Abstract
MiR-145 has been implicated in the progression of non-small cell lung cancer (NSCLC); however, its exact mechanism is not well established. Here, we report that miR-145 expression is decreased in NSCLC cell lines and tumor tissues and that this low level of expression is associated with DNA methylation. MiR-145 methylation in NSCLC was correlated with a more aggressive tumor phenotype and was associated with poor survival time, as shown by Kaplan-Meier analysis. Additional multivariate Cox regression analysis indicated that miR-145 methylation was an independent prognostic factor for poor survival in patients with NSCLC. Furthermore, we found that restoration of miR-145 expression inhibited proliferation, migration and invasion of NSCLC by the direct targeting of mucin 1 by miR-145. Our results indicate that low miR-145 expression, due to methylation, promotes NSCLC cell proliferation, migration and invasion by targeting mucin 1. Therefore, miR-145 may be a valuable therapeutic target for NSCLC.
Keywords: invasion, methylation, migration, miR-145, mucin 1, non–small cell lung cancer, proliferation
Abbreviations
- NSCLC
non–small cell lung cancer
- MSP
Methylation-specific PCR
- HBE
human bronchial epithelial cell line
- CCK-8
Cell Counting Kit-8
- 5-aza-20-deoxycytidine
5-Aza
- GAPDH
glyceraldehydes phosphate dehydrogenase
- SqCC
squamous cell cancer
Introduction
Lung cancer is the leading cause of death from cancer around the world, and presumably 80–85% of lung cancers are non–small cell lung cancer (NSCLC).1 Despite improvements in therapeutic approaches, most patients are diagnosed at advanced stages, and the overall prognosis of NSCLC remains unsatisfactory.2 Therefore, it is important to better understand the mechanisms of disease pathogenesis and to identify new therapeutic targets.
MicroRNAs (miRNAs) are a class of noncoding small RNAs of approximately 22 nucleotides in length that regulate gene expression by suppressing and/or degrading protein-coding mRNA.3 Emerging evidence has shown that miRNAs are involved in cancer initiation and progression.4 MiR-145 has been reported as an important tumor suppressor gene in ovarian carcinoma,5,6,7 malignant pleural mesothelioma,8 breast cancer,9 pancreatic cancer,10,11 liposarcoma,12 colorectal cancer,13,14,15,16 neuroblastoma,17 breast cancer,18,19,20,21 esophageal cancer,22 bladder cancer,23,24 urothelial cancer,25 prostate cancer,26,27 gastric cancer,28 and head and neck cancer,29 and other types. MiR-145 was also implicated in the progression of NSCLC;30,31,32 however, its exact mechanism is not well established. Here, we show that miR-145 methylation, resulting in low-level miR-145 expression, was associated with a more aggressive tumor phenotype in NSCLC. Kaplan-Meier analysis revealed that miR-145 methylation in NSCLC was associated with shorter survival time, and multivariate Cox regression analysis indicated that miR-145 methylation was an independent prognostic factor for poor survival in patients with NSCLC. Furthermore, we found that restoration of miR-145 expression inhibited proliferation, migration and invasion of NSCLC cells through its direct targeting of mucin 1, which is a critical oncogene that promotes NSCLC progression. In summary, our results provide novel mechanistic insights into the NSCLC progression evoked by miR-145 loss and identify a potential clinical biomarker for lung cancer prognosis.
Results
Downregulated miR-145 expression is frequently detected in NSCLC cells and human NSCLC tissues
To explore miR-145 expression in NSCLC, we examined its expression using real-time PCR in a human bronchial epithelial cell line (HBE), 4 NSCLC cell lines and 20 paired human NSCLC and matched adjacent non-tumor tissues. The results showed that all 4 NSCLC cell lines (A549, NCI-H520, NCI-H460 and NCI-H596) had lower miR-145 expression than the normal HBE cell line (Fig. 1A). Furthermore, miR-145 expression was significantly lower in the NSCLC samples than in the matched non-tumor tissues (P < 0.01) (Fig. 1B). In addition, miR-145 expression was examined in 10 primary NSCLC tissue samples with metastasis and 10 primary NSCLC tissues samples without metastasis. We found that miR-145 expression was downregulated in the 10 primary NSCLC tissues with metastasis compared to those without metastasis (P < 0.01) (Fig. 1C). Furthermore, we conducted real-time PCR to examine the expression of miR-145 in NSCLC tissue samples of 122 patients. The median value of all 122 NSCLC samples was chosen as the cutoff point for separating tumor samples with high miR-145 expression from those with low-level miR-145 expression, resulting in 61/122 (50%) NSCLCs with high miR-145 expression and 61/122 (50%) NSCLCs with low miR-145 expression. Kaplan-Meier analysis suggested that the low miR-145 expression in NSCLCs was associated with poor survival time (P < 0.01, Fig. 1D).
Figure 1.
