wide approaches to H3K4me3 binding might identify additional genes not identified by single genes studies.
DNA methylation
DNA methylation has not been extensively investigated in models of METH or AMPH addiction. The maintenance DNMT, DNMT1, is highly expressed in the mouse brain.42 METH was found to differentially modulate DNMT1 expression in the NAc of 2 strains of rats. Fisher 344/N rats exhibit increased DNMT1 expression associated with resistance to METH-induced behavioral sensitization whereas Lewis/N rats show reduced DNMT1 expression and enhanced behavioral sensitization.43 This study had indicated that different genotypes may respond differentially to METH administration. After two weeks of withdrawal from chronic AMPH exposure, DNA methylation was found globally increased in the NAc, in the medial PfC and the orbitofrontal cortex (OfC).44 Such increase in DNA methylation is consistent with a global decrease of transcription after one month of METH withdrawal, as demonstrated with microarray analysis.28 Strikingly, OfC and medial PfC both exhibit a decrease in global DNA methylation pattern, even though they show opposite cellular morphological adaptations.45
Methyl-CpG-binding protein-2 (MeCP2), a “reader” of DNA methylation, is involved in cocaine addiction,46,47 and is also thought to participate in AMPH-triggered behaviors.48,49 Using viral NAc-specific knockdown and overexpression of MeCP2, Deng et al. 48 showed that MeCP2 limits rewarding properties of AMPH as evaluated by CPP. Hypomorphic mutant mice expressing a truncated MeCP2, on the contrary, exhibit no CPP for AMPH.48 These results show that MeCP2 may have distinct, even opposite, roles in the NAc and in other structures. Acute AMPH induces a transient and NAc-specific phosphorylation of MeCP2 at Ser241 and reduces the inhibitory activity of MeCP2.48 MeCP2 phosphorylation is known to regulate brain derived neurotrophic factor (bdnf) expression, a gene important for synaptic plasticity.50 Interestingly, BDNF levels were found to be increased in hippocampus of METH self-administering rats 51 as well as in the plasma of human METH users,52 suggesting that METH-induced changes in MeCP2 phosphorylation may be involved. All taken together, these data suggest that drug exposure activates the inhibitory phosphorylation of MeCP2 in the NAc, which may result in the derepression of various target genes, including bdnf. Consistent with a role of MeCP2 phosphorylation in addiction, Deng et al.49 reported recently that mice expressing a non-phosphorylatable form of MeCP2 display both a reduced threshold for the induction of AMPH-triggered locomotor sensitization and an increased sensitivity to self-administered cocaine.
A recent study has also investigated the effects of METH exposure on the offspring of chronically METH-treated male and female rodents.53 Both methylated-DNA-immunoprecipitation and bisulfite-sequencing approaches demonstrated that hippocampal DNA methylation patterns are altered in the offspring. These observations suggest that in utero exposure to METH can interfere with the epigenetic reprogramming that occurs during embryonic development.54 Interestingly, the offspring exhibited behavioral abnormalities, such as reduced cocaine-CPP for males and reduced response to conditioned fear for both males and females.53
Epigenetic bases of METH-induced alterations in glutamatergic plasticity
The downregulation of glutamate receptors expression in the dorsal striatum following chronic exposure to METH probably stands as one of the best-described example of complex drug-induced epigenetic modifications in the brain.55,56 Psychostimulant drugs produce plastic changes in the striatum that include changes in the expression of AMPA and NMDA glutamate receptors.57 In the case of METH, Jayanthi et al. 55 described epigenetic crosstalks that regulate drug-induced transcriptional downregulation of AMPA and NMDA receptors subunits that included GluA1, GluA2, and GluN1. Downregulation of GluA1 and GluA2 involves METH-induced binding of repressive chromatin remodeling complexes onto the upstream repressive sequence of GluA1 and onto the promoters of GluA2 and GluN1.55,56 Figure 2 depicts these complexes and consequent epigenetic alterations. It is important to note that VPA was found to reduce METH-induced downregulation of these 3 genes by inhibiting HDAC1/2 activity.55,56
Figure 2.
