Abstract
EGFR and p53 are molecular markers which play important role in tumor progression and development. The objective of this study was to assess the association between EGFR and p53 expression and survival, and to determine whether EGFR and p53 expression levels were associated with differences quality of life in OSCC patients undergoing chemoradiation. A total of 120 OSCC patients aged 20–67 y and stage III/IV were recruited. Treatment response was assessed according to W.H.O. (1979). EGFR and p53 expression in tumor tissue was estimated by immunohistochemical (IHC) method and quantified as percentage positive nuclei. Molecular marker expressions of both EGFR and p53 were found significantly (P < 0.01 or P < 0.001) associated with overall response, survivals and quality of life. Neither EGFR nor p53 expression was associated with hematologic or non-hematologic toxicity. EGFR and p53 molecular marker expressions may have significant association with survival and QOL in OSCC patients undergoing chemoradiation.
Keywords: chemoradition, immunohistochemistry, oral squamous cell carcinoma, quality of life, survival endpoints, toxicity
Abbreviations
- IHC
Immunohistochemistry
- QOL
quality of life
- RO resections
microscopically margin-negative resection.
Introduction
Oral cancer is the commonest cancer in India. A large fraction of cases occur in males in the productive years of life. Most of the cases (80%) present in late stages and despite advances in the understanding and treatment of oral squamous cell cancer (OSCC), survival rates are still poor with myriad of disease and treatment related comorbidities imposed by underlying functional or cosmetic compromises which significantly influence quality of life in these patients.1,2
Further, we often come across with the observation of wide interindividual variability in treatment response, toxicity and survival in OSCC patients with same clinical features of primary tumor i.e. same site, size and stage. Treatment responses may be related to differences in underlying tumor biology which in turn may be linked to a greater cancer-related symptom burden and worse patient quality of life, at baseline and after treatment.3,4
Furthermore, poor survival and varied treatment response combined with the severe functional impairment associated with surgery and chemoradiation, underscore the need to identify suitable marker that could provide prognostic assessment of the disease with the ability to predict and identify tumor responses to the treatment, so that the treatment could be targeted and undue treatment can be avoided, which hopefully maximize the response and ameliorate treatment related adverse events as much as possible to achieve the goal of improvement in QOL.
Moreover, many molecular markers are coming up and have been studied and have given new understanding of pathogenesis in oral squamous cell cancer. EGFR and P53 gene have emerged as critical mediators of signal transduction pathways. Overexpressions of these proteins have been known to be associated with tumor cell proliferation, decreased or resistance to apoptosis, angiogenesis, resulting in tumor progression and metastasis.5 Various retrospective studies have correlated their over expression with decreased likelihood of survival and poor prognosis in oral cancers; however, with relatively less emphasis on its association and influence on QOL.6-10
Except for few studies in lung cancer there have been no comprehensive studies evaluating the expression of tumor proteins on traditional treatment outcome of survival and QOL.4,11
The correlation between tumor biology and patient quality of life is poorly understood. A better understanding of the effect of tumor and treatment-related factors on quality of life would facilitate patient counseling and treatment planning.
The current study was hence been planned with the objective to assess the association of the expression of EGFR and p53 with survival and quality of life in OSCC patients undergoing chemoradiation.
Patients and Methods
Sample size
The sample size of the study was priory estimated. The sample size was based on correlation coefficient between molecular markers expressions and QOL domains. Expecting a correlation coefficient of 0.35, significance level 0.01 (Type I error, α-level, 2-sided) and power 0.90 (Type II error, β-level), the minimum sample size required will be 100.
Study population
Total 120 cases of unresectable locally advanced stage (III/IV) oral cancer attending O.P.D. in the year 2009–2011 were enrolled in the study.
Our study population was locally advanced oral cavity cancers (stage III / IV). We included both unresectable and resectable cancers. We included those resectable cases where surgery results in either an unacceptable morbid R0 resection upfront or positive close margin.
We enrolled 134 patients but 14 were excluded from the study due to comorbid conditions and protocol violation.
Tumor was deemed to be unresectable if there was tumor fixation, tumors with features like extensive skin edema up to the zygomatic arch, extensive soft tissue involvement up to the hyoid cartilage and tumors reaching and involving the pterygoid muscles, involvement of retromolar trigone, fixed lymph nodes and low probability of surgical curability. These associated features are generally considered unresectable or resectable with unacceptable morbid curative (R0) resection or positive/close margin with substantially increased functional and cosmetic morbidity.12-14
Patients with histologically proven squamous cell carcinoma, grade 0–1, World Health Organization (W.H.O.) performance status and normal hematological, renal and liver function test were included. Patients with history of previous chemotherapy, radiotherapy and surgery, any comorbid condition and distant metastasis were excluded. The study was approved by the ethics committee of the University, and written informed consent was obtained from all patients before enrollment.
Treatment plan
All the patients received 2 cycles of induction taxol (175 mg/m2 day 1) and cisplatin (50 mg/m2 day 2) chemotherapy followed by radiation along with concurrent cisplatin (35 mg/m2) after 4 weeks of completion of induction chemotherapy. External Beam Radiotherapy (EBRT) was given by Cobalt 60 to a total dose of 70Gy in 35 fractions for 7 weeks to primary tumor site and neck. The treatment was continued despite mucositis or dermatitis. However, chemoradiation was postponed for 1 week or interrupted when patient developed grade 3 mucositis or dermatitis and myelosupression (WBC count < 4000/mm3 or platelets count < 100000/mm3), persistent fever that exceeded 38°C or other clinically apparent infections. The dose of cisplatin was reduced to 50% if the calculated creatinine clearance level 30–50 ml/min and cisplatin was discontinued if the creatinine clearance level less than 30 ml/min.
Response and toxicity assessment
Assessment of tumor response was done by clinical examination and radiological investigations (CT scan) 4–6 weeks after completion of treatment. Tumor responses were mainly assessed by clinical evaluations followed secondarily by imaging studies using CT scan. The definitions of treatment response viz. complete response (CR), partial response (PR), no response (NR), stable disease (SD) and progressive disease (PD) were based on the standard definitions established by W.H.O. (1979).15 Toxicity was graded according to National Cancer Institute Common Toxicity Criteria (NCI-CTC) version 3.0.
Quality of life measures
All patients were asked to complete FACT–H&N version 4 quality of life questionnaires at the commencing of the study, response assessment and end of the study. The FACT–H&N version 4 QOL instrument designed for head and neck cancer patients, consists of 4 domains: physical, social/family, emotional and functional well-being with further supplemented by additional concerns. The component “additional concerns” test symptoms specific to head and neck cancer which provides an opportunity to evaluate site- and treatment-specific issues. Each item is rated on a 0 to 4 Likert type scale, and then combined to produce subscale scores for each domain, as well as a global QOL score. Higher scores represent better QOL.
