Abstract
The Wnt signaling pathway is of central importance in embryogenesis, development and adult tissue homeostasis, and dysregulation of this pathway is associated with cancer and other diseases. Despite the developmental and potential therapeutic significance of this pathway, many aspects of Wnt signaling, including the control of the master transcriptional co-activator β-catenin, remain poorly understood. In order to explore this aspect, a diverse immune llama VHH phagemid library was constructed and panned against β-catenin. VHH antibody fragments from the library were expressed intracellularly, and a number of antibodies were shown to possess function-modifying intracellular activity in a luciferase-based Wnt signaling HEK293 reporter bioassay. Further characterization of one such VHH (named LL3) confirmed that it bound endogenous β-catenin, and that it inhibited the Wnt signaling pathway downstream of the destruction complex, while production of a control Ala-substituted complementarity-determining region (CDR)3 mutant demonstrated that the inhibition of β-catenin activity by the parent intracellular antibody was dependent on the specific CDR sequence of the antibody.
Keywords: Wnt, β-catenin, intrabody, VHH, antibody, phage display
Abbreviations
- APC
adenomatous polyposis coli
- Bcl-9
B-cell CLL/lymphoma 9 protein
- CDR
complementarity-determining region
- CFU
colony forming unit
- CK1
casein kinase 1
- ELISA
enzyme-linked immunosorbant assay
- FAP
familial adenomatous polyposis
- GSK3β
glycogen synthase kinase 3 β
- HCAb
heavy chain only antibody
- HEK
human embryonic kidney
- LRP
Low density lipoprotein receptor related protein
- MOI
multiplicity of infection
- PBMC
peripheral blood mononuclear cell
- scFc
single chain IgG Fc domain
- SEM
standard error of the mean
- TOPflash
luciferase reportergene downstream of TCF/Lef binding sites and a minimal promoter
- VHH
variable domain of a HCAb
Introduction
Most mammalian genomes express 19 known Wnt genes, which encode a family of lipid-modified secreted proteins of approximately 40 kDa in size. Wnt signaling has a central role in adult tissue homeostasis, including control of stem cell proliferation in the gut and the hair follicle cycle, osteoblastogenesis and haematopoietic cell differentiation (for a review see ref 1). In resting cells, β-catenin turnover is regulated by the destruction complex, which contains the tumor suppressor adenomatous polyposis coli (APC), among other proteins. Activation of the canonical Wnt/β-catenin pathway by exogenous Wnt results in the stabilization and activation of β-catenin, which translocates from the cytoplasm into the nucleus where it interacts with numerous partners including the TCF/LEF family. Formation of the bipartite β-catenin/TCF transcription factor2 activates the transcription of Wnt responsive genes3 shown in Figure 1. Studies of β-catenin have revealed it to be a multi-functional protein which also has roles in cell-cell adhesion,4 as a component of the adherens junction, linking E-cadherin to the cell cytoskeleton.5 Structurally, β-catenin contains a series of 12 armadillo repeats, each of which is approximately 40 amino acids, surrounded by unstructured N- and C-terminal domains.6 Protein-protein interaction mapping experiments have demonstrated that all 3 β-catenin domains are involved in both signaling and cytoskeletal interaction.7,8 Over 38 β-catenin interaction partners have been documented (http://www.stanford.edu/group/nusselab/cgi-bin/wnt/protein_interactions). Regulation of intracellular Wnt signaling is achieved through the modification of the interaction of β-catenin with its protein partners, generating multiple intracellular pools, phosphorylation states and conformational forms of β-catenin within the cell, as reviewed in Valenta et al.9
Figure 1.
Canonical Wnt/β-catenin signaling pathway: OFF state. In the absence of Wnt, β-catenin (β-cat) is constantly turned over by the destruction complex. The destruction complex is assembled and maintained by scaffolding proteins Axin and APC. While associated with this complex, β-catenin is sequentially phosphorylated by CK1 and GSK3; phosphorylated β-catenin is recognized by the E3 ubiquitin ligase complex (β-Trcp) and degraded. ON state. Wnt binding to LRP5/6 and Frizzled cell surface Wnt co-receptors initiates canonical signaling, leading to the recruitment of Dishevelled (Dvl) and release of β-catenin from the degradation complex. β-catenin translocates to the nucleus where it binds to the TCF complex. Wnt target genes are actively transcribed. Points of modulation by potential therapeutic molecules are shown in blue for activating molecules and red for inhibitory molecules.
Mutations affecting the Wnt signaling pathway play a role in many diseases including, but not limited to, bone density disorders,10-12 Alzheimer's disease13 and numerous cancers. In fact mutations on this pathway are believed to be present in approximately 20% of all human cancers,14 with the majority of colorectal cancers bearing a mutation in the Wnt signaling pathway. One of the most frequent mutations is found in the APC gene (reviewed by Bienz and Clevers),15 causing the inherited condition familial adenomatous polyposis (FAP), which results from the loss of one allele of APC, leading to the formation of a high number of intestinal polyps and an increased predisposition to colorectal cancer. Additionally, mutations of the β-catenin gene have been identified in ovarian cancers,16 hepatocellular carcinomas,17 endometrial carcinomas18 and prostate cancer.19 Mutations that over-activate the transcription of Wnt/β-catenin target genes are completely dependent on transcriptionally active nuclear β-catenin, hence disrupting the transcriptional activity of β-catenin is of great therapeutic interest. A number of previous studies have identified small molecule20-27 and conventional antibody28 inhibitors of the Wnt signaling pathway, shown in Figure 1.
Intracellular antibodies are antibody or antibody fragments located within a cell, sometimes in a particular cellular compartment, where they interact with their target antigen. Intracellular antibodies have been used in the study of several disease pathways including Ras-expressing tumors,29 Alzheimer's disease30 and Huntington's disease.31 Intracellular antibodies act in a complementary way to other techniques used to study intracellular protein function; for instance they do not prevent the gene protein product from being formed, like RNA interference, or rely on the expression of a mutated form of protein, but instead disrupt or alter the function of the endogenous protein in its native conformation inside the cell. Intracellular antibodies are of particular use in the study of multi-functional proteins, disrupting just one function such as DNA binding32 rather than ablating total protein function. The antigen binding domains of heavy chain only antibodies (HCAbs), referred to as VHHs, are particularly suited for use as intracellular antibodies. VHHs contain ‘hallmark’ substitutions in framework 2, which are proposed to change this predominantly hydrophobic area in a VH where it packs against the VL, into a hydrophilic region in a VHH.33 This helps to explain the monomeric nature and high solubility of many individual VHH domains,34 which are desirable characteristics for intracellular antibodies. Here, we have applied VHH intracellular antibodies to the study of β-catenin biology. We generated and characterized VHH intracellular antibodies capable of specific inhibition of β-catenin's co-activator of transcriptional activity. Such VHHs represent useful tools for the future study of the Wnt signaling pathway, probing novel sites for potential therapeutic intervention.