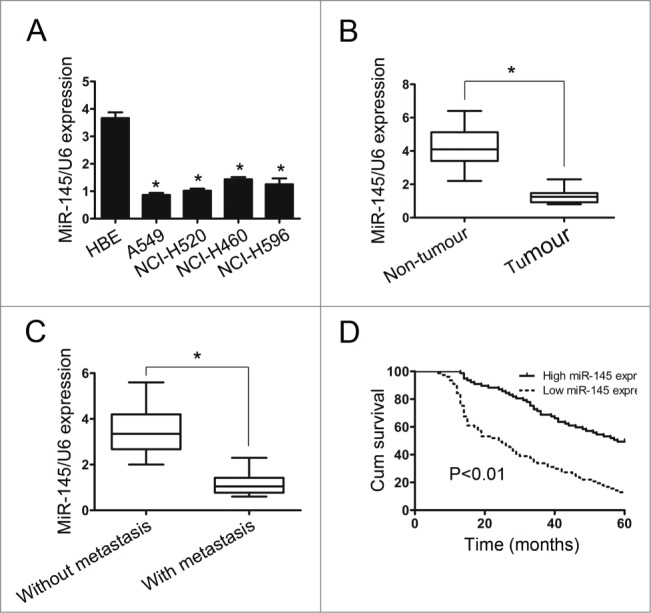
The expression of miR-145 in non-small cell lung cancer cell (NSCLC) lines and tissues. (A) MiR-145 expression was determined in HBE cells and in 4 NSCLC cell lines by real-time PCR (n=3 replicate experiments; P < 0.05). (B) MiR-145 expression in 20 paired NSCLC and adjacent non-tumor tissue samples. (C) MiR-145 expression in NSCLCs with and without distant metastasis. The mean level of miR-145 expression in NSCLCs with distant metastasis was lower than that without distant metastasis. (D) Kaplan-Meier analysis suggested that the low miR-145 expression in NSCLCs was associated with poor survival time.
Restoration of miR-145 expression inhibits NSCLC cell proliferation, migration and invasion
To explore the potential biological function of miR-145 in NSCLC cells, we used Cell Counting Kit-8 (CCK-8) assays and colony formation assays to determine the role of miR-145 in NSCLC cell proliferation. A549 and NCI-H520 cells were transfected with miR145 mimics and miR-control, respectively. CCK-8 assays showed that the OD value difference between the second day and the first day decreased significantly in A549 and NCI-H520 cells transfected with miR-145 mimics compared with miR-control-transfected cells (p = 0.0056, 0.0037) (Fig. 2A). And the similar difference can be found between the third day and the second day in A549 and NCI-H520 cells (p = 0.0475, 0.0459). All the results showed that overexpression of miR-145 suppressed remarkably the proliferation of A549 cells and NCI-H520 cells. Furthermore, colony formation assays showed that miR-145 overexpression suppressed the colony-forming ability of A549 cells and NCI-H520 cells (Fig. 2B).
Figure 2.
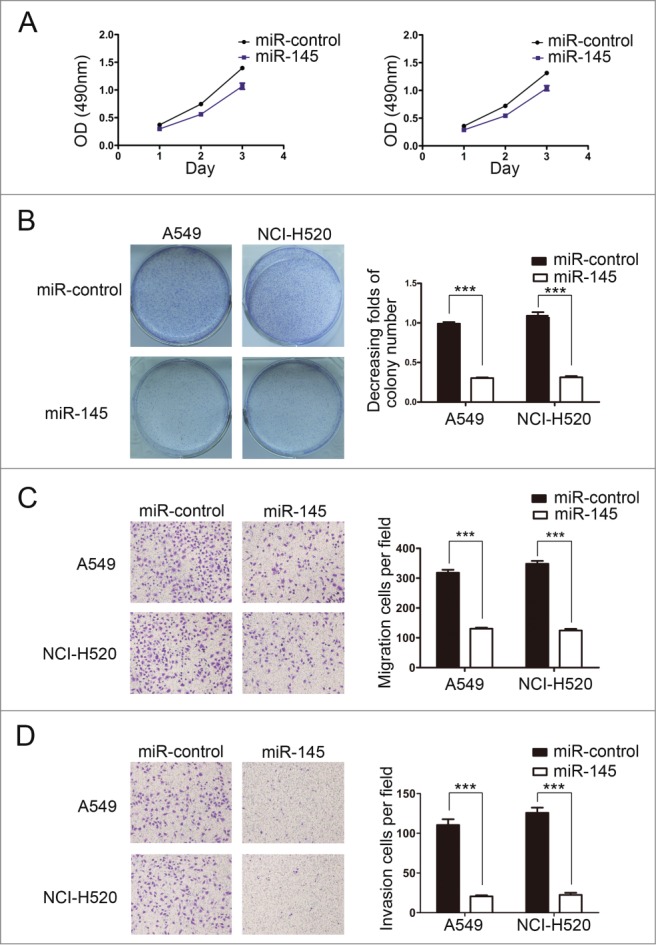
Restoration of miR-145 expression inhibits NSCLC cell proliferation, migration and invasion. (A) Restoration of miR-145 expression in A549 and NCI-H520 cells significantly reduced proliferation compared with the miR-control. (B) Restoration of miR-145 expression in A549 and NCI-H520 cells significantly suppressed colony-forming ability. (C) Restoration of miR-145 expression in A549 and NCI-H520 cells significantly inhibited migration ability. (D) Restoration of miR-145 expression in A549 and NCI-H520 cells significantly reduced invasion ability.
To study whether miR-145 affected the migration and invasion function of NSCLC cells, we performed transwell migration assays and matrigel invasion assays. The results demonstrated that restoration of miR-145 expression dramatically inhibited the migration and invasion function of A549 and NCI-H520 cells (Fig. 2C, D).