The principal investigator performed FACT-H&N QOL survey by interviewing patients in English language. In case of not well versed of English language, the patients were asked in local language Hindi or taken the help of attendant/relatives. The interviewers were given adequate and utmost training so as to elicit non-biased patient responses.16
Immunohistochemistry protocol for p53 and EGFR biomarkers
For immunohistochemical studies, tumor samples from the lesion site were fixed in 10% buffered formalin and then embedded in paraffin. Paraffin embedded formalin fixed tissues were processed routine H&E stained sections evaluated to confirm the diagnosis of squamous cell carcinoma and to grade the lesion. Further sections were processed for p53 and EGFR biomarkers by immunohistochemistry using primary monoclonal antibodies and a polymer based secondary antibody detection kit from Dakopatts, Denmark. Standard Immunohistochemistry protocol was used. In short deparaffinized rehydrated sections were blocked for endogenous peroxidases in 0.3% hydrogen peroxide in methanol, followed by a rinse in distill water. Antigen retrieval was achieved at 121°C in 10mM citrate buffer (pH 6.0) for 10 minutes using Pascal retrieval system from Dakopatts, Denmark. Slides cooled to room temperature were washed thrice with Tris-buffered saline (TBS), and thereafter incubated overnight at 4°C with Primary Antibodies p53 (DO7, Leica Microsystems, Germany) and EGFR (BioGenex, USA). After washing with Tris-buffered saline, the sections were incubated for 30 minutes with secondary antibody. EGFR and p53 were visualized with DAKO Liquid Diaminobenzidine substrate chromogen and counterstained with diluted Mayer's haematoxylin. Sections mounted with DPX were inspected under a Zeiss Z2 imager and photographed at 40x magnification.
The immunohistochemical evaluation was carried out in tumor hotspots including the invasion front, which was regarded as most indicative of the biological activity of the tumor, in 10 high power fields. About 1500–2000 tumor cells were observed in all tumors at a magnification of 40 × 10 selected fields.
The molecular marker expression of both EGFR and p53 were quantifies as percentage positive nuclei and counted in percentages. EGFR tumors were labeled as negative if tumor cell expressed the antigen <10%, moderate positive 10–50% and strongly positive >50 % and p53 expression was evaluated negative if expressed <10%, moderate positive if expressed 11–25% and strongly positive if expressed >25 %.17,18 For analysis purposes, the EGFR (Low ≤ 50%, High >50 %) and p53 (Low ≤ 25%, High >25 %) expressions were sub grouped as low (negative and moderate positive) and high (strong positive).
In the present study, we had designed in house positive and negative controls on the basis of percentage of protein expression. Positive controls were assumed if protein expression was more than 90% and negative if expression less than 10%.
After chemoradiation, patients were followed up to for 2 y The primary end measures of the study were 2 y survivals (overall, disease free and progression free) and QOL (physical well being, social/family well being, emotional well being, functional well being and additional concern). The secondary end point of the study was the treatment related toxicity. Survival time was defined as the interval between the date of initial treatment and the date of the last follow up examination. The QOL domains were assessed at baseline (before treatment), after 1 month of treatment and last follow.
Statistical analysis
Continuous data were summarized as Mean ± SD while discrete (categorical) in no and %. Categorical groups were compared by chi-square (χ2) test. Continuous groups were compared by one way analysis of variance (ANOVA) and the significance of mean difference between the groups was done by Tukey's post hoc test. Pearson correlation analysis was done to assess association between variables. Survival endpoints (overall survival, disease free survival and progression free survival) were compared by Kaplan-Meier method using Log-rank test. Cox multivariate regression analysis was done to assess independent predictors of overall survivals. Multivariate regression analysis was done to assess independent predictors of QOL. A two-tailed (α = 2) P < 0.05 was considered statistically significant.
Results
Basic characteristics
The basic (demographic and clinicopathological) characteristics of OSCC patients are summarized in Table 1. The age of patients ranged from 20–67 yrs with mean (± SD) 49.48 ± 12.19 and median 50 yrs. Among patients, mostly males (80.0%) and mostly had good performance status (56.7%) at presentation (enrollment). In patients, the most prevalent site was buccal mucosa (30.8%), mostly had T4 tumor size (75.8%), N0 node status (35.0%), stage IV (70.8%) and well differentiated histological grade (69.3%).
Table 1.
Basic characteristics of OSCC patients
| Characteristics | No. of OSCC patients (n = 120) (%) |
|---|---|
| Age (yrs): Mean ± SD Range (min to max) |
49.48 ± 12.1920 to 67 |
| Sex: Females Males |
24 (20.0)96 (80.0) |
| Performance status(WHO): Poor Good |
52 (43.3)68 (56.7) |
| Site of lesion: Alveolus Buccal mucosa Hard palate Lip RMT Tongue |
20 (16.7)37 (30.8)16 (13.3)11 (9.2)17 (14.2)19 (15.8) |
| Tumor size: T2 T3 T4 |
2 (1.7)27 (22.5)91 (75.8) |
| Node status: N0 N1 N2 |
38 42 (35.0)(31.7)40 (33.3) |
| Stage: III IV |
35 (29.2)85 (70.8) |
| Histological grade: Poorly differentiated Moderately differentiated Well differentiated |
7 (5.8)30 (25.0)83 (69.2) |
Molecular marker expressions
The expressions of molecular markers EGFR and p53 were shown in Figures 1, 2 respectively. The frequency distributions (Low/High) of expression of EGFR (Low: ≤50% and High: >50 %) and p53 (Low: ≤25% and High: >25 %) are summarized in Table 2. The EGFR expression of 33 (27.5%) patients were low and 87 (72.5%) were high while p53 expression of 29 (24.2%) were low and 91 (75.8%) were high. Comparing the frequency distributions (Low/High) of expressions (%) of 2 markers, χ2 test revealed similar frequency distributions of expression between the 2 markers (χ2 = 0.25, p = 0.555) i.e., not differed statistically.
Figure 1.
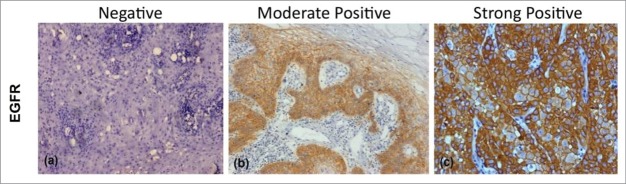
Figure showing Immunohistochemical expression of EGFR in OSCC (A) showing negative cytoplasmic and membranous staining (B) showing moderately positive cytoplasmic and membranous staining (C) showing strongly positive cytoplasmic and membranous staining (DAB x 125 x digital magnification).
Figure 2.
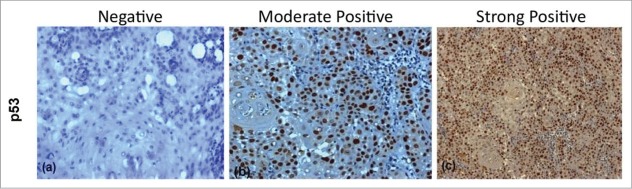
Figure showing Immunohistochemical expression of p53 in OSCC (A) showing negative nuclei (B) showing moderately positive stained nuclei (C) showing strongly positive stained nuclei (DAB x 125 x digital magnification).