Results
Identification of anti-β-catenin VHHs
Generation of VHHs for use as intracellular antibodies targeted to β-catenin was achieved via immunisation of 2 llamas with purified full-length recombinant human β-catenin. A phage display library of VHH domains was then produced from llama peripheral blood mononuclear cells (PBMCs). The library contained approximately 2 × 108 members as determined by dilution titer, and sequence analysis of 158 individual members demonstrated that 78% contained in-frame VHH domains. Each of the VHH subfamilies previously identified in llamas was present,35 along with 8% of conventional VH sequences. The percentage of conventional VH sequences was in line with previous reports of some conventional VH sequences being found as HCAbs.36
We selected β-catenin-specific VHHs from the immune phage library by panning against biotinylated β-catenin both in-solution and adsorbed directly to immunotubes. When panning β-catenin adsorbed to immunotubes, 2 stringencies of washing were used, termed Low and High wash. After only 2 rounds of phage display, incubation of the library with immunotubes coated with β-catenin showed approximately 1000-fold enrichment in phage binding compared with uncoated tubes, indicating that panning had successfully enriched β-catenin-specific VHHs. A single round of panning was carried out in-solution, in an effort to prevent over-selection and loss of VHH sequence diversity. Ninety-six clones were picked from the output of each of the individual rounds of panning that were undertaken. Monoclonal phage rescues were performed and screened by enzyme-linked immunosorbant assay (ELISA) to reveal β-catenin-specific VHHs. Depending on the panning technique and round of panning, the percentage of β-catenin-specific VHHs recorded per screening plate varied from 43 to 100% (Table 1). Only a single binder was observed on an irrelevant protein (streptavidin, data not shown). In total, over 50 different variable regions sequences that bound to β-catenin by ELISA were identified, including representatives from each VHH subfamily described previously.35 The VHHs were further divided based on complementarity-determining region (CDR)3 diversity into 12 different antibody ‘CDR3-families’ and 19 individual sequences that were not similar enough to be defined as family members. In this instance CDR3 families were defined as those containing 90% sequence identity in CDR3. In order to visualize the diversity of the VHH panel produced, we performed a principal components analysis (PCA)37 on a concatenated sequence of all 3 CDRs of each VHH. For the PCA 3 sequence variables values were generated for each CDR residue representing size, electronic nature and hydrophobicity of the amino acid found there. The PCA then maps these sequence variables in a linear manner so that the complexity of the data is reduced but the variance between the data is maximized and described by principal components (PC) 1 and 2. The outcome of this analysis is shown in Figure 2 as a 2-dimensional (2-D) data plot. The distance between 2 points in Figure 2 is directly proportional to CDR sequence identity. Antibodies of identical CDR sequences resulted in the co-localization of data points on the 2-D plot. The wide spread of data points in Figure 2 shows the broad diversity of VHH antibody sequences identified.
Table 1.
Input and output titres for all rounds of phage display
| Panning Round | Input titre (cfu) | Output titre (cfu) | β-catenin binders (%) |
|---|---|---|---|
| Low Wash Round 1 | 6.4 × 1011 | 6.3 × 107 | 42.5 |
| Low Wash Round 2 | 2.5 × 1011 | 1.6 × 108 | 85 |
| Low Wash Round 2 (without β-catenin) | 2.5 × 1011 | 1.5 × 105 | — |
| High Wash Round 1 | 6.4 × 1011 | 3.3 × 107 | 85 |
| High Wash Round 2 | 1.7 × 1011 | 5.5 × 108 | 100 |
| High Wash Round 2 (without β-catenin) | 1.7 × 1011 | 8.0 × 104 | — |
| In-solution Round 1 | 6.4 × 1011 | 4.2 × 107 | 44 |
Figure 2.
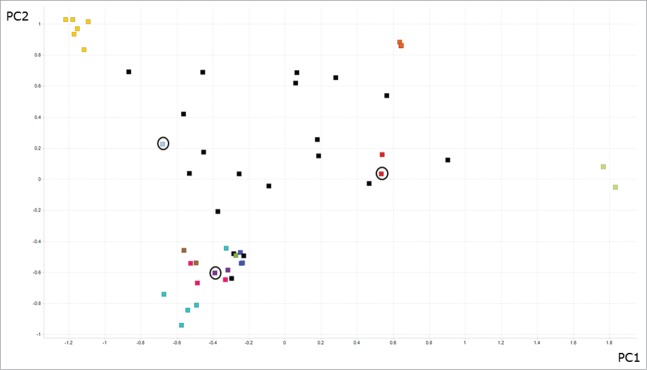
Diversity assessment of llama anti-β catenin VHH antibodies. For each antibody the 3 CDRs were concatenated and aligned in a pairwise, comprehensive manner to generate a sequence distance value. A principal components analysis (PCA) was used to reduce the dimensionality of these data and generate a 2-dimensional data plot. Data for principal component (PC) 1 and 2 are shown on the X and Y axis, respectively. Separately just CDR3 identity was analyzed, and antibodies that demonstrated over 90% sequence identity across CDR3 were deemed to belong to an antibody family. Families have been colored (those data points of the same color are considered to be in a family). All other sequences (black squares) were considered unique. Identical CDR sequences are co-located on the 2-D plot, but indicated with a circle.