The mechanism of low-level miR-145 expression in NSCLC is through hypermethylation: correlation between miR-145 expression and methylation status and its use as a diagnostic marker
To explore the mechanism of the down-regulated miR-141 expression, we performed methylation-specific PCR (MSP) to analyze the methylation status of miR-145 in primary NSCLC tissue samples and in the corresponding non-tumor lung tissue samples of 122 patients. The results showed that the methylation status of miR-145 was significantly higher in NSCLC tissues compared with the corresponding non-tumor lung tissues (Fig. 3A). In addition, analysis of these results indicated that low-level miR-145 expression was associated with RKIP promoter methylation in NSCLC tissues (Fig. 3B). To test whether this epigenetic silencing of miR-145 also occurred in human NSCLC cell lines, real-time PCR was performed to examine miR-145 expression after 5-aza-20-deoxycytidine treatment in NSCLC cells, and an obvious increase in miR-145 levels was detected in these cells (Fig. 3C). These data showed that miR-145 expression is silenced by hypermethylation in NSCLC cells. Overall, these results confirm that methylation is indeed responsible for the downregulation of miR-145 expression in NSCLC.
Figure 3.
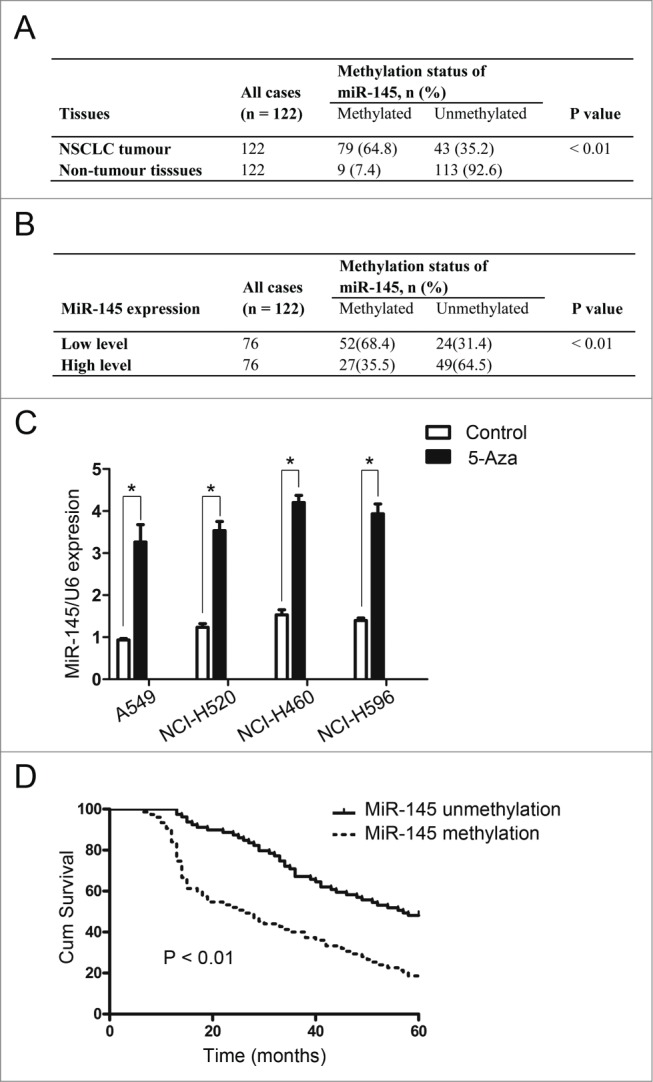
Methylation status of miR-145 in NSCLC. (A) Methylation-specific PCR assays showed that the methylation status of miR-145 was significantly higher in 122 NSCLC tissue samples compared with the corresponding non-tumor lung tissues. (B) Low miR-145 expression was associated with RKIP promoter methylation in 122 NSCLC tissue samples. (C) Real-time PCR was performed to examine the expression of miR-145 after 5-aza-20-deoxycytidine treatment in NSCLC cells, and obvious increases were detected in these cells. (D) Kaplan-Meier analysis revealed that miR-145 methylation in NSCLC was associated with shorter survival time.
To further investigate the clinicopathological and prognostic significance of miR-145 methylation status in patients with NSCLC, we performed correlation analysis. The results showed that miR-145 methylation in NSCLC was associated with a more aggressive tumor phenotype (Table 1). Kaplan-Meier analysis revealed that miR-145 methylation in NSCLC was associated with shorter survival time (P < 0.01, Fig. 3D), and multivariate Cox regression analysis indicated that miR-145 methylation was an independent prognostic factor for poor survival in patients with NSCLC (Table 2).
Table 1.
