Abstract
Dynamic regulation of chromatin structure is an important mechanism for balancing the pluripotency and cell fate decision in embryonic stem cells (ESCs). Indeed ESCs are characterized by unusual chromatin packaging, and a wide variety of chromatin regulators have been implicated in control of pluripotency and differentiation. Genome-wide maps of epigenetic factors have revealed a unique epigenetic signature in pluripotent ESCs and have contributed models to explain their plasticity. In addition to the well known epigenetic regulation through DNA methylation, histone posttranslational modifications, chromatin remodeling, and non-coding RNA, histone variants are emerging as important regulators of ESC identity. In this review, we summarize and discuss the recent progress that has highlighted the central role of histone variants in ESC pluripotency and ESC fate, focusing, in particular, on H1 variants, H2A variants H2A.X, H2A.Z and macroH2A and H3 variant H3.3.
Keywords: chromatin dynamics, embryonic stem cell, histone variant, pluripotency, self-renewal
Abbreviations
- ChIP-seq
Chromatin Immunoprecipitation Sequencing
- EB
embryoid body
- EHP cells
endoderm/hepatic progenitor cells
- ESCs
embryonic stem cells
- iPSCs
induced pluripotent stem cells
- MEFs
mouse embryonic fibroblasts
- NLBs
nucleolar-like bodies
- PcG
Polycomb group
- TSS
transcription start site
Introduction
DNA is wrapped around histone octamers to form nucleosome core particles, which are the basic repeating units of chromatin. A single nucleosome is composed of approximately 2 turns of DNA wrapped around an octamer of core histone proteins formed from a pair of H2A-H2B dimers and an H3-H4 tetramer.1 Although the packaging of DNA into chromatin allows for compaction of large genomes, it also renders the underlying DNA sequence inaccessible to many processes, including transcription factor binding. Consequently, mechanisms have evolved to control chromatin packaging in order to modulate DNA accessibility. Such variations in chromatin states are vital in aiding the differing gene expression patterns that define the hundreds of cell types that form an organism, from the pluripotent stem cell through to the terminally differentiated cell, all of which contain the same DNA sequence.
Dynamic regulation of chromatin structure is an important mechanism for balancing the pluripotency and cell fate decision in embryonic stem cells (ESCs). ESCs are characterized by unusual chromatin packaging,2 and a wide variety of chromatin regulators have been implicated in control of pluripotency and differentiation, as summarized in detail in Christophersen et al.3 Differences in global chromatin configuration can be readily detected by ultrastructural examination of ESC chromatin as well as chromatin from terminally differentiated cells: chromatin of undifferentiated ESCs appears decondensed and highly dynamic in structure, whereas differentiated cells display distinct foci of heterochromatin.4 Overall, ESC differentiation is accompanied by an increase in heterochromatic foci and a decrease in mobility and turnover of chromatin-associated proteins.4-6
An open chromatin configuration may merely be a reflection of the hyper-transcriptional activity found in ESCs. However, the identification of an unique histone modification pattern at the regulatory regions of developmental genes, called ‘bivalent domains’ and consisting of the simultaneous presence of histone H3K27me3 and H3K4me2/me3, led to the idea that developmental genes are bivalently marked and, thus, primed for activation prior to ESC differentiation.7,8 Therefore, an open chromatin configuration and abundance of active histone marks are not only a reflection of high transcriptional activity but also of cell plasticity.
Recent technical advances, particularly in next-generation sequencing technologies, have provided a genome-scale view of epigenetic marks and chromatin modifiers leading to a comprehensive characterization of the ESC epigenome. Genome-wide maps of epigenetic factors have revealed a unique epigenetic signature in pluripotent ESCs and have contributed to explain their plasticity. Also, comparisons of ESCs with cells differentiated from ESCs have proven powerful in understanding the dynamics of epigenetic marks during development. Thus, each specific cell type has not only its own specific expression profile or ‘transcriptome’ but is also featured by its own epigenetic signature or ‘epigenome’ that is transmitted as heritable information through cell divisions.9-11
Epigenetic regulation, including DNA methylation, posttranslational modification of histones, chromatin remodeling, and non-coding RNA, can alter the properties or interacting partners of the nucleosome (for a review see12). Notably, histone variants are emerging as important regulators of ESC identity.13 Variant forms of histones H1, H2A, H2B, and H3, with the exception of H4, have been described. Variants of any one histone type may differ in only a few amino acids (e.g., H3 variants) or may contain significant dissimilarities (e.g., H2 variants) as further discussed in references 14 and 15. Non-canonical or ‘replacement’ histones are expressed throughout the cell cycle and their incorporation is tightly regulated by histone chaperones.
In this review, we summarize and discuss the recent progress that has highlighted the central role of histone variants in ESC pluripotency and ESC fate. We provide a detailed and comprehensive discussion of genome-wide studies that are pertinent to our understanding of mammalian development, focusing on data regarding mouse embryogenesis and mouse ESCs (mESCs) and providing references to other species, when relevant.
H1 Variants
H1 linker histones are abundant chromatin binding proteins that facilitate the formation of higher order chromatin structures. Histone H1 binding to the nucleosomal dyad and the linker DNA entering and exiting the core particle form the chromatosome, which influences chromatin arrangement and compaction.16 In addition to this well-established function of H1 as a chromatin architectural protein, increasing evidence has accumulated indicating that linker histones also act by interacting with many different non-histone nuclear and cytosolic proteins,17 thus functioning both as primary chromatin architectural proteins and simultaneously as recruitment hubs for proteins involved in accessing and modifying the chromatin fiber. The H1 histone family is the most divergent group among the highly conserved histone proteins. Eleven different H1 variants have been characterized in mammals, including somatic H1 variants (H1.1 to H1.5), the replacement H1 (H1.0), germ cell specific H1s (H1t, H1T2, HILS1, and H1oo), as well as the recently characterized variant H1x.18 Deletion of 3 major somatic H1 variants (H1.2, H1.3, and H1.4) together leads to a 50% reduction of the total H1 level and embryonic lethality at midgestation, demonstrating that H1 level is critical for mammalian development.19
H1.3 and H1.2 are among the most abundant linker histones in mESCs, accounting respectively for 32.6% and 16.4% of total H1, whereas the differentiation associated H1, H1.0, accounts for 2% of H1 in undifferentiated ESCs.20,21 These three variants differ significantly in terms of their residence time on chromatin and their ability to promote chromatin condensation in vitro.22,23 They also display different expression patterns during mammalian development.24,25 At present, the mechanisms by which H1 variants modulate chromatin structure and gene expression remain under-explored.
A recent study achieved high resolution mapping of H1.3, H1.2, and H1.0 in mESCs establishing a knock-in system to stringently test the function of tagged H1s and to facilitate the generation of high resolution maps of H1 variants in ESCs by chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-seq).26 On the H1 genome-wide maps they have generated, H1.3 and H1.2 are highly correlated and display similar binding patterns in the ESC genome: both variants are enriched at AT-rich regions, gene deserts, and major satellites, but are depleted at GC-rich, gene-rich regions, and, especially, at active promoters. Thus, H1.3 and H1.2 are quite similar in overall distribution in the genome (Fig. 1). Nevertheless, analyses of the regions that are uniquely enriched for H1.3 or H1.2 reveal some differences in sequence features: H1.2 has a higher enrichment at major satellites than H1.3 but is relatively depleted from long interspersed nuclear element sequences. On the other hand, H1.0, the H1 variant associated with differentiation, is present at very low levels in undifferentiated mESCs and its localization differs significantly from its binding patterns in differentiated cells.