Functional inhibition of β-catenin activity
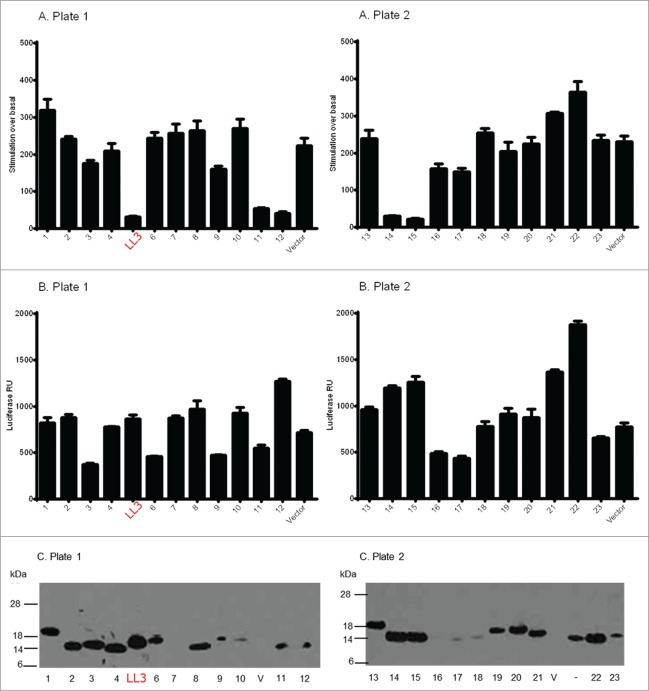
All of the β-catenin binding VHHs were sub-cloned into a mammalian intracellular expression vector containing a myc-HIS tag, and screened for functional modification of β-catenin using a HEK293 bioassay, which permits measurement of Wnt-induced expression of the firefly luciferase reporter. To control for off-target effects of intracellular antibody expression, a vector constitutively expressing Renilla luciferase was co-transfected such that antibodies of interest would be expected to inhibit firefly luciferase expression, but not Renilla luciferase. Western blotting on parallel samples was used to confirm VHH expression levels (Fig. 3). Co-transfection of a Wnt1 expression plasmid and the anti-β-catenin VHHs demonstrated that these intracellular antibodies were generally well tolerated in the HEK293 reporter bioassay and gave firefly and Renilla luciferase activity signals that were comparable to the empty vector control (Fig. 3), indicating that they were neither toxic nor had a functional effect on β-catenin. In contrast, 3 intracellular antibodies (9, 16 and 17) demonstrated potential cellular toxicity since they produced marked decreases in firefly and Renilla luciferase activities.
Figure 3.
Screen for function of VHH intracellular antibodies transiently transfected into HEK293 bioassay cells. Wnt signaling was induced by co-transfection of the Wnt1 gene. (A) Firefly luciferase activity, results are plotted as fold stimulation over empty vector transfected unactivated cells, all conditions performed in triplicate. (B) Renilla luciferase activity, results are plotted as raw signals. Data shows mean and SEM from a representative experiment. (C) Western blots of bioassay cells, cells from replicate bioassay plate wells were resuspended in LDS sample buffer (+DTT), and Western blots were performed with anti-myc antibodies. V refers to empty vector.
In total, 4 active intracellular VHH antibodies (LL3, 12, 14, 15) specifically inhibited the Wnt-induced firefly luciferase signal, but not the constitutive Renilla luciferase signal compared to the vector only control. Expression of these antibodies was confirmed by Western blotting (Fig. 3). Intracellular antibody LL3 was selected for further characterization based on analysis of multiple factors including expression levels of VHH fragments, sequence diversity and amenability to CDR3 mutagenesis for retention of molecular integrity and ablation of specific binding. Only VHH LL3 fulfilled each of these criteria.
To confirm that the inhibition of Wnt signaling mediated by LL3 was a direct result of binding to β-catenin, Ala mutations were introduced to CDR3 to create the non-binding control antibody, LL3.CDRAAA (Fig. 4A). The binding properties of LL3.CDRAAA and the parent antibody, LL3, were compared as VHH fragments with single chain Fc (scFc) tags. LL3-scFc binding to β-catenin was confirmed by ELISA, while LL3.CDRAAA-scFc demonstrated no detectable specific binding to β-catenin when tested by ELISA (Fig. 4B). The affinity of the parent VHH, LL3-scFc, for β-catenin was determined by SPR to be 4nM.
Figure 4.

Intracellular antibody LL3 and CDR3 mutant control intracellular antibody LL3.CDRAAA. (A) CDR3 alignments of LL3 and LL3.CDRAAA. (B) β-Catenin binding ELISA results for LL3 and LL3.CDRAAA with scFc tags. Biotinylated β-catenin was captured on to streptavidin coated plates, and VHH-scFc constructs were then added in half log dilutions from 32 nM and revealed with anti-mouse Fc HRP. The plates were read at 630 and 490 nm and ΔOD recorded. The negative control is a vector only DNA control transfection culture supernatant. Error bars represent 95% confidence limit. (C) Intracellular expression of intracellular antibodies LL3 and LL3.CDRAAA. Expression vectors containing VHH intracellular antibody encoding genes were transfected into HEK293 luciferase bioassay cells, with WNT1-containing vector. Forty hours post transfection, cells were resuspended in LDS sample buffer, 10 μL of each sample was run on a 4–12% bis-tris gel followed by Western blot, probed with mouse anti-myc. Analysis of band density was performed on GE Healthcare ImageQuant analysis software. (D) Relative activities of intracellular antibodies LL3 and LL3.CDRAAA in the HEK293 Wnt-induced luciferase reporter bioassay. LL3 and LL3.CDRAAA were tested for firefly and Renilla luciferase activity in the bioassay. Wnt signaling was induced by co-transfection of the Wnt1 gene. Results plotted as firefly luciferase activity relative to Renilla luciferase activity for each well. All conditions performed in 5 replicates. Data shows mean and SEM from a representative experiment. **** denotes p ≤ 0.0001, *** denotes p of 0.001-0.00011, for a 2 tailed ratio paired Student's t-test.
The LL3.CDRAAA mutant was expressed at a similar level to the parent LL3 antibody in HEK293 reporter bioassay cells (Fig. 4C). To control for general variations in luciferase production caused by factors such as transfection variability and reduced cell viability, the Wnt-driven firefly luciferase signal was divided by the constitutive Renilla luciferase signal. Comparison of LL3 to LL3.CDRAAA demonstrated that inhibition of Wnt signaling was dependent on binding of LL3 to β-catenin (Fig. 4D), and no inhibition of β-catenin dependent firefly luciferase production or the Renilla luciferase signal occurred when LL3.CDRAAA was transfected into HEK293 reporter bioassay cells.
Characterization of anti-β-catenin intracellular antibody LL3
Intracellular antibody LL3 was further analyzed using LL3.CDRAAA as a control. In addition to stimulation by Wnt1 transfection, the HEK293 reporter bioassay was stimulated using Wnt3a conditioned media, or the GSK3β inhibitor LiCl2, which acts downstream of the cell-surface Wnt-Frizzled interaction. LL3 continued to show significant inhibition of Wnt signaling when the HEK293 reporter bioassay cells were stimulated with each of these methods (Fig. 5). The ability of LL3 to bind endogenous cellular β-catenin was confirmed in immunoprecipitation experiments where LL3 and LL3.CDRAAA with scFc tags were used to immunoprecipate β-catenin from lysed HEK293 cells (Fig. 6). β-Catenin was detected from samples immunoprecipated using LL3, but not LL3.CDRAAA.