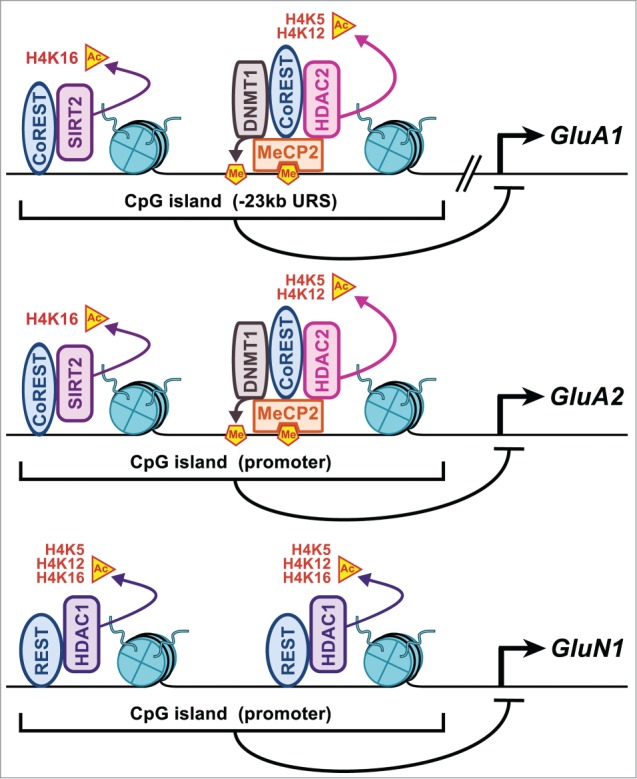
Chronic METH exposure down-regulates GluA1/A2 and GluN1 expression via diverse epigenetic modifications. In the dorsal striatum, chronic METH exposure allows a MeCP2-independent binding of a CoREST:SIRT2 complex and a MeCP2-dependent recruitment of CoREST, HDAC2 and DNMT1 onto the Upstream Repressive Sequence (URS) of GluA1 and on the promoter of GluA2. Together, these enzymes reshape surrounding chromatin into a repressive conformation for gene transcription by catalyzing H4 deacetylation. GluN1 is downregulated through H4 deacetylation on its promoter mediated by a REST:HDAC1 complex.55,56
Concluding remarks
Addiction to AMPH and METH appears to be related to complex epigenetic and transcriptional changes that occur after repeated exposure to these drugs. These alterations include posttranslational histone modifications and DNA methylation. Crosstalks between distinct histone modifications and changes in chromatin structures may serve as determining factors in regulating the expression of gene networks and their protein products. Within these networks, specific genes may trigger subsequent epigenetic cascades that maintain the complex behavioral syndromes that have been labeled addiction. Elucidation of the initial steps involved in the addiction cycle may help to develop preventive pharmacological measures against the progression to compulsive drug taking. On the other hand, characterizing later limbs of addiction could generate therapeutic approaches that may help drive the brain back to homeostasis. These agents might help patients to refrain from further drug use, hence preventing the coincident adverse consequences of AMPH and METH addiction. Because some of the biochemical effects of these 2 drugs appear to be dissimilar, it will be important to study the corresponding epigenetic and transcriptional responses under behavioral conditions that are similar. These types of studies will help to dissect further the molecular substrates of addiction to these 2 psychostimulants, and to identify specific targets for therapeutic interventions. Our present discussion suggests that the development of epigenetic drugs that influence the functions of HDACs, KMTs, and DNMTs might offer important opportunities for future research that goes beyond the dopamine hypothesis of addiction. Finally, the data reviewed in this paper also suggest that more aggressive efforts are needed to translate these observations into clinical practice.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by funds of the Intramural Research Program of the US Department of Health and Human Services/National Institutes of Health/National Institute on Drug Abuse.