Correlation of miR-145 methylation in tissue samples with clinicopathological variables of patients in 122 cases of non-small cell lung cancer
| miR-145 |
||||
|---|---|---|---|---|
| Variables | All cases (n = 122) | Methylated n (%) | Unmethylated n (%) | P Value* |
| Age (years) | ||||
| ≤60 | 55 | 35 | 20 | 0.815 |
| >60 | 67 | 44 | 23 | |
| Gender | ||||
| Male | 78 | 51 | 27 | 0.846 |
| Female | 44 | 28 | 16 | |
| Histological type | ||||
| SqCC | 36 | 25 | 11 | 0.758 |
| A | 71 | 45 | 26 | |
| Other | 15 | 9 | 6 | |
| Differentiation | ||||
| Low | 65 | 48 | 17 | 0.025 |
| Moderate+ high | 57 | 31 | 26 | |
| T factor | ||||
| T1 + T2 | 71 | 49 | 22 | 0.642 |
| T3 + T4 | 41 | 30 | 11 | |
| Lymph node | ||||
| N0 + N1 | 58 | 29 | 29 | 0.001 |
| N2 + N3 | 64 | 50 | 14 | |
| Distant metastasis | ||||
| M0 | 98 | 58 | 40 | 0.009 |
| M1 | 24 | 21 | 3 | |
| Clinical stage | ||||
| = 1/* ROMAN I + = 2/* ROMAN II | 56 | 28 | 28 | 0.002 |
| = 3/* ROMAN III + = 4/* ROMAN IV | 66 | 51 | 15 | |
χ2 test; SqCC, squamous cell cancer; A, adenocarcinoma.
Table 2.
Univariate and multivariate analysis of factors associated with survival time of patients with non-small cell cancer
| Clinical variable | Case number | HR (95% CI) | P Value |
|---|---|---|---|
| Univariate analysis | |||
| miR-145 (Unmethylation vs. Methylation) | 43/79 | 1.851(1.355–2.528) | <0.001 |
| Age ( >60 vs. ≤60) | 67/55 | 1.034(0.777–1.375) | 0.819 |
| Gender (Male vs. Female) | 78/44 | 1.202(0.896–1.613) | 0.220 |
| Histological type (SqCC vs A vs. Other) | 36/71/15 | 1.087(0.847–1.394) | 0.513 |
| Distant metastasis | |||
| (M1 vs. M0) | 24/98 | 5.280(3.552–7.850) | <0.001 |
| T factor (T3 + T4 vs. T1 + T2) | 41/71 | 1.249(0.940–1.660) | 0.126 |
| Lymph node (N2 + N3 vs. N0 + N1) | 64/58 | 5.466(3.942–7.580) | <0.001 |
| Clinical stage ( = 3/* ROMAN III + = 4/* ROMAN IV vs. = 1/* ROMAN I + = 2/* ROMAN II) | 66/56 | 7.424(5.228–10.541) | <0.001 |
| Differentiation | |||
| (Moderate + high vs. Low) | 57/65 | 2.006(1.500–2.684) | <0.001 |
| Multivariate analysis | |||
| miR-145 (Unmethylation vs. Methylation) | 43/79 | 0.414(0.297–0.578) | <0.001 |
| Differentiation | |||
| (Moderate + high vs. Low) | 57/65 | 1.295(0.928–1.806) | 0.128 |
| Distant metastasis | |||
| (M1 vs. M0) | 24/98 | 3.877(2.515–5.976) | <0.001 |
| Lymph node (N2 + N3 vs. N0 + N1) | 64/58 | 1.270(0.703–2.297) | 0.428 |
| Clinical stage ( = 3/* ROMAN III + = 4/* ROMAN IV | |||
| vs. = 1/* ROMAN I + = 2/* ROMAN II) | 66/56 | 6.171(3.248–11.726) | <0.001 |
Mucin 1 is a direct target of miR-145
MiRNAs exert their function by regulating gene expression via targeting protein-encoding mRNAs. A target prediction program, TargetScan, showed that mucin 1 may be a potential target of miR-145. The 3′-UTR of the mucin 1 mRNA contains a conserved binding site for the seed region of miR-145 (Fig. 4A). A549 cells and NCI-H520 cells were therefore transfected with miR145 mimics and miR-control, respectively; however, restoration of miR-145 expression did not elicit the degradation of the mucin mRNA (Fig. 4B). To determine whether mucin 1 was a direct target of miR-145 in NSCLC, a human mucin 1 3´-UTR fragment containing a wild-type or mutant miR-145 binding sequence was cloned downstream of the firefly luciferase reporter gene. Restoration of miR-145 expression reduced the activity of luciferase reporter genes fused to the wild-type mucin 1 3´-UTR in A549 cells and NCI-H520 cells (Fig. 4C). Conversely, in luciferase assays using a plasmid harboring a mutant version of the mucin 1 mRNA 3´-UTR (the miR-145 binding sites were inactivated by site-directed mutagenesis), the luciferase activity of the mutant reporter was unaffected by transfection of miR-145 into A549 and NCI-H520 cells (Fig. 4C). These data suggest that miR-145 may suppress mucin 1 gene expression through the miR-145 binding sequence in its 3´-UTR.
Figure 4.