Figure 5.

Activity of intracellular antibody LL3 in the HEK293 bioassay. HEK293 luciferase bioassay cells expressing VHH intracellular antibody were stimulated with LiCl2 (A) or Wnt3a-conditioned media (B). Firefly luciferase activity was measured and plotted as fold stimulation over unactivated cells, 5 replicates were performed for each condition. Results are representative of 2 independent experiments. **** denotes p ≤ 0.0001 for a 2 tailed unpaired Student's t-test. Data shows mean and SEM.
Figure 6.

Immuno-precipitation of cellular β-catenin from HEK293 bioassay cellular lysate by LL3. LL3-scFc and LL3.CDRAAA-scFc coated beads were tumbled with lysed HEK293 bioassay cells, and specific bound protein was eluted. Western blots for: (A) β-catenin and (B) VHH-scFc construct. An untransfected cell lysate sample was included as a positive control for the detection of endogenous β-catenin.
To compare the relative expression levels of β-catenin and intracellular antibody LL3, we performed quantitative Western blots on lysed HEK293 reporter bioassay cells. From the blots, it was estimated that HEK293 cells contained approximately 3.0 × 108 molecules of intracellular antibody LL3, and approximately 9.5 × 103 molecules of β-catenin per cell. The figure for β-catenin was comparable with other reports of 5.4 × 104 molecules of β-catenin per HEK293 cell.38 Based on these figures, intracellular antibody LL3 was estimated to be in a large excess over β-catenin, in the region of 20,000 molecules of LL3 per β-catenin molecule.
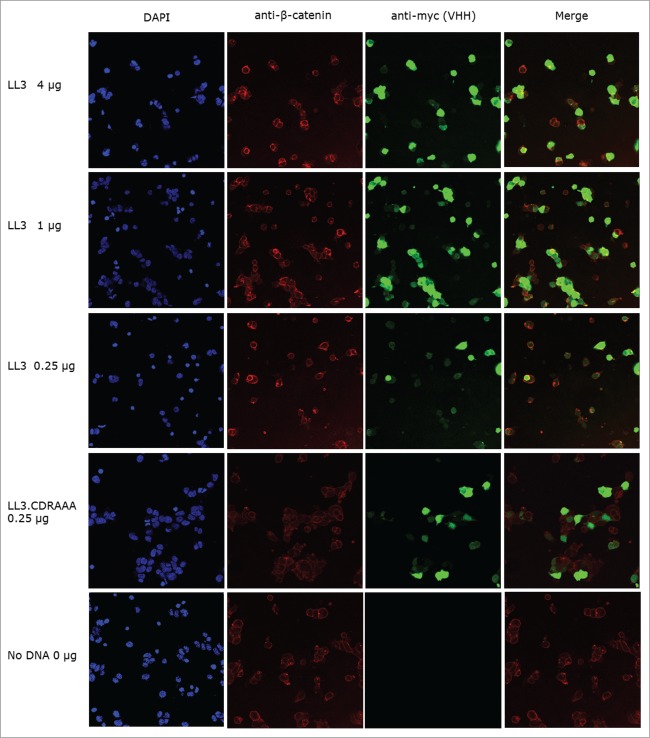
β-Catenin is a multi-functional protein that is found in 3 distinct cellular compartments: the plasma membrane, the cytoplasm and nucleus. The effects of LL3 binding on the cellular localization of β-catenin were investigated via confocal microscopy using the myc-tag present on the intracellular antibody. β-Catenin was predominantly localized at the plasma membrane, where it is known to form part of the adherence junction.4 Expression of intracellular antibody LL3 appeared to be ubiquitous across the cell and did not affect the localization of β-catenin at the plasma membrane, as seen by the yellow rings of β-catenin and LL3 co-localization at the plasma membrane (Fig. 7). These data suggest that inhibition of β-catenin's function as a transcriptional co-activator by intracellular antibody LL3 did not affect the role of β-catenin at the plasma membrane. No discernible difference was detected in nuclear β-catenin levels in cells with or without intracellular antibody LL3 expression, which probably reflects the unstimulated nature of these cells, since Wnt signaling had not been activated during these experiments. The images captured by confocal microscopy demonstrate the difference in VHH protein expression levels between individual cells. It is clear that not all the cells in a sample became successfully transfected with VHH encoding DNA, and this may be why complete inhibition of β-catenin signaling was not achieved by intracellular antibody LL3.
Figure 7.
Confocal microscopy of anti-β-catenin and intracellular antibody stained HEK293 bioassay cells. Fixed HEK293 bioassay cells were stained for intracellular antibody and β-catenin expression; the nuclear stain DAPI was also included. Anti-β-catenin and anti-intracellular antibody images were merged, and yellow donates co-localization. Pictures were taken using Leica TCS SP5 microscope, with 40× magnification.
β-Catenin contains many structural motifs and protein interaction domains. In order to identify which of these were involved in the binding of LL3 to β-catenin, a number of His-tagged-truncated forms of β-catenin (Table 2) were produced. On a reducing Western blot, LL3 bound to truncations 2 and 4 as well as to full-length β-catenin (Fig. 8). The binding pattern indicated that the binding domain of LL3 resided between amino acid residues 164–390 corresponding to the first 6 armadillo repeats of β-catenin and was not a conformation-dependent epitope.
Table 2.
Produced β-catenin fragments
| Fragment description | Amino acids | Primer sequences used in construction | MW (kDa) |
|---|---|---|---|
| F1. N-terminus and first α helix | 1–163 | GTCACAGGATCCATGGCTACTCAAGCTGATTTGATGG CAGGTTGTCGACTCATTACTCGTCATTTAGCAGTTTTGTCAGTTCAGG |
22.0 |
| F2. N-terminus and first 6 armadillo repeats | 1–390 | GTCACAGGATCCATGGCTACTCAAGCTGATTTGATGG CAGGTTGTCGACTCATTATGCAGCATCTGAAAGATTCCTG |
46.7 |
| F3. C-terminus and last 6 armadillo repeats | 391–781 | GGTAAGCAGGATCCGCTGCAACTAAACAGGAAGG CAGGTTGTCGACTCATTACAGGTCAGTATCAAACCAGGCC |
46.8 |
| F4. All 12 armadillo repeats and C-terminus | 138–781 | CTGGTAAGCAGGATCCAACTTGATTAACTATCAAGATGATGCAGAACTTGCC CAGGTTGTCGACTCATTACAGGTCAGTATCAAACCAGGCC |
74 |
| F5. Full length | 1–781 | GTCACAGGATCCATGGCTACTCAAGCTGATTTGATGG CAGGTTGTCGACTCATTACAGGTCAGTATCAAACCAGGCC |
89.5 |
Figure 8.