Figure 1.
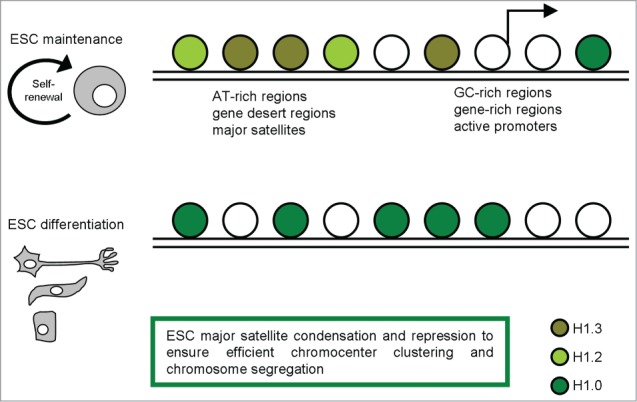
A schematic representation of H1 variants distribution in pluripotent and differentiated cells, with the most relevant functions described.
Since both H1.3 and H1.2 are markedly enriched at major satellites, and considering that major satellite repeats at pericentric heterochromatin from different chromosomes tend to cluster together and form the chromocenter, a nuclear compartment that plays an important role in structural maintenance of the chromosomes,27,28 the effects of H1 depletion on chromocenter clustering in WT and H1 KO ESCs were evaluated.26 The dramatic decrease of total H1 levels led to a decrease in chromocenter numbers, yet having a bigger size, associated with increased expression levels of major satellites. The increase in expression levels at major satellites may be due to an effect of local chromatin decondensation caused by H1 depletion in H1 KO ESCs. Pervasive transcription of repetitive sequences contributes to genome regulation, and aberrant regulation of the expression of satellite sequences interferes with heterochromatin assembly and chromosome segregation.28-31 Thus, these results revealed novel functions of H1 in mammalian genome organization.
Further, Cao et al. recently suggested that the effects of H1 on pericentric major satellites are not restricted to chromatin structure and heterochromatin formation. Loss of H1.3, H1.4, and H1.5 causes a dramatic increase in transcripts from major satellites, but does not change the levels of the repressive epigenetic marks, H3K9me3, H4K20me3, H3K27me3, or DNA methylation at these sequences.26 This suggests that the increase in expression of major satellites in H1 KO ESCs is not mediated by loss of these repressive epigenetic marks, but rather caused by reduced binding of H1 per se or the potential decondensation of local chromatin structure. This study highlights the important role of H1 in driving transcription repression at repetitive DNA, possibly independent of other histone methylation pathway.
The importance of H1 in pluripotent cell renewal has been addressed by Zhang et al., who investigated the role of H1 and chromatin compaction in stem cell pluripotency and differentiation.32 They examined the differentiation of embryonic stem cells that are depleted of multiple H1 subtypes and found that H1.3/H1.4/H1.5 triple null ESCs were more resistant to spontaneous differentiation in adherent monolayer culture upon removal of leukemia inhibitory factor. Further, using an unbiased proteomic approach, Christophorou et al. identified linker histone H1 variants, which are involved in the generation of compact chromatin, as novel PADI4 substrates.33 Citrullination of a single arginine residue within the DNA-binding site of H1 resulted in its displacement from chromatin and global chromatin decondensation. Together, these results uncover a role for citrullination in the regulation of pluripotency and provide new mechanistic insights into how citrullination regulates chromatin compaction.33
In accordance with the mouse model, also hESCs were reported to highly express specific H1 variants, in particular H1.3 and H1.5.34 The authors hypothesized that H1.3 and H1.5 could have a role in maintaining pluripotency/self-renewal, either by producing a more relaxed chromatin structure or by specifically contributing to the regulation of a subset of genes.
Overall, data presented for both mESCs and hESCs reveal that specific H1 variants are involved in heterochromatin assembly; additional genome-wide distribution studies of other H1 variants in pluripotent cells (as well as in differentiated cell types) will likely lead to a better understanding of the role of H1 histone family in higher order chromatin folding, gene expression, and chromatin function in these cells.
H2A.X
H2A.X constitutes about 2.5–25% of total H2A in the mammalian genome35 and is characterized by an extended C-terminal tail containing an evolutionarily conserved serine-glutamine (SQ) motif. In response to DNA double-strand breaks, H2A.X is phosphorylated by ATM and/or DNA-PK36 on a serine residue at position 139 of the SQ motif, yielding a phosphorylated form of the protein known as γH2A.X.35 γH2A.X interacts with several DNA repair proteins, indicating that it is involved in DNA repair processes.37 Recent research has described potentially new and specialized roles for γH2A.X, in addition to the canonical DNA double-strand break response (for a review see ref 38).
A comprehensive histone variant gene expression profiling revealed that H2A.X is among the highest expressed histone variants in undifferentiated mESCs and in preimplantation embryos, from zigotene to blastocyst stage.39 Parallel studies on H2A species incorporation highlighted significant H2A.X incorporation during the early pre-implantation stage. H2A.X signal was detected after fertilization in both male and female pronuclei, the signal intensity remained high at the 2- and 4-cell stages, then it decreased.40 Although Ser139 phosphorylation appeared to be dispensable for H2A.X incorporation,40 H2A.X is highly phosphorylated throughout preimplantation development without any induced DNA damage.41 To support a role of H2A.X in embryogenesis, H2A.X presence during embryonic development was also documented in other species, including Xenopus laevis,42 porcine embryos43 and different human ESC lines.44
In addition to these works describing H2A.X abundance and distribution in ESCs, recent research has described potentially new and specialized roles for γH2A.X mESCs, focusing on specialized functions aimed at the embryonic state maintenance. Banath et al. associated high basal γH2A.X levels in ESCs with global chromatin decondensation rather than pre-existing DNA damage.45 They described that mESCs express about 100 large γH2A.X foci per cell, which decrease during ESC differentiation and are not associated with any DDR factors, such as 53BP1 and RPA.45 High levels of γH2A.X have also been associated with the single-strand breaks occurring in S-phase, and ESC populations, as rapidly dividing cells, have an increased proportion of cells in S-phase, compared to many somatic cells.46 Our group has recently demonstrated that γH2A.X epigenetic modification contributes to sustain the self-renewal ability of ESCs and induced pluripotent stem cells (iPSCs).47 We confirmed previous evidence that γH2A.X levels decrease during ESC differentiation45 and demonstrated, for the first time, that iPSCs behave in the same way.47 We showed that an H2A.X−/− ESC line has a reduced capacity for self-renewal, and demonstrated that its self-renewal ability can be restored through reconstitution with WT H2A.X but not with a mutant form of H2A.X in which the S139 phosphorylation site is abolished.47
Recently, Wu and colleagues explored the new functions of H2A.X in pluripotent stem cells through a ChIP-seq approach aimed at analyzing the genome-wide deposition pattern of H2A.X in mESCs.48 Strikingly, they found that ESC-specific H2A.X deposition regions correlate with the silenced extraembryonic genes targeted by CDX2 in mESCs (Fig. 2). They found that H2A.X deficiency leads to upregulation of some extraembryonic genes, but not of pluripotency genes or germ layer markers. H2A.X deposition thus seems critical in maintaining H3K9me3 level, but not H3K27me3, at extraembryonic gene enhancers. The authors also analyzed the H2A.X deposition pattern in different iPSC lines and determined their developmental potential by tetraploid complementation. Interestingly, the iPSC lines that were capable of tetraploid complementation and the ones that failed were classified into 2 distinct groups with an unsupervised hierarchical cluster analysis of H2A.X deposition.48
Figure 2.