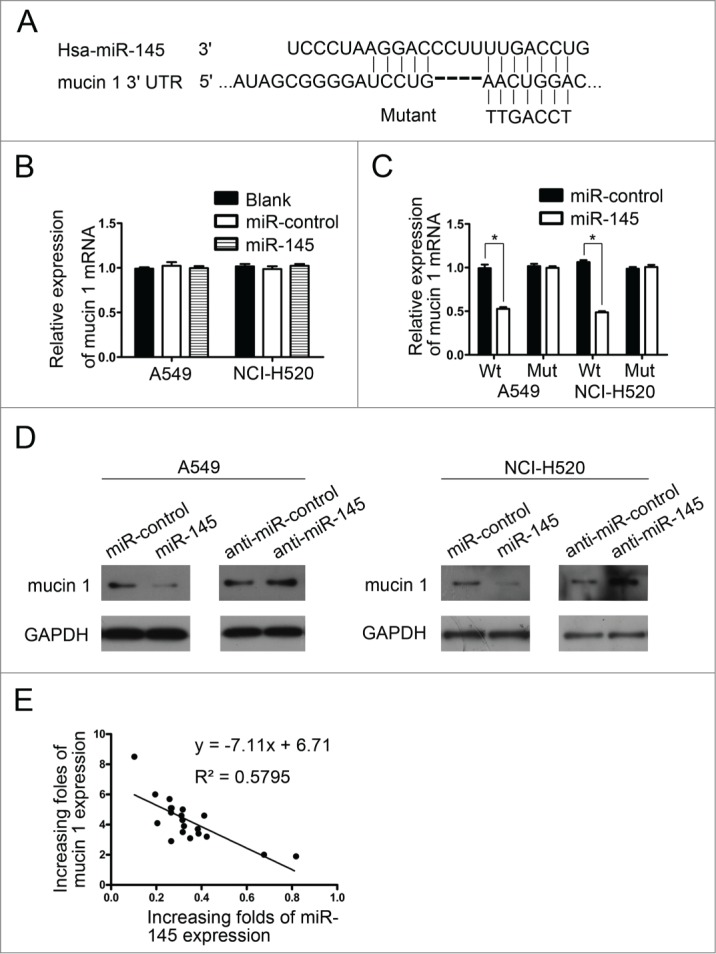
Mucin 1 is target of miR-145. (A) Schematic of the predicted miR-145 binding sequence in the mucin 1 3´-UTR. The complementary site in the mucin 1 3´-UTR for the seed region of miR-145 was mutated as indicated. A human mucin 1 3´-UTR fragment containing a wild-type or mutant miR-145 binding sequence was cloned downstream of the luciferase reporter gene. (B) Real-time PCR was performed to analyze mucin 1 expression in NSCLC cells transfected with miR-145 or the miR-control; the data were normalized to GAPDH mRNA expression. The endogenous levels of mucin 1 mRNA did not change significantly in the miR-145 groups compared with the miR-control. (C) Luciferase activity of the wild-type (Wt) or mutant (Mut) mucin 1 3´ UTR reporter gene in NSCLC cells transfected with miR-145 or miR-control. (D) Mucin 1 immunoblotting of NSCLC cells transfected with miR-145 or miR-control and anti-miR-145 (miR-145 inhibitor) or anti-miR-control. Restoration of miR-145 inhibited mucin 1 expression in A549 and NCI-H520 cells compared with the miR-control. Anti-miR-145 promoted mucin 1 expression in A549 and NCI-H520 cells compared with the anti-miR-control. (E) Mucin 1 protein levels in 20 paired NSCLC and adjacent non-tumor tissue samples were determined by Western blotting, and miR-145 expression was examined by real-time PCR. The degree of mucin 1 upregulation correlated inversely with the extent of miR-145 downregulation.
The effects of miR-145 on the endogenous expression of mucin 1 were determined using Western blotting (Fig. 4D). Restoration of miR-145 expression resulted in a marked decrease in mucin 1 expression in A549 and NCI-H520 cells, whereas miR-145 inhibitor oligonucleotides induced a significant increase in mucin 1 expression.
To examine whether the biological effects of low miR-145 expression correlated with mucin 1 protein levels in clinical NSCLC tissues, mucin 1 protein levels in 20 paired NSCLC and adjacent non-tumor tissue samples were determined by Western blotting, and miR-145 expression was examined by real-time PCR (data not shown). The degree of mucin 1 up-regulation correlated inversely with the extent of miR-145 downregulation (R2 = 0.5795, P < 0.01) (Fig. 4E), indicating that the inhibitory effects of miR-145 on mucin 1 were clinically relevant in NSCLC.
These results indicate that miR-145 inhibits mucin 1 expression at the post-transcriptional level by directly targeting the 3´-UTR of the mucin 1 mRNA.
Restoration of mucin 1 expression reverses the miR-145-mediated inhibition of proliferation, migration and invasion of NSCLC cells
To explore whether mucin 1 inverses the miR-145-mediated inhibition of proliferation, migration and invasion of NSCLC cells, we performed rescue experiments by co-transfecting a constitutively active form of PAK6 or a mock construct with a miR-145 mimic into A549 and NCI-H520 cells. The results of the CCK-8 assays and colony formation assays showed that mucin 1 promoted the proliferation and colony-forming ability of A549-miR-145 and NCI-H520-miR-145 cells compared with mock-transfected cells (Fig. 5A, B). The results of transwell migration assays and matrigel invasion assays indicated that mucin 1 increased the migration and invasion of A549-miR145 and NCI-H520-miR145 cells (Fig. 5C). All of these results showed that restoration of mucin 1 expression could inverse the miR-145-mediated inhibition of proliferation, migration and invasion of NSCLC cells.
Figure 5.