Table 2.
Molecular marker expressions of OSCC patients
| Expression | EGFR (n = 120) (%) | p53 (n = 120) (%) | p value |
|---|---|---|---|
| Low | 33 (27.5) | 29 (24.2) | 0.555 |
| High | 87 (72.5) | 91 (75.8) |
Treatment response
The chemoradiation response (i.e. after 1 month of treatment) of OSCC patients are summarized in Table 3. In tumor, 64 (53.3%) patients had complete response (CR), 41 (34.2%) had partial response (PR) and 15 (12.5%) had no response (NR). In node, 63 (52.5%) patients had complete response, 42 (35.0%) had partial response and 15 (12.5%) had no response (NR). Overall (Tumor + Node), 39 (32.5%) patients had complete response, 63 (52.5%) had partial response and 18 (15.0%) had no response.
Table 3.
Chemoradiation response of OSCC patients
| Characteristics | No. of OSCC patients (n = 120) (%) |
|---|---|
| Tumor response: Complete response Partial response No response |
64 (53.3) 41 (34.2) 15 (12.5) |
| Node response: Complete response Partial response No response |
63 (52.5) 42 (35.0) 15 (12.5) |
| Overall response: Complete response Partial response No response |
39 (32.5)63 (52.5) 18 (15.0) |
The correlation between molecular marker expression levels (%) and overall response (CR, PR and NR) is summarized graphically in Figure 3. Correlating the mean molecular marker expression levels (%) with overall response (CR, PR and NR), EGFR and p53 both showed positive correlation with poor response (CR<PR<NR) i.e., as expression level increases, response decreases. Comparing the mean expression levels of both marker among the 3 response groups, ANOVA revealed significantly different expression levels of both EGFR (F = 82.91, P < 0.001) and p53 (F = 30.59, P < 0.001) among the groups. Further, Tukey test showed significantly different and higher mean (± SD) expressions of both markers in PR (EGFR: 70.95 ± 11.88 vs. 36.38 ± 21.97, P < 0.001; p53: 50.70 ± 19.05 vs. 29.28 ± 25.40, P < 0.001) and NR (EGFR: 84.78 ± 8.06 vs. 36.38 ± 21.97, P <0 .001; p53: 73.33 ± 10.02 vs. 29.28 ± 25.40, P < 0.001) as compared to CR. Moreover, the mean expression of both markers were also significantly higher in NR (EGFR: 84.78 ± 8.06 vs. 70.95 ± 11.88, P < 0.01; p53: 73.33 ± 10.02 vs. 50.70 ± 19.05 P < 0.001) as compared to PR.
Figure 3.

Association of molecular marker levels (%) with overall response in OSCC patients (A) Showing association of PR and NR with increase in levels of EGFR expression as compared to CR (B) showing significant association of NR with strong positive expressions of p53 along with higher mean expression levels of p53 in both PR and NR as compared to CR.
Survival
After chemoradiation, the OSCC patients were followed for 2 y During the period, 26 (21.7%) patients died due to disease, 68 (56.7%) were live and 13 (10.8%) left the treatment (LTF). The overall prevalence of alive (Live + LTF) was 67.5%. The 2 y overall survival of patients ranged from 2 to 24 months with median 11 month. The disease free (CR) and progression free (CR + NR) median survival were 12 month and 10 month, respectively (Table 4) .
Table 4.
Association between molecular marker expression and survival endpoints (n = 120)
| EGFR |
p53 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Survival endpoints | Expression | n | Live(n) (%) | LTF(n) (%) | Death(n) (%) | Median survival (month) | Expression | n | Live(n) (%) | LTF(n) (%) | Death(n) (% | Median survival (month) |
| Overall survival |
Low (≤50%) High (>50%) Total |
33 87 120 |
28 (84.8) 53 (60.9) 81 (67.5) |
2 (6.1) 11 (12.6) 13 (10.8) |
3 (9.1) 23 (26.4) 26 (21.7) |
14 9 11 |
Low (≤25%) High (>25%) Total |
29 91 120 |
27 (93.1) 54 (59.3) 81 (67.5) |
1 (3.4) 12 (13.2) 13 (10.8) |
1 (3.4) 25 (27.5) 26 (21.7) |
12 9 11 |
| Disease free survival | Low (≤50%) High (>50%) Total |
31 8 39 |
27 (87.1) 8 (100.0) 35 (89.7) |
1 (3.2) 0 (0.0) 1 (2.6) |
3 (9.7) 0 (0.0) 3 (7.7) |
14 6 12 |
Low (≤25%) High (>25%) Total |
23 16 39 |
21 (91.3) 14 (87.5) 35 (89.7) |
1 (4.3) 0 (0.0) 1 (2.6) |
1 (4.3) 2 (12.5) 3 (7.7) |
13 12 12 |
| Progression free survival | Low (≤50%) High (>50%) Total |
33 69 102 |
28 (84.8) 53 (76.8) 81 (79.4) |
2 (6.1) 9 (13.0) 11 (10.8) |
3 (9.1) 7 (10.1) 10 (9.8) |
14 8 10 |
Low (≤25%) High (>25%) Total |
29 73 120 |
25 (86.2) 56 (76.7) 81 (79.4) |
2 (6.9) 9 (12.3) 11 (10.8) |
2 (6.9) 8 (11.0) 10 (9.8) |
12 8 10 |
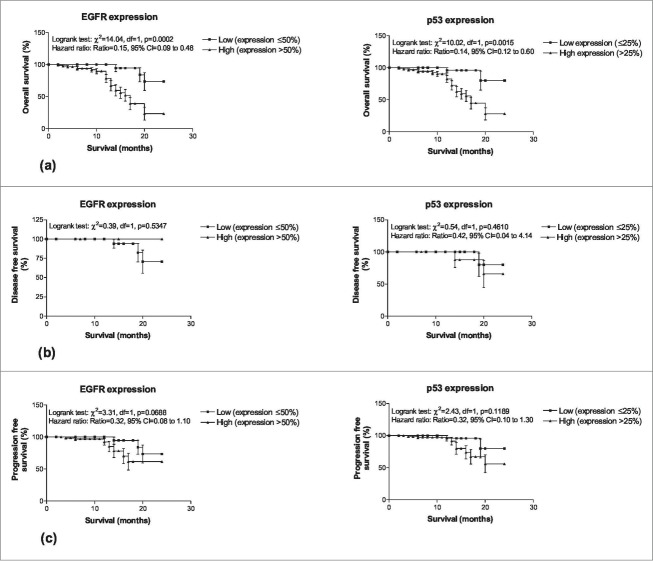
Comparing the low and high molecular markers expression with survival endpoints (Fig. 4), the Log-rank test showed significant association of both markers with overall survivals (EGFR: χ2 = 14.04, P < 0.001; p53: χ2 = 10.02, P < 0.01) indicating that patients with high expression had significantly lower survivals (EGFR: Hazard ratio = 0.15, 95% CI = 0.09–0.48; p53: Hazard ratio = 0.14, 95% CI = 0.12–0.60). However, expression of both the markers did not showed significant associations with both disease free and progression free survival.