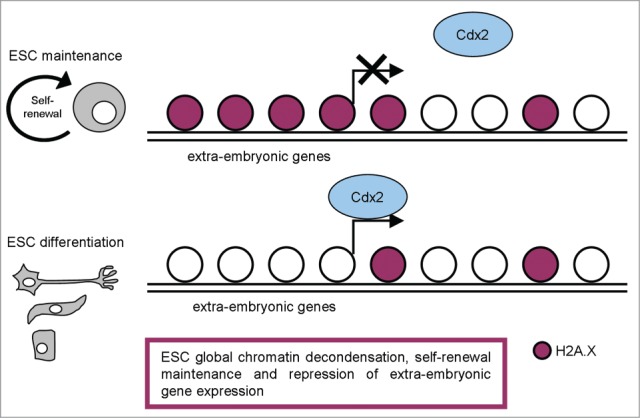
A schematic representation of H2A.X distribution in pluripotent and differentiated cells, with the most relevant functions described.
In line with these observations, Buganim et al. defined the deposition pattern of histone variant H2A.X as a functional epigenetic marker for defining iPSCs quality,49 potentially applicable also to human. Because histones regulate transcription, it is tempting to speculate that the aberrant deposition of H2A.X in poor quality iPSC lines might explain the mild difference in the gene expression between poor- and high-quality iPSCs.
Demonstration that high γH2A.X levels sustain self-renewal and proliferation quality together with developmental potential of ESCs and iPSCs suggests this minor histone modification as another important epigenetic element involved in maintaining the chromatin architecture that contributes to the unique properties of pluripotent stem cells (Fig. 2).
H2A.Z
Histone H2A.Z is an almost universal variant, which evolved early and only once in evolution.50 H2A.Z is only about 60% identical to canonical H2A within the same species,51 but is strikingly more conserved between different species (about 80% identity between most organisms).
A study in Drosophila demonstrated that the essential regions for H2A.Z function are located in its C-terminus (M6 and M7 regions).52 H2A.Z can be post-translationally modified by acetylation, sumoylation and ubiquitination with different functional outcomes (for a review see ref53,54).
Although H2A.Z gene was reported to be expressed during all stages of embryogenesis (zygote, 2-cell, morula, blastocyst, and hatching blastocyst),39 analysis of its nuclear deposition revealed that H2A.Z protein is mainly detected at the blastocyst stage.40
H2A.Z has been implicated in many DNA-mediated processes including gene regulation, with an ability to influence both gene activation and repression.55–60 Recently, several works investigated the essential role of H2A.Z during mammalian development, through genome-wide analysis on mESCs and their differentiated counterparts.
Chreyghton et al. demonstrated that H2A.Z predominantly occupies promoter regions at a defined set of genes in mESCs; in particular, their analysis revealed a striking enrichment for genes connected to the developmental and transcription hierarchies; notably, many of these genes are transcription factors with important roles in a variety of developmental processes.61 These data suggest that H2A.Z predominantly occupies the promoters of genes in ESCs that when expressed, would promote developmental progression and differentiation. They also compared the set of H2A.Z-enriched genes with Polycomb group (PcG) protein binding targets, evaluating in particular the PcG protein Suz12. Their results demonstrated that most of the H2A.Z enriched regions, were also occupied by Suz12. Considering that PcG proteins have been shown to globally maintain their target genes in a silent state7,62,63 they concluded that H2A.Z occupies the promoter regions of a defined set of genes in ESCs that are targets of PcG-mediated repression. Finally, they evaluated if the same pattern was maintained in differentiated cells, examining H2A.Z occupancy in neural precursors. Similarly to its distribution in ESCs, H2A.Z was significantly enriched at the promoter regions, but the set of H2A.Z-enriched genes was remarkably different in neural precursors as compared to ESCs, with H2A.Z predominantly enriched at genes that displayed high expression levels. Their data suggest that co-occupancy of H2A.Z and PcG proteins at promoters is typical of ESCs with a role in arbitrating cell fate transitions and lineage specification during ESC differentiation; then H2A.Z localization changes dramatically during development and differentiation. Its developmental or stage-specific localization and function likely depend on additional modifiers (Fig. 3).
Figure 3.
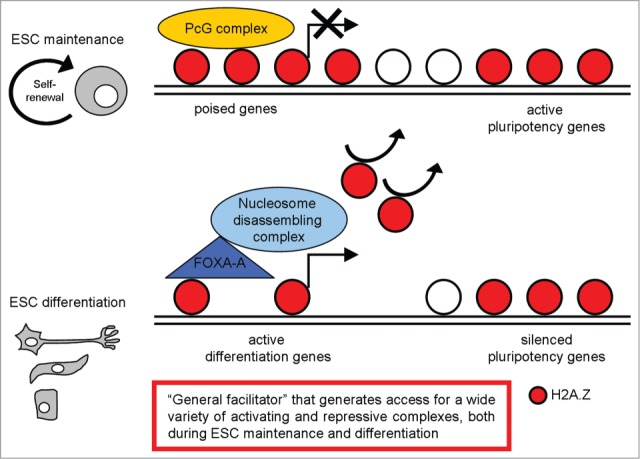
A schematic representation of H2A.Z distribution in pluripotent and differentiated cells, with the most relevant functions described.
To investigate the functional role of H2A.Z, analysis of H2A.Z depleted ESCs was performed and suggests that H2A.Z-depletion per se does not affect maintenance of the ESC state. Differently, H2A.Z is required for lineage specification: embryoid body (EB) analysis showed that H2A.Z-depleted EBs failed to give rise to typical structures representing differentiated cell types and were defective in the correct induction of early differentiation markers such as Brachyury, Pax3 and Wnt3A. These data demonstrates a requirement for H2A.Z to initiate developmental programs, suggesting an important role for H2A.Z in mediating cell fate transitions.61 Hu et al. confirmed the critical H2A.Z contribution in ESC differentiation; however, they also suggested a role of H2A.Z in mediating self-renewal and pluripotency in mESCs.64 The discrepancy between these 2 works could be explained by differences between the genetic background of the ESC lines used in the studies and the different methods used to deplete H2A.Z.