References
- 1.Howell LL, Kimmel HL. Monoamine transporters and psychostimulant addiction. Biochem Pharmacol 2008; 75:196-217; PMID:17825265; http://dx.doi.org/ 10.1016/j.bcp.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 2.Schultz W. Multiple reward signals in the brain. Nat Rev Neurosci 2000; 1:199-207; PMID:11257908; http://dx.doi.org/ 10.1038/35044563 [DOI] [PubMed] [Google Scholar]
- 3.Goodwin JS, Larson GA, Swant J, Sen N, Javitch JA, Zahniser NR, Felice LJD, Khoshbouei H. Amphetamine and methamphetamine differentially affect dopamine transporters in vitro and in vivo. J Biol Chem 2009; 284:2978-89; PMID:19047053; http://dx.doi.org/ 10.1074/jbc.M805298200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadet JL, Krasnova IN, Jayanthi S, Lyles J. Neurotoxicity of substituted amphetamines: molecular and cellular mechanisms. Neurotox Res 2007; 11:183-202; PMID:17449459; http://dx.doi.org/ 10.1007/BF03033567 [DOI] [PubMed] [Google Scholar]
- 5.Bird A. Perceptions of epigenetics. Nature 2007; 447:396-8; PMID:17522671; http://dx.doi.org/ 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- 6.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology 2014; 76 Pt B:259-68; PMID:23643695; http://dx.doi.org/ 10.1016/j.neuropharm.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 2013; 38:94-110; http://dx.doi.org/ 10.1038/npp.2012.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibasaki M, Mizuno K, Kurokawa K, Ohkuma S. L-type voltage-dependent calcium channels facilitate acetylation of histone H3 through PKCγ phosphorylation in mice with methamphetamine-induced place preference. J Neurochem 2011; 118:1056-66; PMID:21781114; http://dx.doi.org/ 10.1111/j.1471-4159.2011.07387.x [DOI] [PubMed] [Google Scholar]
- 9.Jing L, Zhang M, Li J-X, Huang P, Liu Q, Li Y-L, Liang H, Liang J-H. Comparison of single versus repeated methamphetamine injection induced behavioral sensitization in mice. Neurosci Lett 2014; 560:103-6; PMID:24361545; http://dx.doi.org/ 10.1016/j.neulet.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey BN, Andreazza AC, Ceresér KMM, Martins MR, Valvassori SS, Réus GZ, Quevedo J, Kapczinski F. Effects of mood stabilizers on hippocampus BDNF levels in an animal model of mania. Life Sci 2006; 79:281-6; PMID:16460767; http://dx.doi.org/ 10.1016/j.lfs.2006.01.002 [DOI] [PubMed] [Google Scholar]
- 11.Moretti M, Valvassori SS, Varela RB, Ferreira CL, Rochi N, Benedet J, Scaini G, Kapczinski F, Streck EL, Zugno AI, et al.. Behavioral and neurochemical effects of sodium butyrate in an animal model of mania. Behav Pharmacol 2011; 22:766-72; PMID:21989497; http://dx.doi.org/ 10.1097/FBP.0b013e32834d0f1b [DOI] [PubMed] [Google Scholar]
- 12.Arent CO, Valvassori SS, Fries GR, Stertz L, Ferreira CL, Lopes-Borges J, Mariot E, Varela RB, Ornell F, Kapczinski F, et al.. Neuroanatomical profile of antimaniac effects of histone deacetylases inhibitors. Mol Neurobiol 2011; 43:207-14; PMID:21424678; http://dx.doi.org/ 10.1007/s12035-011-8178-0 [DOI] [PubMed] [Google Scholar]
- 13.Kim WY, Kim S, Kim J-H. Chronic microinjection of valproic acid into the nucleus accumbens attenuates amphetamine-induced locomotor activity. Neurosci Lett 2008; 432:54-7; PMID:18164815; http://dx.doi.org/ 10.1016/j.neulet.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 14.Schroeder FA, Lewis MC, Fass DM, Wagner FF, Zhang Y-L, Hennig KM, Gale J, Zhao W-N, Reis S, Barker DD, et al.. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PloS One 2013; 8:e71323; PMID:23967191; http://dx.doi.org/ 10.1371/journal.pone.0071323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalda A, Heidmets L-T, Shen H-Y, Zharkovsky A, Chen J-F. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav Brain Res 2007; 181:76-84; PMID:17477979; http://dx.doi.org/ 10.1016/j.bbr.2007.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coccurello R, Caprioli A, Ghirardi O, Virmani A. Valproate and acetyl-L-carnitine prevent methamphetamine-induced behavioral sensitization in mice. Ann N Y Acad Sci 2007; 1122:260-75; PMID:18077579; http://dx.doi.org/ 10.1196/annals.1403.019 [DOI] [PubMed] [Google Scholar]