Figure 4.
Two years overall survivals (A), disease free survivals (B) and (C) progression free survivals of OSCC patients according to low and high molecular marker expression.
To find out independent predictors of overall survivals, the Cox's multivariate regression analysis was done separately for both the markers and summarized in Table 5. The multivariate Cox regression analysis showed both the makers EGFR (OR = 0.36, 95% CI = 0.22–0.59, P < 0.001) and p53 (OR = 0.56, 95% CI = 0.35–0.59, P < 0.05) are significant and independent predictors of overall survival with higher association with EGFR than p53.
Table 5.
Association of demographic, clinicopathological and p53 expression with overall survival using Cox's multivariate regression analysis (n = 120)
| Predictors | Odds ratio (OR) (95% CI) | p value |
|---|---|---|
| Age (yrs): ≤50 >50 |
Ref 1.39 (0.94–2.07) | 0.101 |
| Sex: Female Male |
Ref 1.01 (0.60–1.70) | 0.959 |
| PS: Good Poor |
Ref 0.80 (0.52–1.22) | 0.301 |
| Lesion site: Buccal mucosa Other sites |
Ref 0.79 (0.52–1.20) | 0.266 |
| Tumor size: T2 + T3 T4 |
Ref 1.48 (0.75–2.90) | 0.255 |
| Node metastasis : N0 N1 + N2 |
Ref 0.85 (0.56–1.29) | 0.437 |
| Stage: III IV |
Ref 0.77 (0.41–1.47) | 0.430 |
| Histological grade: WD MD + PD |
Ref 1.11 (0.71–1.73) | 0.640 |
| Overall response: CR + PR NR |
Ref 1.28 (0.69–2.36) | 0.429 |
| p53 expression: Low (≤25 %) High (>25 %) |
Ref 0.56 (0.35–0.89) | 0.014 |
Odds ratio were calculated with respect to ref group.
Quality of life
The correlation between molecular marker expression levels (%) and QOL of OSCC patients at baseline (pre treatment), after one month of treatment and last follow up are summarized in Table 6. The Pearson correlation analysis revealed a significant (P < 0.001) and negative (inverse) correlation between expression of both the markers and QOL at all periods; indicating as expression levels increases, QOL decreases. Further, at all periods, the p53 showed higher correlation with QOL than EGFR.
Table 6.
Correlation between molecular marker expression levels (%) and quality of life of OSCC patients (n = 120)
| EGFR (%) |
p53 (%) |
|||||
|---|---|---|---|---|---|---|
| Quality of life | Baseline | After 1 month of treatment | Last follow up | Baseline | After 1 month of treatment | Last follow up |
| Physical well being | −0.46*** | −0.48*** | −0.59*** | −0.86*** | −0.72*** | −0.78*** |
| Social/Family well being | −0.34*** | −0.50*** | −0.55*** | −0.67*** | −0.77*** | −0.80*** |
| Emotional well being | −0.58*** | −0.47*** | −0.52*** | −0.88*** | −0.61*** | −0.63*** |
| Functional well being | −0.32*** | −0.49*** | −0.57*** | −0.56*** | −0.72*** | −0.80*** |
| Additional concern | −0.37*** | −0.66*** | −0.51*** | −0.55*** | −0.78*** | −0.64*** |
- P < 0.001.
To find out independent predictors of QOL, the multivariate regression analysis was done separately for both the markers and summarized in Tables 7, 8. At baseline, the QOL domains showed significant (P < 0.05 or P < 0.01 or P < 0.001) and independent associations with EGFR, sex (physical well being, social/family well being and additional concern) and histological grade (physical well being). In contrast, the QOL domains showed significant (P < 0.05 or P < 0.001) and independent associations with p53 and sex (social/family well being).
Table 7.
Association of demographic, clinico-pathological characteristics and EGFR expression levels (%) with QOL domains at baseline, after 1 month of treatment and last follow up using multivariate regression analysis (n = 120)
| Baseline (n = 120) Standardized regression coefficient (b) |
After 1 month of treatment (n = 120) Standardized regression coefficient (b) |
Last follow up (n = 120) Standardized regression coefficient (b) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern |
| Age | −0.013ns | 0.066ns | −0.045ns | 0.026ns | −0.022ns | 0.103ns | 0.090ns | −0.005ns | 0.086ns | −0.026ns | 0.102ns | 0.056ns | 0.049ns | 0.106ns | 0.167* |
| Sex | 0.185* | 0.226* | 0.034ns | 0.149ns | 0.192* | 0.236** | 0.171ns | 0.105ns | 0.135ns | 0.199** | 0.036ns | 0.062ns | −0.041ns | 0.041ns | 0.100ns |
| Performance status | 0.022ns | 0.037ns | 0.121ns | 0.141ns | 0.097ns | 0.089ns | 0.027ns | 0.121ns | 0.121ns | 0.081ns | 0.072ns | 0.071ns | 0.116ns | 0.070ns | 0.053ns |
| Lesion site | 0.017ns | 0.038ns | 0.056ns | 0.025ns | 0.029ns | −0.030ns | 0.024ns | 0.037ns | −0.029ns | −0.041ns | 0.025ns | −0.052ns | 0.006ns | 0.009ns | 0.086ns |
| Tumor size | 0.010ns | −0.008ns | −0.041ns | 0.105ns | −0.022ns | −0.089ns | −0.028ns | −0.017ns | −0.064ns | −0.052ns | −0.104ns | −0.042ns | −0.235ns | −0.077ns | 0.047ns |
| Node metastasis | −0.033ns | 0.030ns | −0.086ns | 0.081ns | 0.147ns | −0.062ns | −0.039ns | 0.039ns | −0.040ns | −0.013ns | −0.120ns | −0.093ns | −0.093ns | −0.153* | −0.141* |
| Stage | 0.034ns | 0.033ns | 0.159ns | −0.088ns | −0.012ns | −0.023ns | 0.041ns | 0.077ns | 0.057ns | −0.069ns | 0.073ns | 0.109ns | 0.333* | 0.108ns | −0.079ns |
| Histological grade | 0.218* | 0.086ns | 0.065ns | 0.017ns | −0.001ns | 0.092ns | 0.098ns | −0.051ns | 0.053ns | 0.018ns | 0.032ns | 0.031ns | −0.051ns | 0.056ns | −0.024ns |
| Overall response | NA | −0.233** | −0.203* | −0.269** | −0.067ns | −0.304*** | −0.289** | −0.292** | −0.272** | −0.385*** | −0.596*** | ||||
| EGFR | −0.457*** | −0.364*** | −0.569*** | −0.347*** | −0.424*** | −0.333*** | −0.409*** | −0.400*** | −0.429*** | −0.512*** | −0.403*** | −0.399*** | −0.384*** | −0.352*** | −0.216** |
- P > 0.05,
- P < 0.05,
- P < 0.01,
- P < 0.001.
NA: Overall response not included in the analysis as it is not available at baseline.