Western blots of small scale β-catenin fragment protein inductions. (A) Anti-His blot. (B) LL3 blot. F refers to fragment; the molecular weights for which are given in Table 2.
Discussions
To modify intracellular function, an intracellular antibody must be stably expressed inside a cell and must retain the ability to engage the target antigen with sufficient affinity to exert a functional effect. We identified the β-catenin-specific VHH, LL3, which was characterized in depth and demonstrated these attributes. LL3 exhibited specific function modifying activity when tested in the HEK293 Wnt-induced luciferase reporter bioassay, and while the stability of LL3 in the cell was not directly measured, the specific activity, expression and diffuse cellular staining indicated that the VHH fragment was stable and did not form visible aggregates inside the cell. These observations validated the selection of llama VHH domains for use as intracellular antibodies in this study and were in accordance with previous reports of VHHs as intracellular antibodies.39-44 Mutation of 3 residues in CDR3 of LL3 was sufficient to disrupt antigen binding; this was in line with another intracellular domain antibody study where mutation of 4 residues in CDR1 was sufficient to disrupt intracellular function.29
A number of studies have investigated the effect of intracellular antibody pI on intracellular aggregation,45 and concluded that an overall negative charge at pH7.4 was beneficial to intracellular antibody solubility. Consistent with this work the intracellular antibody LL3 studied here was predicted to have a pI value of 6.7.
Intracellular antibody LL3 showed functional inhibition of β-catenin co-transcriptional activity in the HEK293 bioassay stimulated with Wnt1, Wnt3a, and LiCl2. Wnt3a and Wnt1 are both thought to signal in a similar canonical fashion, but may use different sets of Frizzled co-receptors.46 LiCl2 is a potent inhibitor of GSK3β, which prevents β-catenin phosphorylation, hence acting as a stimulator of canonical Wnt signaling. The fact that LL3 could inhibit LiCl2 induction of firefly luciferase signal indicates that the interaction of intracellular antibody LL3 and β-catenin modulates signaling downstream of the GSK3β phosphorylation of β-catenin, and thus downstream of the destruction complex (Fig. 1).
The epitope on β-catenin recognized by LL3 was shown to lay between residues 164 to 390. This is a broad region, and the actual LL3 binding epitope is likely to comprise considerably fewer residues. However, it is of interest that the 164–390 region coincides with the binding epitopes of Axin,47 Pontin52,48 Bcl-9,49 E-cadherin,50 and TCF,51,52 and, consequently, LL3 may work by interfering with one or more of these β-catenin interactions. LL3 inhibited β-catenin transcriptional co-activation activity; hence, it is most likely that LL3 disrupts the interaction of β-catenin with a transcriptional enhancer such as TCF, Bcl-9 or Pontin52. The potential sites of other molecules intervention in the Wnt signaling pathway are shown schematically in Figure 1. LL3 may inhibit a similar axis to small molecule compounds PKF 115–854 and CGP049090,25 in disrupting the β-catenin/TCF complex, or as carnosic acid inhibiting the interaction of β-catenin and Bcl-9,21 or alternatively present a novel inhibitory axis between β-catenin and Pontin52.
Although disruption of the β-catenin-Axin interaction cannot be ruled out, disruption of this axis would be expected to increase β-catenin transcriptional co-activation activity, rather than inhibit it when tested by the HEK293 reporter bioassay. Additionally disruption of the β-catenin-E-cadherin interaction would be expected to have modulated β-catenin accumulation at the plasma membrane, which was not witnessed by confocal microscopy of LL3 transfected cells.
Further study of the Wnt signaling pathway intracellular antibodies, such as LL3, may aid the discovery of drugs to target this pathway. The use of antibodies to guide small molecule drug discovery was recently reviewed.53 Analysis of the LL3-β-catenin complex crystal structure could identify biologically relevant conformations of β-catenin and opportunities for small molecule blockade of Wnt signaling.
In summary, we have described a novel VHH fragment that functions inside the cell to disrupt the transcriptional activating activity of β-catenin. Such well-tolerated tools will enable further study of the Wnt signaling pathway, with potential to validate sites for therapeutic intervention in multiple diseases.
Materials and Methods
Generation of full length human β-catenin
The full-length human β-catenin cDNA clone was obtained from Origene. β-Catenin was expressed and purified essentially as described.6
Generation of immune VHH phage library
Two male llamas were immunised over 3 months with 5 doses of 0.25mg recombinant β-catenin in Freund's adjuvant. PBMCs were collected from these animals and RNA purified using RNeasy kit (Qiagen). VH genes were amplified using primers ‘llimmlibfor’(SAGGTGCAGCTGGTRGAGTCTGGGGGAG) and ‘CH2Ig primer’54 by PCR following RT of purified RNA using oligodT primers. VHH genes were specifically distinguished from the resulting PCR product based on size, re-amplified using primer ‘FR1for’ with ‘FR4rev’36 and cloned into pASTT. The ligated product was electroporated into XL-1 Blue E.coli (Stratagene), and the library was estimated to contain 2 × 108 members. Phage particles were prepared essentially as described.55
Phage panning
In the first panning experiment, the immune library was incubated in β-catenin-conjugated immunotubes blocked in 3% bovine serum albumin (BSA). Two separate schemes of washing were then performed: a Low wash protocol (5 washes in round 1 and 10 washes in round 2) and a High wash protocol (20 washes in round 1 and 40 washes in round 2), prior to elution of phage with 0.1 M HCl. In the second round of panning, input phage from both wash strategies were panned against both β-catenin conjugated and unconjugated immunotubes. One round of panning was also completed on antigen in solution. Recombinant biotinylated β-catenin was incubated (rolling) with 6 × 1011 library phage particles, previously blocked in phosphate-buffered saline (PBS) containing 2.5% milk. β-Catenin and associated phage were then captured out of solution using M280 streptavidin-conjugated Dynabeads® (Invitrogen). The beads were washed by magnetic capture 4 times, and bound phage were eluted as above. Small scale phage rescues were performed for individual E. coli colonies picked from output phage infected colonies from each round of all panning conditions. Each sample was tested for binding to β-catenin or streptavidin by ELISA (further detailed below). A positive ‘hit’ was determined to be any sample with an antigen binding signal greater than 3 times the negative signal.