A more recent work defined the mechanism by which H2A.Z may mediate ESC differentiation.65 Considering that Foxa1/2 acts in chromatin remodeling during early development66-69 and H2A.Z has been suggested to be critical for ESCs differentiation,61,63,70,71 the authors investigated their possible role in controlling global nucleosome dynamics during ESC differentiation, determining the genome-wide localization of H2A.Z and Foxa during ESC differentiation into endoderm/hepatic progenitor (EHP) cells. They found strong nucleosome depletion near Foxa2 binding sites during ESC differentiation. Nucleosome depletion regions were enriched near H2A.Z binding sites in ESCs and Foxa2 binding sites in EHP cells, which suggests that nucleosomes containing H2A.Z in ESCs were preferentially lost during differentiation as compared to non-H2A.Z nucleosomes. Next, they identified the nucleosome disassembly/assembly chaperone protein Nap1l1 (the mouse homolog of NAP1), the SWI/SNF complex component Smarca4, and the SWR1 component Kat5 as the major chromatin remodeling complexes and chaperones that might mediate the Foxa2/H2A.Z-dependent nucleosome depletion process. They support a 3-step model for ESC differentiation into EHP cells: (1) growth factor-induced cell differentiation initiates Foxa2 expression; (2) Foxa2 binds to nucleosomal DNA on H2A.Z-containing nucleosomes; (3) Foxa2 and H2A.Z recruit nucleosome disassembly complexes (Nap1l1/SWI/SNF/INO80), enabling nucleosome depletion and cell differentiation (Fig. 3).
H2A.Z role in destabilizing the local nucleosome structure and facilitating nucleosome removal was confirmed by Hu et al.64 They provided data showing differential roles of H2A.Z at active genes and repressed genes in the self-renewal and differentiation of ESCs. H2A.Z facilitates expression of many pluripotency genes and also the repression of differentiation genes by generating chromatin accessibility and thereby facilitating the efficient targeting of activating and repressive complexes, respectively. During differentiation of ESCs, optimal induction of differentiation genes and the complete silencing of pluripotency genes also requires H2A.Z to facilitate access of the appropriate complexes. Another recent paper identified the H2A.Z C-terminal acidic patch as the critical domain that couples control of chromatin dynamics to the regulation of developmental gene expression patterns during lineage commitment.72
There are several possibilities to explain how H2A.Z can mediate seemingly opposing functions, such as repressing genes in ESCs and activating genes in differentiated cells: H2A.Z can be post-translationally modified and its incorporation correlates with particular histone modification patterns; post-translational modification of H2A.Z may underlay its differential distribution in ESCs compared to differentiated cells; the function of H2A.Z may depend on the nucleosome into which it is incorporated.
In conclusion, H2A.Z is an important player in regulating ESC fate, whose localization and functions change dramatically during development and differentiation; it may be defined as a ‘general facilitator’ that generates access for a wide variety of activating and repressive complexes (Fig. 3).
MacroH2A Variants
The histone variant macroH2A plays a key role in cell state stabilization in mammals. Mammals encode 3 macroH2A variants, macroH2A.1.1 and macroH2A.1.2, which are alternatively spliced isoforms of a single gene, and the distinct gene product macroH2A.2.73 All three macroH2A variants are distinguished by the presence of the unusual ‘macro’ domain fused to their relatively well-conserved H2A cores. This domain protrudes from the compact structure of the nucleosome and is likely to affect the function and organization of the surrounding chromatin.74 MacroH2A occupancy generally correlates with gene repression and low transcriptional activity.75,76 Accordingly, inhibiting macroH2A increased the transcription levels, or facilitated the activation, of target genes.74,77,78 The view of macroH2A as a pure repressor was challenged by numerous examples where macro incorporation is associated with increased gene expression,76,79 including during early lineage specification after embryoid body formation from ESCs.80
All the 3 macroH2A isoforms are expressed in mESCs and at the blastocyst stage of embryogenesis40,80,81 and studies that have analyzed macroH2A.1 and macroH2A.2 in parallel found no difference in the pattern of distribution.82 At the protein level, macroH2A.1 is expressed to a greater extent than macroH2A.2 and quantifying the relative abundance of mRNAs, mESCs turned out to contain 7 times more messenger for macroH2A.1 than macroH2A.2.80
Creppe et al. performed ChIP-seq analysis on macroH2A.1 distribution in ESCs and evaluated its role in mESC self renewal and differentiation.80 Their study revealed that macroH2A.1 occupies the promoter regions of differentiation genes with a minimum occupancy at the immediate transcription start sites (TSS); genes with increasing expression levels were progressively underrepresented among macroH2A.1 target genes. They reported that in mESCs macroH2A deficiency did not affect cell proliferation, viability, and self-renewal capability. Differently, during EB formation, macroH2A.1 loss resulted in reduced or delayed activation of differentiation genes, many of which were shown to be direct target genes of macroH2A.1, together with an incomplete inactivation of the pluripotency genes that encode the key regulatory transcription factors Sox2, Nanog, and Oct4. Overall their data provide evidence for the physiological relevance of the still poorly recognized proactivating function of macroH2A in ESC differentiation, showing that the presence of macroH2A.1 on differentiation genes is required for their adequate induction (Fig. 4).
Figure 4.
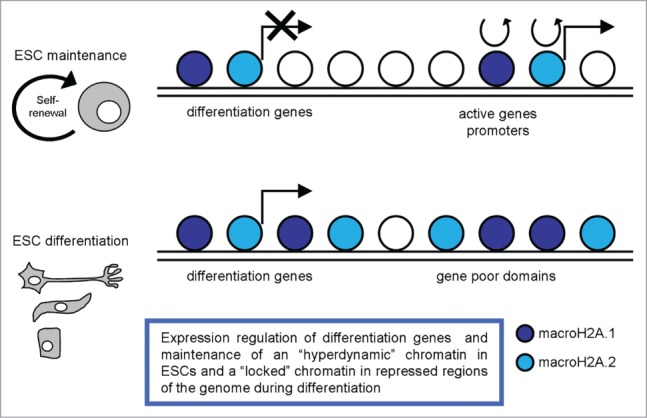
A schematic representation of macroH2A variants distribution in pluripotent and differentiated cells, with the most relevant functions described.
A recent work described a pulse chase system for studying histone dynamics in mESCs, and reported on the dynamics of several histone variants, including macroH2A.2, in both pluripotent ESCs and committed mouse embryonic fibroblasts (MEFs).83 They observed extensive differences in the dynamics and localization of this variant between mESCs and MEFs. Notably, macroH2A.2 is highly dynamic in ESCs, with rapid exchange occurring over gene promoters, alongside much more stably bound domains that cover large blocks of the genome. Upon differentiation to MEFs macroH2A.2 becomes much more stably-bound to the genome, consistent with the idea that this histone variant plays a role in ‘locking down’ repressed regions of the genome (Fig. 4). In ESCs they observed widespread localization of macroH2A.2 across gene rich domains, along with a strong TSS-proximal peak of macroH2A.2. They also documented much more stably bound domains that cover large blocks of the genome. Upon differentiation to MEFs, macroH2A.2 is generally lost from those promoters where it is most dynamic in ESCs and is broadly gained over gene poor domains and at genes associated with alternative cell fates, such as neural or immune cell differentiation. Their results are broadly consistent with the idea that pluripotent cells are characterized by ‘hyperdynamic’ chromatin,4 observing local, rather than global, dynamic macroH2A.2 behavior at a small fraction of loci-promoters of highly expressed genes. Their data on macroH2A.2 association with active promoters in mESCs further argue against a simple model for a universally repressive function of macroH2A; they speculated that stable association of macroH2A, rather than average macroH2A occupancy per se, is more likely to play a role in gene repression.