Standardized regression coefficient (b): − sign indicates negative (inverse) effect, + sign indicates positive (direct) effect.
Table 8.
Association of demographic, clinico-pathological characteristics and p53 expression levels (%) with QOL domains at baseline, after 1 month of treatment and last follow up using multivariate regression analysis (n = 120)
| Baseline (n = 120) Standardized regression coefficient (b) |
After 1 month of treatment (n = 120) Standardized regression coefficient (b) |
Last follow up (n = 120) Standardized regression coefficient (b) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern | Physical well being | Social/family well being | Emotional well being | Functional well being | Additional concern |
| Age | 0.004ns | 0.078ns | −0.020ns | 0.040ns | −0.002ns | 0.112ns | 0.102ns | 0.013ns | 0.100ns | −0.003ns | 0.115ns | 0.067ns | 0.066ns | 0.115* | 0.173** |
| Sex | 0.100ns | 0.158* | −0.058ns | 0.089ns | 0.129ns | 0.142ns | 0.061ns | 0.026ns | 0.028ns | 0.101ns | −0.065ns | −0.044ns | −0.115ns | −0.055ns | 0.044ns |
| Performance status | −0.069ns | −0.037ns | 0.041ns | 0.083ns | 0.049ns | 0.047ns | −0.021ns | 0.095ns | 0.076ns | 0.049ns | 0.030ns | 0.025ns | 0.092ns | 0.028ns | 0.029ns |
| Lesion site | 0.009ns | 0.031ns | 0.044ns | 0.018ns | 0.020ns | −0.034ns | 0.018ns | 0.030ns | −0.035ns | −0.051ns | 0.019ns | −0.058ns | −0.001ns | 0.004ns | 0.083ns |
| Tumor size | 0.059ns | 0.033ns | −0.012ns | 0.132ns | −0.013ns | −0.074ns | −0.013ns | −0.023ns | −0.055ns | −0.063ns | −0.094ns | −0.029ns | −0.242* | −0.063ns | 0.054ns |
| Node metastasis | 0.031ns | 0.083ns | −0.049ns | 0.116ns | 0.159ns | −0.048ns | −0.026ns | 0.026ns | −0.034ns | −0.033ns | −0.113ns | −0.082ns | −0.107ns | −0.140* | −0.136* |
| Stage | −0.080ns | −0.059ns | 0.036ns | −0.169ns | −0.097ns | −0.095ns | −0.045ns | 0.006ns | −0.030ns | −0.159ns | −0.009ns | 0.026ns | 0.265* | 0.033ns | −0.123ns |
| Histological grade | 0.083ns | −0.024ns | −0.062ns | −0.072ns | −0.080ns | 0.022ns | 0.017ns | −0.103ns | −0.025ns | −0.047ns | −0.041ns | −0.047ns | −0.100ns | −0.015ns | −0.065ns |
| Overall response | NA | −0.087ns | −0.035ns | −0.176* | 0.089ns | −0.191** | −0.140* | −0.131* | −0.186* | −0.236*** | −0.512*** | ||||
| p53 | −0.856*** | −0.693*** | −0.883*** | −0.591*** | −0.588*** | −0.629*** | −0.741*** | −0.543*** | −0.725*** | −0.677*** | −0.686*** | −0.716*** | −0.510*** | −0.650*** | −0.378** |
- P > 0.05,
- P < 0.05,
- P < 0.01,
- P < 0 .001.
NA: Overall response not included in the analysis as it is not available at baseline.
Standardized regression coefficient (b): − sign indicates negative (inverse) effect, + sign indicates positive (direct) effect.
After 1 month of treatment, the QOL domains showed significant (P < 0.05 or P < 0.01 or P < 0.001) and independent associations with EGFR, sex (physical well being), age (additional concern) and overall response (physical well being, social/family well being, emotional well being and additional concern). In contrast, the QOL domains showed significant (P < 0.05 or P < 0.01 or P < 0.001) and independent associations with p53 and overall response (emotional well being and additional concern).
At last follow up, the QOL domains showed significant (P < 0.05 or P < 0.01 or P < 0.001) and independent associations with EGFR, overall response, stage (emotional well being), lymph node metastasis (functional well being), age and stage (additional concern). In contrast, the QOL domains showed significant (P < 0.05 or P < 0.01 or P < 0.001) and independent associations with p53, overall response, tumor size and stage (emotional well being), age and lymph node metastasis (functional well being and additional concern). Further, at all periods, QOL domains showed higher associations (regression coefficient: b value) with the p53 than EGFR.
Toxicity
The association between molecular marker expression levels (%) and treatment related hematological and non hematological toxicity of OSCC patients after one month of chemoradiation treatment are summarized in Tables 9, 10, respectively. The expression of both the markers did not show significant associations with both the toxicities.
Table 9.
Association between molecular marker expressions and hematological toxicity of OSCC patients after one month of treatment (n = 120)
| EGDR |
p53 |
|||||
|---|---|---|---|---|---|---|
| Hematological toxicity | Low (Expression ≤50%) (n = 33) (%) | High (Expression >50%) (n = 87) (%) | p value | Low (Expression ≤50%) (n = 29) (%) | High (Expression >50%) (n = 91) (%) | p value |
| Anaemia: 0 1 2 3 4 |
2 (6.1)9 (27.3) 8 (24.2) 8 (24.2) 6 (18.2) |
11 (12.6)21 (24.1) 15 (17.2) 21 (24.1) 19 (21.8) |
0.769 |
3 (10.3)5 (17.2) 6 (20.7) 10 (34.5) 5 (17.2) |
10 (11.0)25 (27.5) 17 (18.7) 19 (20.9) 20 (22.0) |
0.575 |
| Leukopenia: 0 1 2 3 4 |
4 (12.1)7 (21.2) 7 (21.2) 9 (27.3) 6 (18.2) |
8 (9.2)29 (33.3) 21 (24.1) 17 (19.5) 12 (13.8) |
0.657 |
5 (17.2)7 (24.1) 7 (24.1) 5 (17.2) 5 (17.2) |
7 (7.7)29 (31.9) 21 (23.1) 21 (23.1) 13 (14.3) |
0.570 |
| Neutropenia: 0 1 2 3 4 |
5 (15.2)9 (27.3) 8 (24.2) 4 (12.1) 7 (21.2) |
19 (21.8)30 (34.5) 17 (19.5) 16 (18.4) 5 (5.7) |
0.114 |
4 (13.8)8 (27.6) 9 (31.0) 3 (10.3) 5 (17.2) |
20 (22.0)31 (34.1) 16 (17.6) 17 (18.7) 7 (7.7) |
0.210 |
| Thrombocytopenia: 0 1 2 3 4 |
7 (21.2)7 (21.2) 7 (21.2) 9 (27.3) 3 (9.1) |
28 (32.2)20 (23.0) 18 (20.7) 11 (12.6) 10 (11.5) |
0.377 |
6 (20.7)6 (20.7) 9 (31.0) 6 (20.7) 2 (6.9) |
29 (31.9)21 (23.1) 16 (17.6) 14 (15.4) 11 (12.1) |
0.430 |
| Febrile neutropenia: 0 1 2 3 |
23 (69.7)3 (9.1) 4 (12.1) 3 (9.1) |
69 (79.3)9 (10.3) 5 (5.7) 4 (4.6) |
0.484 |
18 (62.1)4 (13.8) 4 (13.8) 3 (10.3) |
74 (81.3)8 (8.8) 5 (5.5) 4 (4.4) |
0.174 |
Table 10.