Bioassay screening
HEK293 cells stably transfected with Tcf-firefly Luciferase reporter construct (as described by Holdsworth et al.)56 were grown in DMEM (Gibco® Life Technologies) supplemented with 10% fetal bovine serum, 2 mM glutamine and non-essential amino acids (Gibco® Life Technologies). Cells were seeded at 5 × 104/well in white poly-D-lysine coated 96-well plates and were transfected with expression plasmids (including the Renilla Luciferase pGL4.74 plasmid from Promega) using Lipofectamine 2000 (Gibco® Life Technologies) according to the manufacturer's instructions. Plates were incubated for 40 hours at 37°C in 5% CO2. When required Wnt3a conditioned media or LiCl2 were added, 18 hours post transfection. Following the incubation, firefly and Renilla luciferase activity were measured using Dual-Glo® (Promega) as manufacturer's instructions. Luminescence was recorded using a Biotek Synergy plate reader.
ELISAs
ELISA plates were coated overnight at 4°C. Washes were performed between each step of the assay, and consisted of 4 washes in PBS (containing 0.1% Tween20). All plates were blocked in PBS containing 3% BSA, and all samples were blocked in PBS containing 2.5% milk, for at least an hour prior to addition of screening samples to ELISA plates. Following the final wash, TMB (Calbiochem) was added, and the OD of plates was read at 630 and 490nm, with ΔOD recorded using a Biotek Synergy plate reader.
β-Catenin binding ELISA
ELISA plates were coated with streptavidin (2 μg/mL). Biotinylated β-catenin (1 μg/mL) was added in PBS, for 30 minutes. Blocked samples were added (phage samples or transient VHH-scFc samples). After an hour, bound samples were detected using a suitable antibody at 1 in 5,000 dilution in PBS (containing 3% BSA). Detection antibodies were goat anti-M13-HRP (GE Healthcare) or goat anti-mouse Fc HRP (Jackson ImmunoResearch).
Streptavidin binding ELISA
ELISA plates were coated with streptavidin (2 μg/mL). Blocked samples were added. After an hour bound samples were detected using a suitable antibody at 1 in 5,000 dilution in PBS (containing 3% BSA).
Confocal microscopy
HEK293 cells transfected with Myc-His tagged VHH constructs were attached to poly-D-lysine coated 8 well culture microscope slides in complete media. The cells were fixed in 4% paraformaldehyde and blocked/permeabilised in TBS block (containing 0.3% Triton 100, 1% BSA and 5% goat serum) for 1 hour. Anti-myc (9E10) Alexa Fluor® 488 (Gentaur), and anti-β-catenin (6B3) (Cell Signalling Technologies) were then added and incubated overnight at 4°C in a moist chamber. The anti-β-catenin detection antibody, anti-rabbit Alexa Fluor® 647 (Jackson ImmunoResearch), was added at a 1 in 1000 dilution, and incubated for 1 hour in the dark. Finally DAPI ProLong® Gold Antifade (Invitrogen) was added with a coverslip. Images were captured using a Leica TCS SP5 microscope.
VHH-scFc protein production
HEK293 cells were transfected with VHH-scFc expression vectors using 293fectin™ (Invitrogen), in accordance with the manufacturer's instructions. The single chain mouse IgG Fc constructs comprised VHH, hinge, CH2 and CH3 domains, linked to another hinge, CH2, CH3 sequence through a 59 amino acid linker (GGSSTASGSGSGGSGTAGSSGGAGSSGGSTTAGGSASGSGSTGSGTGGASSGGASGASG). This construct had previously been shown to aid extracellular expression, purification and orientated immobilisation, and as a tag for VHHs. Transfected HEK293 cells were incubated at 37°C, with 5% CO2 on a shaking platform for 5–6 d. The cells were then harvested from the cultures by centrifugation at 423×g, and the VHH-scFc containing supernatant was stored at 4°C.
Immunoprecipitation
HEK293 bioassay cells, 2 × 106, were washed in ice-cold PBS, and resuspended in 200 μL of ice-cold lysis buffer (including protease inhibitor cocktail). The samples were then rolled at 4°C for 30 minutes, followed by centrifugation for 15 minutes at 10,000×g at 4°C. The HEK293 cell lysate supernatant was then removed into a fresh chilled microfuge tube. VHH-scFcs were conjugated to sheep anti-mouse Fc Dynabeads® (Invitrogen) as manufacturer's instructions. The beads were then each added to 500 μL of HEK293 cell lysate and rolled at 4°C overnight. Each set of beads were magnetically captured out of solution and individually washed 3 times in fresh cell lysis buffer. Protein was then eluted from the beads by boiling in LDS sample buffer including dithiothreitol (DTT) (Invitrogen) for 5 minutes. Samples were then analyzed by anti-β-catenin and anti-VHH-scFc Western blots.
SDS-PAGE and immunological detection of proteins (Western blotting)
Appropriate amounts of protein samples were separated by SDS-PAGE on 4–12% bis-tris gels (Invitrogen), before being electro-blotted using iBLOT (Invitrogen) onto PVDF membranes. Membranes were probed by incubation with appropriate antibodies. These included mouse anti-myc (9e10, Gentuar) and goat anti-mouse Fc HRP (Jackson ImmunoResearch) for the detection of VHHs with and without scFc tags; biotinylated-LL3 (produced in-house), rabbit anti-β-catenin (6B3, Cell Signalling Technologies) and rabbit anti-penta HIS (Bethyl laboratories) followed by streptavidin HRP (Jackson ImmunoResearch) or anti-rabbit Fc HRP (Jackson ImmunoResearch) respectively for the detection of β-catenin. All membranes were developed with Pico Super Signal ECL (Pierce) for 5 minutes, and images captured using ImageQuant LAS 4000 (GE Healthcare) and densitometry of resulting bands analyzed by ImageQuant TL analysis software.
β-catenin fragment generation
BL21 Star™ DE3 cells (Invitrogen) expressing each of the β-catenin constructs (shown in Table 2) were grown in antibiotic selective 2xTY media supplemented with IPTG (final concentration of 0.3 mM), shaking overnight. Cells from the overnight culture were harvested by centrifugation at 4,000×g for 5 minutes. The pelleted cells were lysed with 1× Bugbuster® (Novagen) containing Lysonase (Novagen) in 20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, 2 mM DTT) by rolling at room temperature for 20 minutes.