These data highlighted that on the one hand macroH2A.1 has a role in regulating differentiation gene expression, on the other hand macroH2A.2 variant is important in maintaining a ‘hyperdynamic’ chromatin in pluripotent cells and in ‘locking down’ repressed regions of the genome during differentiation (Fig. 4).
H3.3
In mammals, 5 H3 variants have been identified: 2 canonical variants, H3.1 and H3.2 (also referred to as H3) and 3 replacement variants, H3.3, the centromere-specific variant CenH384 and the testis-specific histone H3t.85 In addition, recently other 2 primate-specific H3 variants, H3.X and H3.Y, were characterized.86 They are expressed throughout the cell cycle, in quiescence, and are enriched in various stages of differentiation compared with their canonical counterparts.7,88 H3.3 has only 4 amino acid differences with H3.2 (at positions 31, 87, 89 and 90) and 5 with H3.1 (with an additional difference at amino acid 96). Histone H3.3 presents a unique genetic paradigm in that 2 conventional intron-containing genes (H3f3a and H3f3b) encode the identical protein. In spite of the high sequence similarity between H3.3 and H3, these specific residues have been proposed to account for particular properties of histone H3.3; specifically, H3.3 has been associated with active transcription in somatic cells.86,89 More recently, its role in undifferentiated cells has been investigated.
Almost 10 y ago, histone variant H3.3 expression and localization in early mouse embryogenesis were examined.90 This work highlighted that H3.3 is present in the oocyte as a maternal factor and is then incorporated preferentially into the male pronucleus before genome activation. Then H3.3 can be detected in the nuclei of mouse embryos in all of the stages analyzed, from the zygote to the blastocyst stage, suggesting that the H3.3 replacement is among the epigenetic mechanisms involved in the early embryogenesis. A more recent paper investigated which particular role H3.3 can execute during embryogenesis.91 To address the contribution of specific residues within the histone H3.3 to the establishment and subsequent reprogramming of chromatin after fertilization, authors expressed this variant harboring point mutations in zygotes and assessed the embryo development ability. Of the residues analyzed on H3.3, only K27 seemed to be important for early embryonic development and indeed, embryos expressing H3.3 K27R exhibited a reduced rate of development. Additionally, authors analyzed whether embryos overexpressing H3.3 K27R showed defects in epigenetic marks and observed altered distribution and reduced global levels of both H3K27me3 and H3K27me1 in embryos expressing H3.3 K27R, in comparison to H3.3 WT. Further, they observed that in zygotes H3.3 localizes to the heterochromatic DAPI-intense rings surrounding the nucleolar-like bodies (NLBs) in the male pronucleus but not in the female one.
These dense ring-like structures around NLBs in the zygote are formed by the chromocenters that contain pericentromeric and centromeric chromatin. Expression of H3.3 K27R mutant induces a spatial relocalization of chromocenters at the 2-cell stage. Because the association of chromocenters to NLBs has been suggested to reflect their heterochromatic nature,92 their mislocalization in H3.3 K27R-mutant embryos suggests defects in heterochromatin. Functionally, they observed that mutation of H3.3 K27 results in the accumulation of major satellite transcripts after the zygote first mitotic division, supporting the idea that the mutation of H3.3 K27 led to derepression, or lack of silencing, of these repeats. It is possible that H3.3 K27R leads to inefficient production of major satellite dsRNA, which in turn would lead to an inability to initiate effective heterochromatinization of the repeats, suggesting defects in silencing and/or paternal heterochromatin formation. Thus, this work uncovered a novel function for H3.3 in the initial establishment of heterochromatin in the mouse embryo.
Several other works evaluated H3.3 localization and roles in mESC model. Allis group reported genome-wide profiles of H3 variants in mESCs cells.93 They found that H3.3 was enriched around TSS of bivalent genes, while mutation of H3.3 toward H3.2 or H3.1 abolished this enrichment. After differentiation, the profile of H3.3 changed with resolution of bivalent domains and, in bivalent ES genes that resolved to H3K4me3 thus becoming transcriptionally active, H3.3 was maintained around the TSS and also incorporated into the gene body, in correlation with H3K36me3 and H3K4me1. In contrast, for bivalent genes that remained transcriptionally repressed and resolved to either H3K27me3 or no mark, H3.3 enrichment was reduced at the TSS upon differentiation. Additionally H3.3 was localized around the TSS of both active and repressed high CpG content promoters, and both H3.3 and phosphorylated RNA polymerase II were significantly enriched beyond the transcriptional end sites of highly expressed genes Their data also highlighted that H3.3 enrichment in the gene body correlates with markers of transcription, including Ser-5 phosphorylated RNA polymerase II, H3K4me3, H3K36me3, and H3K4me1. Then they determined whether this pattern was dependent on the specific H3.3 chaperon Hira and demonstrated that the vast majority of H3.3 enrichment at both active and repressed genes in ES cells is Hira-dependent.
Requirement of H3.3 incorporation to fully maintain the pluripotent nature of ESCs was highlighted in the work of Meshorer et al.94 Consistent with a globally highly transcriptionally active genome in ESCs, they found that the only structural chromatin protein without an increased hyperdynamic pool was H3.3, which preferentially associates with transcriptionally active regions. This finding was also consistent with the observed accelerated differentiation of Hira null cells, since, in those cells, less H3.3 was incorporated into the open chromatin regions, thus facilitating the formation of heterochromatin regions and promoting differentiation.
Differently, H3.3 presence at most known regulatory elements (i.e., genic and intergenic transcription factor binding sites) is Hira-independent (Fig. 5) and will be mediated by other factors. Among these, the chromatin remodeler Chd195 is a H3.3 depositing molecule reported to be required for ESC self-renewal and maintenance of Oct4 expression, and depletion of Chd1 led to the loss of open chromatin.96
Figure 5.
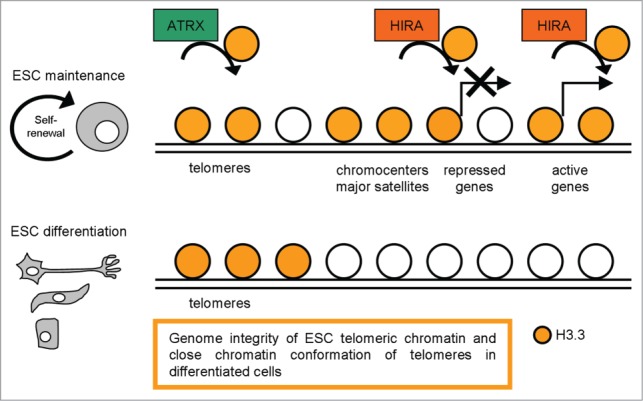
A schematic representation of H3.3 distribution in pluripotent and differentiated cells, with the most relevant functions described.
A requirement for H3.3 chaperones other that Hira also emerged from the recent work of Tang et al.97 They found that an important requirement for H3.3 during the early phase of oocyte growth was seen in the developmental failure of H3f3a, H3f3b double-mutant primary oocytes. However, given that Hira-deficient oogonia could develop and be ovulated, their result clearly demonstrated a requirement for other H3.3 chaperones, possibly DAXX and ATRX, in regulating folliculogenesis.