Association between molecular markers expressions and non hematological toxicity of OSCC patients after one month of treatment (n = 120)
| EGFR |
p53 |
|||||
|---|---|---|---|---|---|---|
| Non hematological toxicity | Low (Expression ≤50%) (n = 33) (%) | High (Expression >50%) (n = 87) (%) | p value | Low (Expression ≤50%) (n = 29) (%) | High (Expression >50%) (n = 91) (%) | p value |
| Nausea: 0 1 2 3 4 |
2 (6.1)12 (36.4) 12 (36.4) 5 (15.2) 2 (6.1) |
9 (10.3)35 (40.2) 33 (37.9) 7 (8.0) 3 (3.4) |
0.702 |
2 (6.9)11 (37.9) 11 (37.9) 4 (13.8) 1 (3.4) |
9 (9.9)36 (39.6) 34 (37.4) 8 (8.8) 4 (4.4) |
0.934 |
| Vomiting: 0 1 2 3 4 |
6 (18.2)9 (27.3) 14 (42.4) 3 (9.1) 1 (3.0) |
13 (14.9)35 (40.2) 31 (35.6) 4 (4.6) 4 (4.6) |
0.642 |
5 (17.2)10 (34.5) 12 (41.4) 2 (6.9) 0 (0.0) |
14 (15.4)34 (37.4) 33 (36.3) 5 (5.5) 5 (5.5) |
0.751 |
| Renal dysfunction: 0 1 2 3 |
21 (63.6)7 (21.2) 3 (9.1) 2 (6.1) |
66 (75.9)14 (16.1) 2 (2.3) 5 (5.7) |
0.320 |
20 (69.0)6 (20.7) 3 (10.3) 0 (0.0) |
67 (73.6)15 (16.5) 2 (2.2) 7 (7.7) |
0.111 |
| Mucositis: 0 1 2 3 4 |
2 (6.1)7 (21.2) 7 (21.2) 7 (21.2) 10 (30.3) |
2 (2.3)19 (21.8) 29 (33.3) 23 (26.4) 14 (16.1) |
0.301 |
1 (3.4)6 (20.7) 7 (24.1) 6 (20.7) 9 (31.0) |
3 (3.3)20 (22.0) 29 (31.9) 24 (26.4) 15 (16.5) |
0.546 |
| Dermatitis (in field) 0 1 2 3 4 |
1 (3.0)14 (42.4) 13 (39.4) 4 (12.1) 1 (3.0) |
2 (2.3)44 (50.6) 36 (41.4) 3 (3.4) 2 (2.3) |
0.472 |
0 (0.0)13 (44.8) 12 (41.4) 3 (10.3) 1 (3.4) |
3 (3.3)45 (49.5) 37 (40.7) 4 (4.4) 2 (2.2) |
0.640 |
| Diarrhea: 0 1 2 3 4 |
25 (75.8)4 (12.1) 1 (3.0) 3 (9.1) 0 (0.0) |
64 (73.6)8 (9.2) 10 (11.5) 2 (2.3) 3 (3.4) |
0.209 |
22 (75.9)5 (17.2) 0 (0.0) 2 (6.9) 0 (0.0) |
67 (73.6)7 (7.7) 11 (12.1) 3 (3.3) 3 (3.3) |
0.127 |
Discussion
Squamous cell carcinoma is by far the most common cancer type of the oral cavity, representing more than 90% of all oral cancer. Although combined multimodality treatment, including surgery, chemotherapy, and radiation, have increased disease control for locally-advanced OSCC but it comes at the expense of increased acute and late toxicities which affects functional ability such as chewing, swallowing, taste sensation, speech and quality of voice, having a more profound effect on function and quality of life (QOL) than has been previously recognized.19,20
Apart from treatment related toxicities most of the studies on QOL issues in head and neck cancer have analyzed and concluded that notable impairment in QOL imposed by underlying functional or cosmetic compromises may also depend on post treatment response; which has been known to be varied in same stage disease, possibly due to variation in genetic signature likely to evoke expression of various biomarkers which might significantly affect therapeutic procedure and outcome.3,4,21,22
Recently research efforts have been focused on searching and estimation of molecular markers that are prognostic, predictive, effective and appropriate in customizing the treatment to achieve favorable and more desirable outcome.23,24
EGFR phosphorylation activate cascade of molecular chain reaction which stimulate multitude of oncogenic downstream signaling pathways like Ras-MAPK-ERK and PI3K/AKT which are associated with event of cell proliferation, cell cycle progression and inhibition of apoptosis leading to invasion, angiogenesis, metastasis.25
The expression of EGFR has been identified as noteworthy and an independent predictor of locoregional relapses. It is expressed in or highly expressed in approximately 90% of OSCC tumors along with variety of tumors including non-small cell lung cancer, breast, head and neck, gastric, colorectal, esophageal, prostate, bladder, renal, pancreatic and ovarian cancer.26,27
A lower level of EGFR has been associated with an improved disease-free interval after treatment with chemoradiation in patients with OSCC.28
p53 inactivation has also been correlated with worse prognosis, increased recurrence and poor survival rates in oral cancer. Mutations in the TP53 gene are found in up to 40–50 % of SCCHN tumors.29 Mutations of p53 have also been associated with decreased survival in patients with various other types of cancer, and serve as an independent prognostic factor.
Mutations of the tumor suppressor gene p53 are the most significant events in several human cancers. Various studies have documented that more than 90% of the p53 gene mutations in SCCHN in general are missense mutations, which are caused by change in an amino acid and a probable increase in stability of the protein which can be detected by immunohistochemical analysis due to stability of the protein.30,31
Studies have shown a strong correlation between immunohistochemical overexpression of p53 protein and the presence of missense mutations within the p53 gene.32-34 In breast cancer, accumulation of p53 protein is correlated with both p53 mutation and shortened survival, and has predicted decreased overall survival in node-negative patients.28 In colorectal cancer, accumulation of p53 correlated with high risk for disease recurrence and decreased survival.35,36 Overexpression of p53 have been associated with poor survival in a number of studies.32-34
Various retrospective and prospective studies have shown the relationship of p53 protein expression and prognosis among patients receiving radiation therapy in OSCC. It was concluded that overexpression of p53 protein is an indication of poor response to radiation therapy as compared with patients whose tumors did not accumulate p53 protein.37,38
In the present study we have found poor survival in patients where p53 was overexpressed (>25 %) as compared to moderate or negative expressions (≤25%), indicating possibility of occurrence of missense mutation in these patients and suggesting a prognostic role in OSCC patients undergoing chemoradiation.