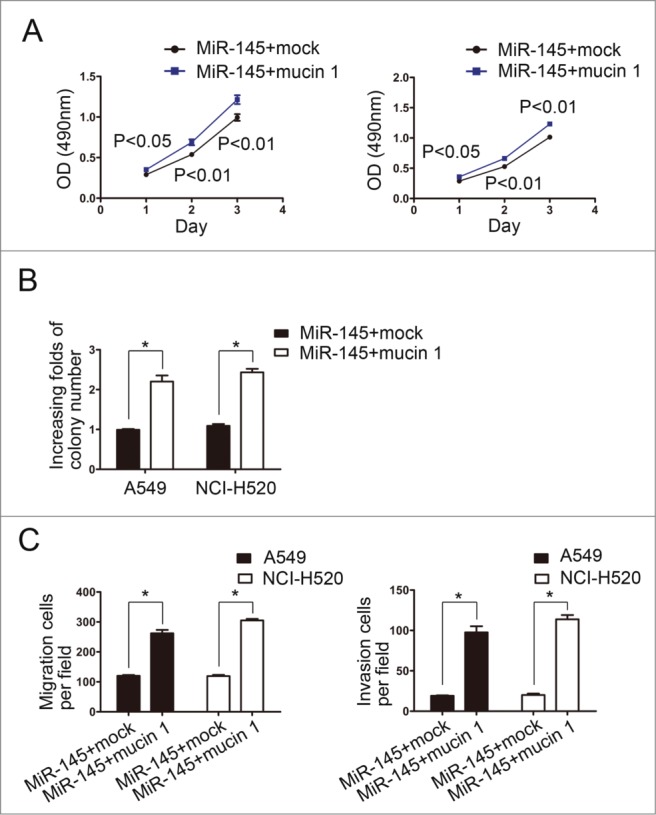
Restoration of mucin 1 expression reverses miR-145-suppressed proliferation, migration and invasion. (A) CCK-8 assays showed that mucin 1 promoted the proliferation of A549-miR-145 and NCI-H520-miR-145 cells compared with mock groups. (B) The colony-forming abilities of A549-miR-145 and NCI-H520-miR-145 cells transfected with the mucin 1 vector were promoted compared with cells transfected with the control vector. (C) Transwell migration assays and matrigel invasion assays indicated that mucin 1 increased the migration and invasion abilities of A549-miR145 and NCI-H520-miR145 cells.
Discussion
In this study, we found that miR-145 acts as a tumor-suppressor marker in NSCLC. Restoration of miR-145 expression inhibited NSCLC cell proliferation, migration and invasion. Meanwhile, low miR-145 expression was associated with miR-145 methylation status and was correlated with a more aggressive tumor phenotype, indicating that miR-145 is an independent predictor of reduced survival time in patients with NSCLC.
Previous studies have shown that miR-145 is downregulated in different types of cancer and is an important tumor suppressor that inhibits cancer cell proliferation, migration and invasion. Some groups have also reported that low miR-145 expression in NSCLC is associated with poor survival time and is implicated in the progression of NSCLC.30,31 Cho et al implicated mutation of epidermal growth factor receptor in low miR-145 expression;32 however, the exact mechanism for the low-level expression of miR-145 had been largely unknown.
DNA methylation is a critical epigenetic signature that is involved in transcriptional regulation, genomic imprinting, and silencing of repetitive DNA elements.34 One recently recognized and growing key, novel role for abnormal promoter DNA hypermethylation is the fostering of pathway disruption, which correlates with the transcriptional repression of multiple miRNAs. This could lead to the upregulation of oncogenic targets of microRNAs and to constitutive activation of signaling that could increase invasion and migration activities.35 Suh et al showed that miR-145 was regulated by DNA methylation in prostate cancer.27 However, to our knowledge, there were no reports about low miR-145 expression associated with DNA methylation in NSCLC. In this study, we found that DNA methylation was indeed responsible for the downregulated miR-145 expression in NSCLC, indicating a novel molecular mechanism for low miR-145 expression.
Mucin 1 is a member of a family of extensively O-glycosylated proteins that are predominantly expressed by epithelial cells. Mucin 1 and other mucins create a physical barrier that protects epithelia from damage induced by exposure to the external environment.36 The mucin 1 oncoprotein is aberrantly overexpressed in diverse human malignancies, including lung cancer, conferring a poor prognosis in NSCLC patients.37,38 However, the mechanism of mucin 1 overexpression in NSCLC patients has remained largely unknown. In our study, we found that mucin 1 was targeted by miR-145 in NSCLC and that overexpression of mucin 1 caused by low miR-145 levels promoted cancer cell proliferation, migration and invasion.
In summary, our data show that low miR-145 expression, due to methylation, promotes NSCLC cell proliferation, migration and invasion by targeting mucin 1. Therefore, miR-145 may be a valuable therapeutic target for NSCLC.
Materials and methods
Tissue specimens
All NSCLC tissue samples and adjacent normal tissue samples were obtained after attaining informed consent from the patients at the Third Affiliated Hospital, Sun Yat-sen University (Guangzhou, China), between January 2005 and January 2008. All patients' diagnoses were histopathologically confirmed. This study was approved by the institutional research ethics committee.