Choo group contributed to clarify H3.3 function in regulating telomeric chromatin structure in mESCs.98 They examined the epigenetic factors that regulate telomere chromatin in mESCs and changes in telomere chromatin properties during cellular differentiation. They reported the enrichment of Ser31-phosphorylated H3.3 at metaphase telomeres and localization of H3.3 at interphase telomeres in ESCs. They also studied the functional role of H3.3 at the telomeres by H3.3 RNAi knockdown experiments and highlighted that H3.3 RNAi knockdown ESCs present an increased number of telomere dysfunctional-induced foci (TIFs, cytological telomeric foci containing DNA damage response factors, such as γH2A.X),99 suggesting that H3.3 contributes to the functional integrity of telomere chromatin in ESCs (Fig. 5). In accordance with this work H3.3 presence at telomeric regions was described in the previously cited paper,94 demonstrating that H3.3 presence at telomeres is Hira-independent and Atrx is the specific chaperon required for H3.3 enrichment at telomeres and repression of telomeric repeat-containing RNA. Upon ESC differentiation, Ser31 phosphorylated H3.3 signals are greatly reduced at the telomeres without any observable telomere-length shortening; in parallel, the telomere chromatin concomitantly becomes more ‘repressed/closed’, as indicated by increased association of H3K9me3 and H4K20me3 and decreased MNase sensitivity following differentiation.93 Blasco suggested that the assembly of heterochromatin at the telomeres forms a ‘closed’ conformation that results in less accessibility to telomere-elongating machinery/telomerase activity.100 A relatively more ‘open’ telomeric chromatin structure in ESCs compared with their differentiating counterparts should favor a greater accessibility to telomere-elongating activities. The loading and unloading of the chromatin-remodeling histone H3.3 variant at the telomeres may potentially serve as a primary epigenetic reprogramming cue operating at the nucleosomal level that controls telomere-length homeostatic equilibrium and structural organization in pluripotent ESCs.
In conclusion, the dynamic presence of H3.3 in both genic and intergenic regions, in active and inactive genes, suggests that its function in regulating chromatin structure may depend on the different post-transcriptionally modifications and/or may act in concert with other epigenetic modifications.
Conclusions
We have presented the complex scenario linking histone variants, chromatin regulation and ESC maintenance/differentiation. This overview on H1 variants, the H2A variants H2A.X, H2A.Z and macroH2A, and the H3 variant H3.3 highlights that histone variants can be involved in a multitude of functions, sometimes seemingly opposite, ranging from chromatin condensation to decondensation, from transcription activation to repression, chromatin dynamic and locked maintenance, genomic telomere maintenance, self renewal and differentiation regulation. Whenever possible, we have taken into account the knowledge about expression levels in self-renewing ESCs and differentiating cells of all mentioned histone variants (Fig. 6).
Figure 6.
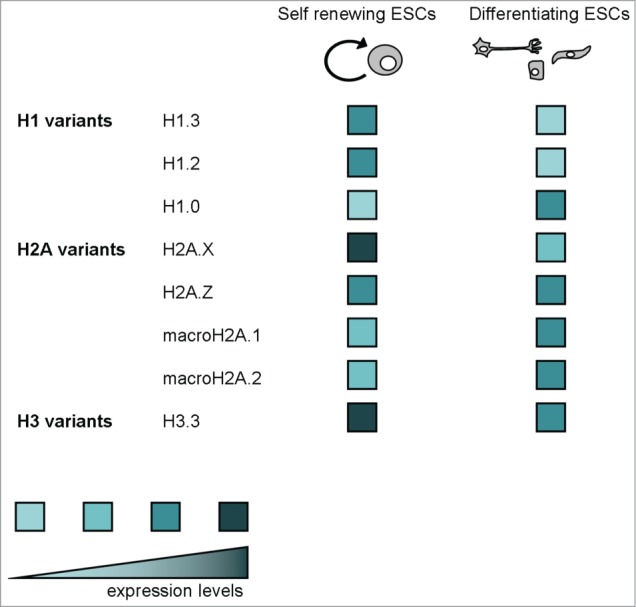
A schematic representation of expression levels in self-renewing ESCs and differentiating cells of all mentioned histone variants.
There are several possibilities to explain this complex scenario: many of these variants can be post-translationally modified and their incorporation and function may correlate with particular histone modification patterns; both ESCs and differentiated cells may involve different chaperons that regulate histone incorporation; and finally, the differential histone distribution and/or function may depend on other epigenetic factors. Advanced technology has recently helped clarifying how do extrinsic signals, transcription factors, and chromatin modifications collaborate and become integrated into a fine-tuned network to maintain ESC identity. Yet, other fundamental questions remain open, like how precisely do transcriptional machineries and chromatin-binding proteins find histone modification marks at specific loci. Future work will need to better understand the dynamic nature of nucleosomes in ESCs, how this is used to regulate the accessibility of the underlying DNA and how the described histone variant functions can be modulated by the complex ESC epigenetic landscape.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank previous and current lab members for their contributions.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251-60; PMID:9305837; http://dx.doi.org/ 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315-26; PMID:16630819; http://dx.doi.org/ 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 3.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med 2010; 207:2287-95; PMID:20975044; http://dx.doi.org/ 10.1084/jem.20101438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10:105-16; PMID:16399082; http://dx.doi.org/ 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci 2005; 118:3861-8; PMID:16105879; http://dx.doi.org/ 10.1242/jcs.02500 [DOI] [PubMed] [Google Scholar]
- 6.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A 2007; 104:15619-24; PMID:17893336; http://dx.doi.org/ 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al.. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349-53; PMID:16625203; http://dx.doi.org/ 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315-26; PMID:16630819; http://dx.doi.org/ 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev 2009; 19:127-34; PMID:19299119; http://dx.doi.org/ 10.1016/j.gde.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 2011; 12:7-18; PMID:21116306; http://dx.doi.org/ 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
- 11.Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol 2013; 25:387-94; PMID:23465541; http://dx.doi.org/ 10.1016/j.ceb.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 2009; 10:526-37; PMID:19603040; http://dx.doi.org/ 10.1038/nrm2727 [DOI] [PubMed] [Google Scholar]
- 13.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007; 128:635-8; PMID:17320500; http://dx.doi.org/ 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 14.Biterge B, Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014; 356:457-66; PMID:24781148; http://dx.doi.org/ 10.1007/s00441-014-1862-4 [DOI] [PubMed] [Google Scholar]
- 15.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014; 15:259-71; PMID:24614311; http://dx.doi.org/ 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res 2013; 41:9593-609; PMID:23945933; http://dx.doi.org/ 10.1093/nar/gkt700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010; 20:519-28; PMID:20309017; http://dx.doi.org/ 10.1038/cr.2010.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 2009; 431:1-12; PMID:19059319; http://dx.doi.org/ 10.1016/j.gene.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 19.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 2003; 23:4559-72; PMID:12808097; http://dx.doi.org/ 10.1128/MCB.23.13.4559-4572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005; 123:1199-212; PMID:16377562; http://dx.doi.org/ 10.1016/j.cell.2005.10.028 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu Z, Medrzycki M, Cao K, Fan Y. Reduction of Hox gene expression by histone H1 depletion. PLoS One 2012; 7:e38829; PMID:22701719; http://dx.doi.org/ 10.1371/journal.pone.0038829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One 2009; 4:e0007243; PMID:19794910; http://dx.doi.org/ 10.1371/journal.pone.0007243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Th'ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem 2005; 280:27809-14; PMID:15911621; http://dx.doi.org/ 10.1074/jbc.M501627200 [DOI] [PubMed] [Google Scholar]