Therefore expression of EGFR and p53 may serve as prognostic markers in patients of locally advanced oral squamous cell carcinoma and foremost need for such biomarkers interpretation and awareness is evident to improve the treatment response, survival and quality of life in oral cancer through more rational patient selection for curative regimens as well as palliative treatment.
Further, broader implementation of incorporation of these biomarkers into routine clinical, investigative, therapeutic procedures and therapeutic decisions in patients with these tumors would facilitate in procuring better treatment outcome with reformed QOL possibly by maximizing the response, diminishing interrupts and further easing and increasing the tolerability to get over to treatment related toxicities during the treatment period.
Although various studies have shown convincing evidences which indicate association of expression of molecular markers with treatment outcome, there are a limited number of studies that have analyzed and accentuated exact and absolute impact of correlation between expression of molecular markers, response and QOL.38
Yang et al. study was the pioneer work in linking genetic variations to QOL measures. The authors studied the role of glutathione metabolic genes on QOL in advanced lung cancer patients treated with platinum based chemotherapy and observed that inherited factors had a potential to predict patient's QOL. However, authors also claimed that plausible mechanism of association between genomic variations and QOL are underdeveloped at this stage.4
The findings from our studies suggest that a primary determinant of patient QOL is related to primary tumor biology, rather than treatment-related toxicity.
In our study we also found that patients with overexpressed p53 and EGFR had poor response with low QOL. Improvement in QOL in the patients with lower levels of p53 and EGFR expression might reciprocate potentiality to better tolerability due to escalation and maximization of response.
Therefore understanding the relationship between primary tumor biology, patient QOL and survival would be helpful in patient counseling and treatment planning.
Hence, deliberate and strategic effort should be focused to understand the more specific and sensitive markers aiding in tumor diagnosis, selection of treatment modality, monitoring of response to therapeutic interventions, early detection of tumor recurrence, prediction of the results from treatment modality and identification of subsets of patients with unfavorable outcome during the therapeutic interventions and follow-up, avoid undue treatment to achieve beneficial outcome in response and survival to achieve the goal of improvement in QOL along with cure in patients with these tumors. Thereby the role of molecular markers in diagnosis, treatment selection and surveillance for oral cancer requires additional investigation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Murphy BA, Ridner S, Wells N, Dietrich M. Quality of life research in head and neck cancer: a review of the current state of the science. Crit Rev Oncol Hematol 2007; 62(3):251-67; PMID:17408963; http://dx.doi.org/ 10.1016/j.critrevonc.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 2.Murphy BA. Late treatment effects: reframing the questions. Lancet Oncol 2009; 10(6):530-1; PMID:19482238; http://dx.doi.org/ 10.1016/S1470-2045(09)70118-4 [DOI] [PubMed] [Google Scholar]
- 3.Gold KA, Kim ES. Role of molecular markers and gene profiling in Head and neck cancer. Curr Opin Oncol 2009; 21(3):206-11; PMID:19346943; http://dx.doi.org/ 10.1097/CCO.0b013e328329ac00 [DOI] [PubMed] [Google Scholar]
- 4.Yang P, Mandrekar SJ, Hillman SH, Allen Ziegler KL, Sun Z, Wampfler JA, Cunningham JM, Sloan JA, Adjei AA, Perez E, et al.. Evaluation of glutathione metabolic genes on outcomes in advanced non-small cell lung cancer patients after initial treatment with platinum-based chemotherapy: an NCCTG-97-24-51 based study. J Thorac Oncol 2009; 4(4):479-85; PMID:19347979; http://dx.doi.org/ 10.1097/JTO.0b013e31819c7a2c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arteaga CL. The epidermal growth factor receptor: From mutant oncogene in nonhuman cancers to therapeutic target in human neoplasia. J Clin Oncol 2001; 19(18 Suppl):32S-40S; PMID:11560969 [PubMed] [Google Scholar]
- 6.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst 1998; 90(11):824-32; PMID:9625170; http://dx.doi.org/ 10.1093/jnci/90.11.824 [DOI] [PubMed] [Google Scholar]
- 7.Boyle JO, Hakim J, Koch W, van der Riet P, Hruban RH, Roa RA, Correo R, Eby YJ, Ruppert JM, Sidransky D. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res 1993; 53(19):4477-80; PMID:8402617 [PubMed] [Google Scholar]
- 8.Warnakulasuriya S. Prognostic and predictive markers for oral squamous cell carcinoma: The importance of clinical, pathological and molecular markers. Saudi J Med Med Sci 2014; 2:12-6; http://dx.doi.org/ 10.4103/1658-631X.128400 [DOI] [Google Scholar]
- 9.Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest 2012; 122(6):1951-7; PMID:22833868; http://dx.doi.org/ 10.1172/JCI59889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denaro N, Lo Nigro C, Natoli G, Russi EG, Adamo V, Merlano MC. The Role of p53 and MDM2 in Head and Neck Cancer. ISRN Otolaryngol 2011; 931813; PMID:23724261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprangers MA, Sloan JA, Veenhoven R, Cleeland CS, Halyard MY, Abertnethy AP, Baas F, Barsevick AM, Bartels M, Boomsma DI, et al.. The establishment of the GENEQOL consortium to investigate the genetic disposition of patient-reported quality-of-life outcomes. Twin Res Hum Genet 2009; 12(3):301-11; PMID:19456223; http://dx.doi.org/ 10.1375/twin.12.3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posner MR, Hershock DM, Blajman CR, Mickiewicz E, Winquist E, Gorbounova V, Tjulandin S, Shin DM, Cullen K, Ervin TJ, et al.. TAX 324 Study Group. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N Engl J Med 2007; 357(17):1705-15; PMID:17960013; http://dx.doi.org/ 10.1056/NEJMoa070956 [DOI] [PubMed] [Google Scholar]
- 13.Vermorken JB, Remenar E, van Herpen C, Gorlia T, Mesia R, Degardin M, Stewart JS, Jelic S, Betka J, Preiss JH, et al.. EORTC 24971/TAX 323 Study Group. Cisplatin, fluorouracil, and docetaxel in unresectable head and neck cancer. N Engl J Med 2007; 357(17):1695-704; PMID:17960012; http://dx.doi.org/ 10.1056/NEJMoa071028 [DOI] [PubMed] [Google Scholar]