Disclosure of Potential Conflicts of Interest
Some of the authors hold shares and share options in UCB Pharma.
Acknowledgment
We would like to thank James Snowdon for his help on variable region diversity assessment and visualization.
Funding
This work was partially funded by the European Union FP7 ANTICARB program (HEALTH-2007-201587).
References
- 1. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006; 127:469-80; PMID:17081971; http://dx.doi.org/ 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 2. Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 2005; 12:364-71. [DOI] [PubMed] [Google Scholar]
- 3. Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci 2008; 99:631-7; PMID:18177486; http://dx.doi.org/ 10.1111/j.1349-7006.2007.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J 1989; 8:1711-7; PMID:2788574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ben-Ze'ev A, Geiger B. Differential molecular interactions of beta-catenin and plakoglobin in adhesion, signaling and cancer. Curr Opin Cell Biol 1998; 10:629-39; PMID:9818174 [DOI] [PubMed] [Google Scholar]
- 6. Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Crystal structure of a full-length beta-catenin. Structure 2008; 16:478-87; PMID:18334222; http://dx.doi.org/ 10.1016/j.str.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aberle H, Schwartz H, Hoschuetzky H, Kemler R. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to alpha-catenin. J Biol Chem 1996; 271:1520-6; PMID:8576147 [DOI] [PubMed] [Google Scholar]
- 8. Hecht A, Litterst CM, Huber O, Kemler R. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem 1999; 274:18017-25; PMID:10364252 [DOI] [PubMed] [Google Scholar]
- 9. Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J 2012; 31:2714-36; PMID:22617422; http://dx.doi.org/ 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum Mol Genet 2001; 10:537-43; PMID:11181578; http://dx.doi.org/ 10.1093/hmg/10.5.537 [DOI] [PubMed] [Google Scholar]
- 11. Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001; 107:513-23; PMID:11719191; http://dx.doi.org/ 10.1016/S0092-8674(01)00571-2 [DOI] [PubMed] [Google Scholar]
- 12. Zheng HF, Tobias JH, Duncan E, Evans DM, Eriksson J, Paternoster L, Yerges-Armstrong LM, Lehtimaki T, Bergstrom U, Kahonen M, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet 2012; 8:e1002745; PMID:22792071; http://dx.doi.org/ 10.1371/journal.pgen.1002745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci U S A 2007; 104:9434-9; PMID:17517621; http://dx.doi.org/ 10.1073/pnas.0603523104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cadigan KM. Wnt-beta-catenin signaling. Curr Biol 2008; 18:R943-R7; PMID:18957245; http://dx.doi.org/ 10.1016/j.cub.2008.08.017 [DOI] [PubMed] [Google Scholar]
- 15. Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell 2000; 103:311-20; PMID:11057903; http://dx.doi.org/ 10.1016/S0092-8674(00)00122-7 [DOI] [PubMed] [Google Scholar]
- 16. Palacios J, Gamallo C. Mutations in the beta-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res 1998; 58:1344-7; PMID:9537226 [PubMed] [Google Scholar]
- 17. Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res 1998; 58:2524-7; PMID:9635572 [PubMed] [Google Scholar]
- 18. Fukuchi T, Sakamoto M, Tsuda H, Maruyama K, Nozawa S, Hirohashi S. Beta-catenin mutation in carcinoma of the uterine endometrium. Cancer Res 1998; 58:3526-8; PMID:9721853 [PubMed] [Google Scholar]
- 19. Voeller HJ, Truica CI, Gelmann EP. Beta-catenin mutations in human prostate cancer. Cancer Res 1998; 58:2520-3; PMID:9635571 [PubMed] [Google Scholar]
- 20. Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 2009; 5:100-7; PMID:19125156; http://dx.doi.org/ 10.1038/nchembio.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de la Roche M, Rutherford TJ, Gupta D, Veprintsev DB, Saxty B, Freund SM, Bienz M. An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid. Nat Commun 2012; 3:680; PMID:22353711; http://dx.doi.org/ 10.1038/ncomms1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, et al. An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res 2007; 67:573-9; PMID:17234765; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-2726 [DOI] [PubMed] [Google Scholar]
- 23. Grandy D, Shan J, Zhang X, Rao S, Akunuru S, Li H, Zhang Y, Alpatov I, Zhang XA, Lang RA, et al. Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J Biol Chem 2009; 284:16256-63; PMID:19383605; http://dx.doi.org/ 10.1074/jbc.M109.009647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009; 461:614-20; PMID:19759537; http://dx.doi.org/ 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- 25. Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell 2004; 5:91-102; PMID:14749129; http://dx.doi.org/ 10.1016/S1535-6108(03)00334-9 [DOI] [PubMed] [Google Scholar]
- 26. Teo JL, Ma H, Nguyen C, Lam C, Kahn M. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A 2005; 102:12171-6; PMID:16093313; http://dx.doi.org/ 10.1073/pnas.0504600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol 2010; 6:829-36; PMID:20890287; http://dx.doi.org/ 10.1038/nchembio.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, Fischer M, Chaudhari A, Ji M, Kapoun AM, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A 2012; 109:11717-22; PMID:22753465; http://dx.doi.org/ 10.1073/pnas.1120068109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanaka T, Williams RL, Rabbitts TH. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J 2007; 26:3250-9; PMID:17568777; http://dx.doi.org/ 10.1038/sj.emboj.7601744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sudol KL, Mastrangelo MA, Narrow WC, Frazer ME, Levites YR, Golde TE, Federoff HJ, Bowers WJ. Generating differentially targeted amyloid-beta specific intrabodies as a passive vaccination strategy for Alzheimer's disease. Mol Ther 2009; 17:2031-40; PMID:19638957; http://dx.doi.org/ 10.1038/mt.2009.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Colby DW, Chu Y, Cassady JP, Duennwald M, Zazulak H, Webster JM, Messer A, Lindquist S, Ingram VM, Wittrup KD. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Nat Acad Sci U S A 2004; 101:17616-21; PMID:15598740; http://dx.doi.org/ 10.1073/pnas.0408134101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strube RW, Chen SY. Characterization of anti-cyclin E single-chain Fv antibodies and intrabodies in breast cancer cells: enhanced intracellular stability of novel sFv-F(c) intrabodies. J Immunol Methods 2002; 263:149-67; PMID:12009211; http://dx.doi.org/ 10.1016/S0022-1759(02)00035-2 [DOI] [PubMed] [Google Scholar]