- 17.Shen H-Y, Kalda A, Yu L, Ferrara J, Zhu J, Chen J-F. Additive effects of histone deacetylase inhibitors and amphetamine on histone H4 acetylation, cAMP responsive element binding protein phosphorylation and DeltaFosB expression in the striatum and locomotor sensitization in mice. Neuroscience 2008; 157:644-55; PMID:18848971; http://dx.doi.org/ 10.1016/j.neuroscience.2008.09.019 [DOI] [PubMed] [Google Scholar]
- 18.Harkness JH, Hitzemann RJ, Edmunds S, Phillips TJ. Effects of sodium butyrate on methamphetamine-sensitized locomotor activity. Behav Brain Res 2013; 239:139-47; PMID:23137698; http://dx.doi.org/ 10.1016/j.bbr.2012.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J-X, Han R, Deng Y-P, Chen S-Q, Liang J-H. Different effects of valproate on methamphetamine- and cocaine-induced behavioral sensitization in mice. Behav Brain Res 2005; 161:125-32; PMID:15904719; http://dx.doi.org/ 10.1016/j.bbr.2005.01.015 [DOI] [PubMed] [Google Scholar]
- 20.Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 2010; 35:870-80; http://dx.doi.org/ 10.1038/npp.2009.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens MJ, Nemeroff CB. Pharmacology of valproate. Psychopharmacol Bull 2003; 37 Suppl 2:17-24; PMID:14624230 [PubMed] [Google Scholar]
- 22.Cadet JL, Jayanthi S, McCoy MT, Ladenheim B, Saint-Preux F, Lehrmann E, De S, Becker KG, Brannock C. Genome-wide profiling identifies a subset of methamphetamine (METH)-induced genes associated with METH-induced increased H4K5Ac binding in the rat striatum. BMC Genomics 2013; 14:545; PMID:23937714; http://dx.doi.org/ 10.1186/1471-2164-14-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, Lehrmann E, Becker KG, Cadet JL. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PloS One 2012; 7:e34236; PMID:22470541; http://dx.doi.org/ 10.1371/journal.pone.0034236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319:110-20; PMID:18501342; http://dx.doi.org/ 10.1016/j.ydbio.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Li F, Wu N, Su R-B, Li J. Methamphetamine induces dynamic changes of histone deacetylases in different phases of behavioral sensitization. CNS Neurosci Ther 2014; 20:874-6; PMID:24954603; http://dx.doi.org/ 10.1111/cns.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han M-H, et al.. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci 2013; 16:434-40; PMID:23475113; http://dx.doi.org/ 10.1038/nn.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al.. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 2007; 56:517-29; PMID:17988634; http://dx.doi.org/ 10.1016/j.neuron.2007.09.032 [DOI] [PubMed] [Google Scholar]
- 28.Cadet JL, Brannock C, Jayanthi S, Krasnova IN. Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat. Mol Neurobiol 2014; http://dx.doi.org/ 10.1007/s12035-014-9040-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SGE. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep 2012; 2:462-9; PMID:22939983; http://dx.doi.org/ 10.1016/j.celrep.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol 2001; 21:5041-9; PMID:11438660; http://dx.doi.org/ 10.1128/MCB.21.15.5041-5049.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 2004; 117:5965-73; PMID:15564374; http://dx.doi.org/ 10.1242/jcs.01589 [DOI] [PubMed] [Google Scholar]