Cell lines and cell culture
Cell lines used in this study included human bronchial epithelial cells (HBE) and NSCLC cells (A549, NCI-520, NCI-460, NCI-H596), all of which were purchased from Cell Bank, Chinese Academy of Sciences (Shanghai, China). The NSCLC cell lines were maintained in RPMI 1640 (Gibco, Invitrogen Life Technologies, Carlsbad, CA USA) supplemented with 10% newborn calf serum (Gibco, Invitrogen Life Technologies, Carlsbad, USA), and the HBE cells were cultured in keratinocyte serum–free medium with 25 µg/ml bovine pituitary extract and 0.2 ng/ml recombinant epidermal growth factor (Invitrogen Life Technologies, Carlsbad, CA USA). The cells were transfected with DNA constructs for 5 min using siPORT™ NeoFX™ Transfection Agent (Ambion, USA).
RNA isolation and quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA USA). cDNA was synthesized using the PrimeScript RT Reagent Kit (Promega, Madison, WI USA) according to the manufacturer's instructions, and real-time PCR was performed using the ABI 7900HT Fast Real-Time PCR system (Applied Biosystems, CA, USA).
Vector construction
The pre-miR-23a, pre-miR-23a-sponge-inhibitor sequence and mucin 1 plasmids were synthesized by GenePharma (Shanghai, China).
Luciferase reporter assay
The putative miR-145 binding site and mutated 3′ UTR sequences of the mucin 1 mRNA were cloned and named Wt mucin 1 3′ UTR and Mut mucin 1 3′ UTR, respectively. For the reporter assays, cells grown in a 48-well plate were cotransfected with Wt mucin 1 3′ UTR or Mut mucin 1 3′ UTR and miR-145 or miR-control. A dual luciferase assay was then conducted 48 h after transfection. Transfections were performed in duplicate and repeated at least 3 times in independent experiments.
Western blotting
The following primary antibodies were used: anti-mucin 1 (1:1000; Santa Cruz Biotechnology) or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:8000, Cell Signaling). The intensities of protein fragments were examined using GeneTools software (version 3.03; Syngene, Cambridge, UK).
Methylation-specific PCR (MSP)
MSP assays were performed as described previously.33
CCK-8 proliferation assays
Cell proliferation capability was determined using CCK-8 proliferation assays. Cells (3 × 103) were incubated in 96-well culture plates with 10 μl CCK-8 solution on days 1, 2, and 3. Then the optical density was examined using a spectrophotometer (Bio-Rad).
Colony formation assays
Cells were seeded in 6-well plates at 5 × 102 cells per well, and after 14 days, the colonies were stained with 0.01% crystal violet. Each experiment was performed in triplicate and in 2 independent assays.
Matrigel invasion assays and transwell migration assays
For matrigel invasion assays, 5 × 104 cells were plated in a matrigel invasion chamber (BD Biosciences) without serum; medium with serum was used as a chemoattractant. Twenty-four hours later, the non-invading cells were removed with cotton swabs. Invasive cells located on the lower side of the chamber were then fixed in formaldehyde and stained with crystal violet. Transwell migration assays were performed in a similar manner as the matrigel invasion assays, but without matrigel on the filter.
Statistical analysis
Statistical analyses were performed using SPSS software (version 17.0). Differences between variables were assessed using the χ2 test. Kaplan-Meier analysis was used for the survival analysis, and differences in overall survival were determined using the log-rank test. Using the Cox regression model, multivariate survival analysis was conducted on all variables that were significant by univariate analysis. P-values <0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Science and Technology Planning Project of Guangdong Province (2012B031800063).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates ofworldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127:2893-917; PMID:21351269; http://dx.doi.org/ 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009; 59:225-49; PMID:19474385; http://dx.doi.org/ 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- 3.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012; 148:1172-87; PMID:22424228; http://dx.doi.org/ 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH, Kang H, Kim G, An HJ. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett 2015; 356:937-45; PMID:25444913; http://dx.doi.org/ 10.1016/j.canlet.2014.11.011 [DOI] [PubMed] [Google Scholar]
- 6.Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei Y, Li Y, Yang Q, Liu J, Wei JJ, et al.. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget 2014; 5:10816-29; PMID:25333261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, Li Y, Xie C, Yin X, Liu Y, Cao Y, Fang Y, Lin X, Xu Y, Xu W, et al.. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer 2014; 135:1286-96; PMID:24510775; http://dx.doi.org/ 10.1002/ijc.28774 [DOI] [PubMed] [Google Scholar]