- 24.Wang ZF, Sirotkin AM, Buchold GM, Skoultchi AI, Marzluff WF. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol 1997; 271:124-38; PMID:9300059; http://dx.doi.org/ 10.1006/jmbi.1997.1166 [DOI] [PubMed] [Google Scholar]
- 25.Meergans T, Albig W, Doenecke D. Varied expression patterns of human H1 histone genes in different cell lines. DNA Cell Biol 1997; 16:1041-9; PMID:9324306; http://dx.doi.org/ 10.1089/dna.1997.16.1041 [DOI] [PubMed] [Google Scholar]
- 26.Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, Lee EK, Wu H, Medrzycki M, Pan C, et al.. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet 2013; 9:e1003417; PMID:23633960; http://dx.doi.org/ 10.1371/journal.pgen.1003417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu TC, Cooper JE, Mace ML Jr., Brinkley BR. Arrangement of centromeres in mouse cells. Chromosoma 1971; 34:73-87; PMID:4105528; http://dx.doi.org/ 10.1007/BF00285517 [DOI] [PubMed] [Google Scholar]
- 28.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 2004; 166:493-505; PMID:15302854; http://dx.doi.org/ 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome 1995; 6:76-83; PMID:7767009; http://dx.doi.org/ 10.1007/BF00303248 [DOI] [PubMed] [Google Scholar]
- 30.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 2009; 10:973-82; PMID:19680288; http://dx.doi.org/ 10.1038/embor.2009.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 2006; 103:8709-14; PMID:16731634; http://dx.doi.org/ 10.1073/pnas.0508006103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, Medrzycki M, Berhe DT, Pan C, McDevitt TC, et al.. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012; 8:e1002691; PMID:22589736; http://dx.doi.org/ 10.1371/journal.pgen.1002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, et al.. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014; 507:104-8; PMID:24463520; http://dx.doi.org/ 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terme JM, Sese B, Millan-Arino L, Mayor R, Izpisua Belmonte JC, Barrero MJ, Jordan A. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem 2011; 286:35347-57; PMID:21852237; http://dx.doi.org/ 10.1074/jbc.M111.281923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858-68; PMID:9488723; http://dx.doi.org/ 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 36.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 2001; 15:2177-96; PMID:11544175; http://dx.doi.org/ 10.1101/gad.914401 [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004; 3:959-67; PMID:15279782; http://dx.doi.org/ 10.1016/j.dnarep.2004.03.024 [DOI] [PubMed] [Google Scholar]
- 38.Turinetto V, Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucl Acids Res 2015; 43(5):2489-98; PMID:25712102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kafer GR, Lehnert SA, Pantaleon M, Kaye PL, Moser RJ. Expression of genes coding for histone variants and histone-associated proteins in pluripotent stem cells and mouse preimplantation embryos. Gene Expr Patterns 2010; 10:299-305; PMID:20601166; http://dx.doi.org/ 10.1016/j.gep.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 40.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 2010; 137:3785-94; PMID:20943707; http://dx.doi.org/ 10.1242/dev.051805 [DOI] [PubMed] [Google Scholar]
- 41.Ziegler-Birling C, Helmrich A, Tora L, Torres-Padilla ME. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol 2009; 53:1003-11; PMID:19598117; http://dx.doi.org/ 10.1387/ijdb.082707cz [DOI] [PubMed] [Google Scholar]
- 42.Shechter D, Chitta RK, Xiao A, Shabanowitz J, Hunt DF, Allis CD. A distinct H2A.X isoform is enriched in Xenopus laevis eggs and early embryos and is phosphorylated in the absence of a checkpoint. Proc Natl Acad Sci U S A 2009; 106:749-54; PMID:19131518; http://dx.doi.org/ 10.1073/pnas.0812207106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohrer RC, Che L, Goncalves PB, Duggavathi R, Bordignon V. Phosphorylated histone H2A.x in porcine embryos produced by IVF and somatic cell nuclear transfer. Reproduction 2013; 146:325-33; PMID:23858475; http://dx.doi.org/ 10.1530/REP-13-0271 [DOI] [PubMed] [Google Scholar]
- 44.Shaw ML, Williams EJ, Hawes S, Saffery R. Characterisation of histone variant distribution in human embryonic stem cells by transfection of in vitro transcribed mRNA. Mol Reprod Dev 2009; 76:1128-42; PMID:19606468; http://dx.doi.org/ 10.1002/mrd.21077 [DOI] [PubMed] [Google Scholar]
- 45.Banath JP, Banuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 2009; 315:1505-20; PMID:19154734; http://dx.doi.org/ 10.1016/j.yexcr.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 46.Chuykin IA, Lianguzova MS, Pospelova TV, Pospelov VA. Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle 2008; 7:2922-8; PMID:18787397; http://dx.doi.org/ 10.4161/cc.7.18.6699 [DOI] [PubMed] [Google Scholar]
- 47.Turinetto V, Orlando L, Sanchez-Ripoll Y, Kumpfmueller B, Storm MP, Porcedda P, Minieri V, Saviozzi S, Accomasso L, Cibrario Rocchietti E, et al.. High basal gammaH2AX levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells 2012; 30:1414-23; PMID:22628289; http://dx.doi.org/ 10.1002/stem.1133 [DOI] [PubMed] [Google Scholar]
- 48.Wu T, Liu Y, Wen D, Tseng Z, Tahmasian M, Zhong M, Rafii S, Stadtfeld M, Hochedlinger K, Xiao A. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell 2014; 15:281-94; PMID:25192463; http://dx.doi.org/ 10.1016/j.stem.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 49.Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al.. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014; 15:295-309; PMID:25192464; http://dx.doi.org/ 10.1016/j.stem.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thatcher TH, Gorovsky MA. Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res 1994; 22:174-9; PMID:8121801; http://dx.doi.org/ 10.1093/nar/22.2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure 2008; 16:166-79; PMID:18275809; http://dx.doi.org/ 10.1016/j.str.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 52.Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature 1999; 399:694-7; PMID:10385122; http://dx.doi.org/ 10.1038/21436 [DOI] [PubMed] [Google Scholar]
- 53.Thambirajah AA, Li A, Ishibashi T, Ausio J. New developments in post-translational modifications and functions of histone H2A variants. Biochem Cell Biol 2009; 87:7-17; PMID:19234519; http://dx.doi.org/ 10.1139/O08-103 [DOI] [PubMed] [Google Scholar]
- 54.Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev 2006; 16:119-24; PMID:16503125; http://dx.doi.org/ 10.1016/j.gde.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 55.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 2007; 446:572-6; PMID:17392789; http://dx.doi.org/ 10.1038/nature05632 [DOI] [PubMed] [Google Scholar]
- 56.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 2005; 3:e384; PMID:16248679; http://dx.doi.org/ 10.1371/journal.pbio.0030384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 2005; 102:18385-90; PMID:16344463; http://dx.doi.org/ 10.1073/pnas.0507975102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 2003; 112:725-36; PMID:12628191; http://dx.doi.org/ 10.1016/S0092-8674(03)00123-5 [DOI] [PubMed] [Google Scholar]