- 14.Patil VM, Noronha V, Joshi A, Muddu VK, Gulia S, Bhosale B, Arya S, Juvekar S, Chatturvedi P, Chaukar DA, et al.. Induction chemotherapy in technically unresectable locally advanced oral cavity cancers: does it make a difference?. Indian J Cancer 2013; 50(1):1-8; PMID:23713035; http://dx.doi.org/ 10.4103/0019-509X.112263 [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization WHO handbook for reporting results of cancer treatment. http://whqlibdoc.who.int/publications/9241700483.pdf. [Google Scholar]
- 16.List MA, D'Antonio LL, Cella DF, Siston A, Mumby P, Haraf D, Vokes E. The Performance Status Scale for Head and Neck Cancer Patients and the Functional Assessment of Cancer Therapy-Head and Neck Scale. A study of utility and validity. Cancer 1996; 77(11):2294-301; PMID:8635098; http://dx.doi.org/ 10.1002/(SICI)1097-0142(19960601)77:11%3c2294::AID-CNCR17%3e3.0.CO;2-S [DOI] [PubMed] [Google Scholar]
- 17.Shiraki M, Odajima T, Ikeda T, Sasaki A, Satoh M, Yamaguchi A, Noguchi M, Nagai I, Hiratsuka H. Combined expression of p53, cyclin D1 and epidermal growth factor receptor improves estimation of prognosis in curatively resected oral cancer. Mod Pathol 2005; 18:1482-9; PMID:16007067; http://dx.doi.org/ 10.1038/modpathol.3800455 [DOI] [PubMed] [Google Scholar]
- 18.Cruz I, Napier SS, van der Waal I, Snijders PJ, Walboomers JM, Lamey PJ, Cowan CG, Gregg TA, Maxwell P, Meijer CJ. Suprabasal p53 Immunoexpresssion is strongly assicated with high grade dysplasia and risk for maliganant transformation in potentially malignant oral lesion Nothern Ireland. J Clin Pathol 2002; 55:98-104; PMID:11865002; http://dx.doi.org/ 10.1136/jcp.55.2.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah JP, Singh B: Keynote comment: why the lack of progress for oral cancer? Lancet Oncol 2006; 7:356-357; PMID:16648036; http://dx.doi.org/ 10.1016/S1470-2045(06)70667-2 [DOI] [PubMed] [Google Scholar]
- 20.Williams HK. Molecular pathogenesis of oral squamous carcinoma. Mol Pathol 2000; 53(4):165-72; PMID:11040937; http://dx.doi.org/ 10.1136/mp.53.4.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sloan JA, Zhao CX. Genetics and quality of life. Curr Probl Cancer 2006; 30(6):255-60; PMID:17123877; http://dx.doi.org/ 10.1016/j.currproblcancer.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 22.Forastiere AA. Another look at induction chemotherapy for organ preservation in patients with head and neck cancer. J Natl Cancer Inst 1996; 88(13):855-6; PMID:8656431; http://dx.doi.org/ 10.1093/jnci/88.13.855 [DOI] [PubMed] [Google Scholar]
- 23.Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncology, Oral Oncol 2009; 45(4–5):301-8; http://dx.doi.org/ 10.1016/j.oraloncology.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 24.Morton RP, Izzard ME. Quality-of-life outcomes in head and neck cancer patients. World J Surg 2003; 27(7):884-9; PMID:14509523; http://dx.doi.org/ 10.1007/s00268-003-7117-2 [DOI] [PubMed] [Google Scholar]
- 25.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol 2006; 24(17):2666-72; PMID:16763281; http://dx.doi.org/ 10.1200/JCO.2005.04.8306 [DOI] [PubMed] [Google Scholar]
- 26.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 1995; 19(3):183-232; PMID:7612182; http://dx.doi.org/ 10.1016/1040-8428(94)00144-I [DOI] [PubMed] [Google Scholar]
- 27.Thomas GR, Nadiminti H, Regalado J. Molecular predictors of clinical outcome in patients with head and neck squamous cell carcinoma. Int J Exp Pathol 2005; 86(6):347-63; PMID:16309541; http://dx.doi.org/ 10.1111/j.0959-9673.2005.00447.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan JJ, Inga A, Conway K, Edmiston S, Carey LA, Wu L, Resnick MA. Altered-Function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol Cancer Res 2010; 8(5):701-16; PMID:20407015; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temam S, Flahault A, Perie S, Monceaux G, Coulet F, Callard P, Bernaudin JF, St Guily JL, Fouret P. p53 gene status as a predictor of tumor response to induction chemotherapy of patients with locoregionally advanced squamous cell carcinomas of the head and neck. J Clin Oncol 2000; 18(2):385-94; PMID:10637254 [DOI] [PubMed] [Google Scholar]
- 30.Harris CC. p53: at the crossroads of molecular carcinogenesis and risk assessment. Science 1993; 262:1980-1; PMID:8266092; http://dx.doi.org/ 10.1126/science.8266092 [DOI] [PubMed] [Google Scholar]
- 31.Nylander K, Nilsson P, Mehle C, Roos G. p53 mutations, protein expression and cell proliferation in squamous cell carcinomas of the head and neck. Br J Cancer 1995; 71:826-30; PMID:7710950; http://dx.doi.org/ 10.1038/bjc.1995.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baas IO, Mulder JW, Offerhaus GJ, Vogelstein B, Hamilton SR. An evaluation of six antibodies for immunohistochemistry of mutant p53 gene product in archival colorectal neoplasms. J Pathol 1994; 172:5-12; PMID:7931827; http://dx.doi.org/ 10.1002/path.1711720104 [DOI] [PubMed] [Google Scholar]
- 33.Kressner U, Inganas M, Byding S, Blikstad I, Pahlman L, Glimelius B, Lindmark G. Prognostic value of p53 genetic changes in colorectal cancer. J Clin Oncol 1999; 17:593-9; PMID:10080604 [DOI] [PubMed] [Google Scholar]
- 34.Havrilesky L, Darcy KM, Hamdan H, Priore RL, Leon J, Bell J, Berchuck A. Prognostic significance of p53 mutation and p53 overexpression in advanced epithelial ovarian cancer: a Gynecologic Oncology Group Study. Gynecologic Oncology Group Study. J Clin Oncol 2003; 21:3814-25; PMID:14551300; http://dx.doi.org/ 10.1200/JCO.2003.11.052 [DOI] [PubMed] [Google Scholar]
- 35.Tortola S, Marcuello E, Gonzalez I, Reyes G, Arribas R, Aiza G, Sancho FJ, Peinado MA, Capella G. p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. J Clin Oncol 1999; 17(5):1375-81; PMID:10334521 [DOI] [PubMed] [Google Scholar]
- 36.Vousden KH. Outcomes of p53 activation-spoilt for choice. J Cell Sci 2006; 119:5015-20; PMID:17158908; http://dx.doi.org/ 10.1242/jcs.03293 [DOI] [PubMed] [Google Scholar]
- 37.Quon H, Liu F, Cummings B. Potential molecular prognos-tic markers in head and neck squamous cell carcinomas. Head Neck 2001; 23(2):147-59; PMID:11303632; http://dx.doi.org/ 10.1002/1097-0347(200102)23:2%3c147::AID-HED1010%3e3.0.CO;2- [DOI] [PubMed] [Google Scholar]
- 38.Raybaud-Diogene H, Fortin A, Morency R, Roy J, Moteil R, Tetu B. Markers of radioresistance in squamous cell carcinomas of the head and neck: a clinicopathologic and immunohistochemical study. J Clin Oncol 1997; 15(3):1030-8.4; PMID:9060543 [DOI] [PubMed] [Google Scholar]