- 32.Pitchers KK, Vialou V, Nestler EJ, Laviolette SR, Lehman MN, Coolen LM. Natural and Drug Rewards Act on Common Neural Plasticity Mechanisms with ΔFosB as a Key Mediator. J Neurosci 2013; 33:3434-42; PMID:23426671; http://dx.doi.org/ 10.1523/JNEUROSCI.4881-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nestler EJ, Barrot M, Self DW. ΔFosB: A sustained molecular switch for addiction. Proc Natl Acad Sci 2001; 98:11042-6; PMID:11572966; http://dx.doi.org/ 10.1073/pnas.191352698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy MT, Jayanthi S, Wulu JA, Beauvais G, Ladenheim B, Martin TA, Krasnova IN, Hodges AB, Cadet JL. Chronic methamphetamine exposure suppresses the striatal expression of members of multiple families of immediate early genes (IEGs) in the rat: normalization by an acute methamphetamine injection. Psychopharmacology (Berl) 2011; 215:353-65; PMID:21229349; http://dx.doi.org/ 10.1007/s00213-010-2146-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornish JL, Hunt GE, Robins L, McGregor IS. Regional c-Fos and FosB/ΔFosB expression associated with chronic methamphetamine self-administration and methamphetamine-seeking behavior in rats. Neuroscience 2012; 206:100-14; PMID:22266344; http://dx.doi.org/ 10.1016/j.neuroscience.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 36.Renthal W, Carle TL, Maze I, Covington HE, Truong H-T, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci Off J Soc Neurosci 2008; 28:7344-9; http://dx.doi.org/ 10.1523/JNEUROSCI.1043-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074-80; PMID:11498575; http://dx.doi.org/ 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 38.Ikegami D, Narita M, Imai S, Miyashita K, Tamura R, Narita M, Takagi S, Yokomizo A, Takeshima H, Ando T, et al.. Epigenetic modulation at the CCR2 gene correlates with the maintenance of behavioral sensitization to methamphetamine. Addict Biol 2010; 15:358-61; PMID:20624155; http://dx.doi.org/ 10.1111/j.1369-1600.2010.00219.x [DOI] [PubMed] [Google Scholar]
- 39.Aguilar-Valles A, Vaissière T, Griggs EM, Mikaelsson MA, Takács IF, Young EJ, Rumbaugh G, Miller CA. Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biol Psychiatry 2014; 76:57-65; PMID:24183790; http://dx.doi.org/ 10.1016/j.biopsych.2013.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krasnova IN, Chiflikyan M, Justinova Z, McCoy MT, Ladenheim B, Jayanthi S, Quintero C, Brannock C, Barnes C, Adair JE, et al.. CREB phosphorylation regulates striatal transcriptional responses in the self-administration model of methamphetamine addiction in the rat. Neurobiol Dis 2013; 58:132-43; PMID:23726845; http://dx.doi.org/ 10.1016/j.nbd.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Shin S, Subramaniam M, Bruinsma E, Kim T-D, Hawse JR, Spelsberg TC, Janknecht R. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res Commun 2010; 401:412-6; PMID:20863814; http://dx.doi.org/ 10.1016/j.bbrc.2010.09.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differ Res Biol Divers 1994; 56:39-44; http://dx.doi.org/ 10.1046/j.1432-0436.1994.56120039.x [DOI] [PubMed] [Google Scholar]
- 43.Numachi Y, Shen H, Yoshida S, Fujiyama K, Toda S, Matsuoka H, Sora I, Sato M. Methamphetamine alters expression of DNA methyltransferase 1 mRNA in rat brain. Neurosci Lett 2007; 414:213-7; PMID:17254711; http://dx.doi.org/ 10.1016/j.neulet.2006.12.052 [DOI] [PubMed] [Google Scholar]