- 59.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J 2003; 22:1599-607; PMID:12660166; http://dx.doi.org/ 10.1093/emboj/cdg160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 2005; 123:219-31; PMID:16239141; http://dx.doi.org/ 10.1016/j.cell.2005.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 2008; 135:649-61; PMID:18992931; http://dx.doi.org/ 10.1016/j.cell.2008.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20:1123-36; PMID:16618801; http://dx.doi.org/ 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al.. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006; 125:301-13; PMID:16630818; http://dx.doi.org/ 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013; 12:180-92; PMID:23260488; http://dx.doi.org/ 10.1016/j.stem.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 2012; 151:1608-16; PMID:23260146; http://dx.doi.org/ 10.1016/j.cell.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat Struct Mol Biol 2011; 18:742-6; PMID:21623366; http://dx.doi.org/ 10.1038/nsmb.2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McPherson CE, Shim EY, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 1993; 75:387-98; PMID:8402920; http://dx.doi.org/ 10.1016/0092-8674(93)80079-T [DOI] [PubMed] [Google Scholar]
- 68.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol 1999; 209:1-10; PMID:10208738; http://dx.doi.org/ 10.1006/dbio.1999.9228 [DOI] [PubMed] [Google Scholar]
- 69.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature 2005; 435:944-7; PMID:15959514; http://dx.doi.org/ 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- 70.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al.. Nucleosome organization in the Drosophila genome. Nature 2008; 453:358-62; PMID:18408708; http://dx.doi.org/ 10.1038/nature06929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004; 303:343-8; PMID:14645854; http://dx.doi.org/ 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- 72.Subramanian V, Mazumder A, Surface LE, Butty VL, Fields PA, Alwan A, Torrey L, Thai KK, Levine SS, Bathe M, et al.. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet 2013; 9:e1003725; PMID:23990805; http://dx.doi.org/ 10.1371/journal.pgen.1003725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buschbeck M, Di Croce L. Approaching the molecular and physiological function of macroH2A variants. Epigenetics. 2010;5:118-23; PMID:20160488; http://dx.doi.org/ 10.4161/epi.5.2.11076 [DOI] [PubMed] [Google Scholar]
- 74.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol 2005; 25:7616-24; PMID:16107708; http://dx.doi.org/ 10.1128/MCB.25.17.7616-7624.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 2009; 16:1074-9; PMID:19734898; http://dx.doi.org/ 10.1038/nsmb.1665 [DOI] [PubMed] [Google Scholar]
- 76.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 2010; 24:21-32; PMID:20008927; http://dx.doi.org/ 10.1101/gad.1876110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J 2006; 25:4843-53; PMID:17036053; http://dx.doi.org/ 10.1038/sj.emboj.7601364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol 2007; 27:2758-64; PMID:17242180; http://dx.doi.org/ 10.1128/MCB.02334-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev 2006; 20:3324-36; PMID:17158748; http://dx.doi.org/ 10.1101/gad.396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Creppe C, Janich P, Cantarino N, Noguera M, Valero V, Musulen E, Douet J, Posavec M, Martin-Caballero J, Sumoy L, et al.. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol Cell Biol 2012; 32:1442-52; PMID:22331466; http://dx.doi.org/ 10.1128/MCB.06323-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yukawa M, Akiyama T, Franke V, Mise N, Isagawa T, Suzuki Y, Suzuki MG, Vlahovicek K, Abe K, Aburatani H, et al.. Genome-wide analysis of the chromatin composition of histone H2A and H3 variants in mouse embryonic stem cells. PLoS One 2014; 9:e92689; PMID:24658136; http://dx.doi.org/ 10.1371/journal.pone.0092689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pehrson JR, Changolkar LN, Costanzi C, Leu NA. Mice without macroH2A histone variants. Mol Cell Biol. 2014; 34:4523-33; PMID:25312643; http://dx.doi.org/ 10.1128/MCB.00794-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yildirim O, Hung JH, Cedeno RJ, Weng Z, Lengner CJ, Rando OJ. A system for genome-wide histone variant dynamics in ES cells reveals dynamic MacroH2A2 replacement at promoters. PLoS Genet 2014; 10:e1004515; PMID:25102063; http://dx.doi.org/ 10.1371/journal.pgen.1004515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 2008; 9:923-37; PMID:19002142; http://dx.doi.org/ 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Witt O, Albig W, Doenecke D. Testis-specific expression of a novel human H3 histone gene. Exp Cell Res 1996; 229:301-6; PMID:8986613; http://dx.doi.org/ 10.1006/excr.1996.0375 [DOI] [PubMed] [Google Scholar]
- 86.Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al.. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol 2010; 190:777-91; PMID:20819935; http://dx.doi.org/ 10.1083/jcb.201002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frank D, Doenecke D, Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene 2003; 312:135-43; PMID:12909349; http://dx.doi.org/ 10.1016/S0378-1119(03)00609-7 [DOI] [PubMed] [Google Scholar]
- 88.Filipescu D, Szenker E, Almouzni G. Developmental roles of histone H3 variants and their chaperones. Trends Genet 2013;29:630-40; PMID:23830582; http://dx.doi.org/ 10.1016/j.tig.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 89.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics 2002; 80:487-98; PMID:12408966; http://dx.doi.org/ 10.1006/geno.2002.6850 [DOI] [PubMed] [Google Scholar]
- 90.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol 2006; 50:455-61; PMID:16586346; http://dx.doi.org/ 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]
- 91.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 2010; 12:853-62; PMID:20676102; http://dx.doi.org/ 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 2007; 116:403-15; PMID:17447080; http://dx.doi.org/ 10.1007/s00412-007-0106-8 [DOI] [PubMed] [Google Scholar]
- 93.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al.. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140:678-91; PMID:20211137; http://dx.doi.org/ 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006; 7:540-6; PMID:16723974; http://dx.doi.org/ 10.1038/nrm1938 [DOI] [PubMed] [Google Scholar]
- 95.Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al.. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007; 317:1087-90; PMID:17717186; http://dx.doi.org/ 10.1126/science.1145339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al.. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009; 460:863-8; PMID:19587682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang MC, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, Lim SL, Hudson DF, Kalitsis P, O'Bryan MK, et al.. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 2015; 11:e1004964; PMID:25675407; http://dx.doi.org/ 10.1371/journal.pgen.1004964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al.. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res 2009; 19:404-14; PMID:19196724; http://dx.doi.org/ 10.1101/gr.084947.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 2005; 24:2667-78; PMID:15973431; http://dx.doi.org/ 10.1038/sj.emboj.7600733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 2007; 8:299-309; PMID:17363977; http://dx.doi.org/ 10.1038/nrg2047 [DOI] [PubMed] [Google Scholar]