- 8.Cioce M, Ganci F, Canu V, Sacconi A, Mori F, Canino C, Korita E, Casini B, Alessandrini G, Cambria A, et al.. Protumorigenic effects of mir-145 loss in malignant pleural mesothelioma. Oncogene 2014; 33:5319-31; PMID:24240684; http://dx.doi.org/ 10.1038/onc.2013.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al.. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer 2014; 13:220; PMID:25248370; http://dx.doi.org/25277192 10.1186/1476-4598-13-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, et al.. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget 2014; 5:7599-609; PMID:25277192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 2010; 24:2754-9; PMID:21159816; http://dx.doi.org/ 10.1101/gad.1950610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gits CM, van Kuijk PF, Jonkers MB, Boersma AW, Smid M, van Ijcken WF, Coindre JM, Chibon F, Verhoef C, Mathijssen RH, et al.. MicroRNA expression profiles distinguish liposarcoma subtypes and implicate miR-145 and miR-451 as tumor suppressors. Int J Cancer 2014; 135:348-61; PMID:24375455; http://dx.doi.org/ 10.1002/ijc.28694 [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Zhu J, Ou C, Deng Z, Chen M, Huang W, Li L. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer 2014; 110:2300-9; PMID:24642628; http://dx.doi.org/ 10.1038/bjc.2014.122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Pagliuca A, Valvo C, Fabrizi E, di Martino S, Biffoni M, Runci D, Forte S, De Maria R, Ricci-Vitiani L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013; 32:4806-13; PMID:23128394; http://dx.doi.org/ 10.1038/onc.2012.495 [DOI] [PubMed] [Google Scholar]
- 15.Ibrahim AF, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res 2011; 71:5214-24; PMID:21690566; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4645 [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res 2012; 40:761-74; PMID:21917858; http://dx.doi.org/ 10.1093/nar/gkr730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, Huang K, Zheng L, Tong Q. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 α. Oncogene 2014; 33:387-97; PMID:23222716; http://dx.doi.org/ 10.1038/onc.2012.574 [DOI] [PubMed] [Google Scholar]
- 18.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 2009; 106:3207-12; PMID:19202062; http://dx.doi.org/ 10.1073/pnas.0808042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 2010; 70:378-87; PMID:19996288; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al.. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-α in human breast cancer cells. Cell Death Differ 2010; 17:246-54; PMID:19730444; http://dx.doi.org/ 10.1038/cdd.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al.. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 2010; 29:6569-80; PMID:20818426; http://dx.doi.org/ 10.1038/onc.2010.386 [DOI] [PubMed] [Google Scholar]
- 22.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer 2010; 127:2804-14; PMID:21351259; http://dx.doi.org/ 10.1002/ijc.25284 [DOI] [PubMed] [Google Scholar]
- 23.Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sorensen KD, Ulhoi B, Borre M, Kjems J, Dyrskjot L, et al.. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene 2010; 29:1073-84; PMID:19915607; http://dx.doi.org/ 10.1038/onc.2009.395 [DOI] [PubMed] [Google Scholar]
- 24.Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M, Orntoft TF, et al.. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer 2012; 106:366-74; PMID:22108519; http://dx.doi.org/ 10.1038/bjc.2011.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sorensen KD, Ulhoi B, Borre M, Kjems J, Dyrskjot L, et al.. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene 2010; 29:1073-84; PMID:19915607; http://dx.doi.org/ 10.1038/onc.2009.395 [DOI] [PubMed] [Google Scholar]
- 26.Zaman MS, Chen Y, Deng G, Shahryari V, Suh SO, Saini S, Majid S, Liu J, Khatri G, Tanaka Y, et al.. The functional significance of microRNA-145 in prostate cancer. Br J Cancer 2010; 103:256-64; PMID:20588276; http://dx.doi.org/ 10.1038/sj.bjc.6605742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, et al.. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 2011; 32:772-8; PMID:21349819; http://dx.doi.org/ 10.1093/carcin/bgr036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao P, Xing AY, Zhou GY, Zhang TG, Zhang JP, Gao C, Li H, Shi DB. The molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancer. Oncogene 2013; 32:491-501; PMID:22370644; http://dx.doi.org/23548270 10.1038/onc.2012.61 [DOI] [PubMed] [Google Scholar]
- 29.Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC, Chiou GY, Chou MY, Chiou SH. miR145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res 2013; 73:3425-40; PMID:23548270; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3840 [DOI] [PubMed] [Google Scholar]
- 30.Campayo M, Navarro A, Vinolas N, Diaz T, Tejero R, Gimferrer JM, Molins L, Cabanas ML, Ramirez J, Monzo M, et al.. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J 2013; 41:1172-8; PMID:22835608; http://dx.doi.org/ 10.1183/09031936.00048712 [DOI] [PubMed] [Google Scholar]
- 31.Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol 2011; 8:125-31; PMID:21289483; http://dx.doi.org/ 10.4161/rna.8.1.14259 [DOI] [PubMed] [Google Scholar]
- 32.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 2009; 45:2197-206; PMID:19493678; http://dx.doi.org/ 10.1016/j.ejca.2009.04.039 [DOI] [PubMed] [Google Scholar]
- 33.Li DX, Cai HY, Wang X, Feng YL, Cai SW. Promoter methylation of Raf kinase inhibitory protein: A significant prognostic indicator for patients with gastric adenocarcinoma. Exp Ther Med 2014; 8:844-50; PMID:25120612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo PL, Alberich-Jorda M, Zhang P, et al.. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013; 503:371-6; PMID:24107992; http://dx.doi.org/ 10.1038/nature12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duraisamy S, Kufe T, Ramasamy S, Kufe D. Evolution of the human MUC1 oncoprotein. Int J Oncol 2007; 31:671-7; PMID:17671696 [PubMed] [Google Scholar]
- 36.Khodarev NN, Pitroda SP, Beckett MA, MacDermed DM, Huang L, Kufe DW, Weichselbaum RR. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res 2009; 69:2833-7; PMID:19318547; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong KK, Kufe D. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res 2014; 20:5423-34; PMID:25189483; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-3168 [DOI] [PMC free article] [PubMed] [Google Scholar]