- 33. Spinelli S, Frenken L, Bourgeois D, de RL, Bos W, Verrips T, Anguille C, Cambillau C, Tegoni M. The crystal structure of a llama heavy chain variable domain. Nat Struct Biol 1996; 3:752-7; PMID:8784347 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen VK, Desmyter A, Muyldermans S. Functional heavy-chain antibodies in Camelidae. Adv Immunol 2001; 79:261-96; PMID:11680009 [DOI] [PubMed] [Google Scholar]
- 35. Harmsen MM, Ruuls RC, Nijman IJ, Niewold TA, Frenken LG, de GB. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol Immunol 2000; 37:579-90; PMID:11163394 [DOI] [PubMed] [Google Scholar]
- 36. Deschacht N, De Groeve K, Vincke C, Raes G, De Baetselier P, Muyldermans S. A novel promiscuous class of camelid single-domain antibody contributes to the antigen-binding repertoire. J Immunol 2010; 184:5696-704; PMID:20404276; http://dx.doi.org/ 10.4049/jimmunol.0903722 [DOI] [PubMed] [Google Scholar]
- 37. Pearson K. On lines and planes of closest fit to systems of points in space. Philos Mag 1901; 2:559-72; http://dx.doi.org/ 10.1080/14786440109462720 [DOI] [Google Scholar]
- 38. Tan CW, Gardiner BS, Hirokawa Y, Layton MJ, Smith DW, Burgess AW. Wnt signalling pathway parameters for mammalian cells. PLoS One 2012; 7:e31882; PMID:22363759; http://dx.doi.org/ 10.1371/journal.pone.0031882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doyle PJ, Saeed H, Hermans A, Gleddie SC, Hussack G, Arbabi-Ghahroudi M, Seguin C, Savard ME, Mackenzie CR, Hall JC. Intracellular expression of a single domain antibody reduces cytotoxicity of 15-acetyldeoxynivalenol in yeast. J Biol Chem 2009; 284:35029-39; PMID:19783651; http://dx.doi.org/ 10.1074/jbc.M109.045047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gueorguieva D, Li S, Walsh N, Mukerji A, Tanha J, Pandey S. Identification of single-domain, Bax-specific intrabodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J 2006; 20:2636-8; PMID:17060401; http://dx.doi.org/ 10.1096/fj.06-6306fje [DOI] [PubMed] [Google Scholar]
- 41. Jobling SA, Jarman C, Teh MM, Holmberg N, Blake C, Verhoeyen ME. Immunomodulation of enzyme function in plants by single-domain antibody fragments. Nat Biotechnol 2003; 21:77-80; PMID:12483224 [DOI] [PubMed] [Google Scholar]
- 42. Serruys B, Van HF, Verbrugghe P, Leroux-Roels G, Vanlandschoot P. Llama-derived single-domain intrabodies inhibit secretion of hepatitis B virions in mice. Hepatology 2009; 49:39-49; PMID:19085971; http://dx.doi.org/ 10.1002/hep.22609 [DOI] [PubMed] [Google Scholar]
- 43. Summanen M, Granqvist N, Tuominen RK, Yliperttula M, Verrips CT, Boonstra J, Blanchetot C, Ekokoski E. Kinetics of PKCepsilon activating and inhibiting llama single chain antibodies and their effect on PKCepsilon translocation in HeLa cells. PLoS One 2012; 7:e35630; PMID:22536418; http://dx.doi.org/ 10.1371/journal.pone.0035630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 rev prevents viral production. J Biol Chem 2010; 285:21768-80; PMID:20406803; http://dx.doi.org/ 10.1074/jbc.M110.112490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kvam E, Sierks MR, Shoemaker CB, Messer A. Physico-chemical determinants of soluble intrabody expression in mammalian cell cytoplasm. Protein Eng Des Sel 2010; 23:489-98; PMID:20378699; http://dx.doi.org/ 10.1093/protein/gzq022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garcia-Morales C, Liu CH, Abu-Elmagd M, Hajihosseini MK, Wheeler GN. Frizzled-10 promotes sensory neuron development in Xenopus embryos. Dev Biol 2009; 335:143-55; PMID:19716814; http://dx.doi.org/ 10.1016/j.ydbio.2009.08.021 [DOI] [PubMed] [Google Scholar]
- 47. Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev 2003; 17:2753-64; PMID:14600025; http://dx.doi.org/ 10.1101/gad.1142603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci U S A 1998; 95:14787-92; PMID:9843967; http://dx.doi.org/ 10.1073/pnas.95.25.14787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol Cell 2006; 24:293-300; PMID:17052462 [DOI] [PubMed] [Google Scholar]
- 50. Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 2001; 105:391-402; PMID:11348595; http://dx.doi.org/ 10.1016/S0092-8674(01)00330-0 [DOI] [PubMed] [Google Scholar]
- 51. Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol 2001; 8:1048-52; PMID:11713475 [DOI] [PubMed] [Google Scholar]
- 52. Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell 2000; 103:885-96; PMID:11136974; http://dx.doi.org/ 10.1016/S0092-8674(00)00192-6 [DOI] [PubMed] [Google Scholar]
- 53. Lawson AD. Antibody-enabled small-molecule drug discovery. Nat Rev Drug Disc 2012; 11:519-25; PMID:22743979; http://dx.doi.org/ 10.1038/nrd3756 [DOI] [PubMed] [Google Scholar]
- 54. Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng 1994; 7:1129-35; PMID:7831284; http://dx.doi.org/ 10.1093/protein/7.9.1129 [DOI] [PubMed] [Google Scholar]
- 55. Loset GA, Lobersli I, Kavlie A, Stacy JE, Borgen T, Kausmally L, Hvattum E, Simonsen B, Hovda MB, Brekke OH. Construction, evaluation and refinement of a large human antibody phage library based on the IgD and IgM variable gene repertoire. J Immunol Methods 2005; 299:47-62; PMID:15914190 [DOI] [PubMed] [Google Scholar]
- 56. Holdsworth G, Slocombe P, Doyle C, Sweeney B, Veverka V, Le Riche K, Franklin RJ, Compson J, Brookings D, Turner J, et al. Characterization of the interaction of sclerostin with the low density lipoprotein receptor-related protein (LRP) family of Wnt co-receptors. J Biol Chem 2012; 287:26464-77; PMID:22696217; http://dx.doi.org/ 10.1074/jbc.M112.350108 [DOI] [PMC free article] [PubMed] [Google Scholar]