- 44.Mychasiuk R, Muhammad A, Ilnytskyy S, Kolb B. Persistent gene expression changes in NAc, mPFC, and OFC associated with previous nicotine or amphetamine exposure. Behav Brain Res 2013; 256:655-61; PMID:24021241; http://dx.doi.org/ 10.1016/j.bbr.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 45.Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 2004; 47, Supplement 1:33-46; PMID:15464124; http://dx.doi.org/ 10.1016/j.neuropharm.2004.06.025 [DOI] [PubMed] [Google Scholar]
- 46.Pol Bodetto S, Carouge D, Fonteneau M, Dietrich J-B, Zwiller J, Anglard P. Cocaine represses protein phosphatase-1Cβ through DNA methylation and Methyl-CpG Binding Protein-2 recruitment in adult rat brain. Neuropharmacology 2013; 73:31-40; PMID:23688924; http://dx.doi.org/ 10.1016/j.neuropharm.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 47.Pol Bodetto S, Romieu P, Sartori M, Tesone-Coelho C, Majchrzak M, Barbelivien A, Zwiller J, Anglard P. Differential regulation of MeCP2 and PP1 in passive or voluntary administration of cocaine or food. Int J Neuropsychopharmacol Off Sci J Coll Int Neuropsychopharmacol CINP 2014; 17:2031-44. [DOI] [PubMed] [Google Scholar]
- 48.Deng JV, Rodriguiz RM, Hutchinson AN, Kim I-H, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci 2010; 13:1128-36; PMID:20711186; http://dx.doi.org/ 10.1038/nn.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng JV, Wan Y, Wang X, Cohen S, Wetsel WC, Greenberg ME, Kenny PJ, Calakos N, West AE. MeCP2 Phosphorylation Limits Psychostimulant-Induced Behavioral and Neuronal Plasticity. J Neurosci 2014; 34:4519-27; PMID:24671997; http://dx.doi.org/ 10.1523/JNEUROSCI.2821-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Z, Hong EJ, Cohen S, Zhao W-N, Ho H-YH, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al.. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 2006; 52:255-69; PMID:17046689; http://dx.doi.org/ 10.1016/j.neuron.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McFadden LM, Vieira-Brock PL, Hanson GR, Fleckenstein AE. Methamphetamine self-administration attenuates hippocampal serotonergic deficits: role of brain-derived neurotrophic factor. Int J Neuropsychopharmacol Off Sci J Coll Int Neuropsychopharmacol CINP 2014; 17:1315-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D-J, Roh S, Kim Y, Yoon S-J, Lee H-K, Han C-S, Kim Y-K. High concentrations of plasma brain-derived neurotrophic factor in methamphetamine users. Neurosci Lett 2005; 388:112-5; PMID:16039058; http://dx.doi.org/ 10.1016/j.neulet.2005.06.042 [DOI] [PubMed] [Google Scholar]
- 53.Itzhak Y, Ergui I, Young JI. Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol Psychiatry 2014; 20:232-9; PMID:24535458 [DOI] [PubMed] [Google Scholar]
- 54.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol 2013; 20:282-9; PMID:23463313; http://dx.doi.org/ 10.1038/nsmb.2489 [DOI] [PubMed] [Google Scholar]
- 55.Jayanthi S, McCoy MT, Chen B, Britt JP, Kourrich S, Yau H-J, Ladenheim B, Krasnova IN, Bonci A, Cadet JL. Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol Psychiatry 2014; 76:47-56; PMID:24239129; http://dx.doi.org/ 10.1016/j.biopsych.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cadet JL, Jayanthi S. Epigenetics of methamphetamine-induced changes in glutamate function. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 2013; 38:248-9; http://dx.doi.org/ 10.1038/npp.2012.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf ME, Ferrario CR. AMPA receptor plasticity in the nucleus accumbens after repeated exposure to cocaine. Neurosci Biobehav Rev 2010; 35:185-211; PMID:20109488; http://dx.doi.org/ 10.1016/j.neubiorev.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]


