Abstract
Glioma is the most common malignant brain tumors with poor prognosis. The molecular events involved in the development and progression of glioma remain unclear. In this study, the expression levels of miR-302c-3p were examined in glioma tissues by qRT-PCR. The in vitro and in vivo functional effects of miR-302c-3p were examined further. Luciferase reporter assays were conducted to confirm the targeting associations. Results showed that the expression level of miR-302c-3p in glioma tissues was significantly lower than those in normal brain tissues (P < 0.001). The decreased expression of mi-302c-3p in glioma was positively associated with WHO grade (P < 0.001). Up-regulation of MTDH was also detected in glioma tumors compared with normal brain tissues (P = 0.0027) and is inversely correlated with miR-302c-3p expression (P = 0.003, R2 = 0.4065). MTDH mRNA is a direct target of miR-302c-3p, whose ectopic expression decreases MTDH expression through binding to its 3′-untranslated region. Overexpression of miR-302c-3p results in a dramatic inhibition of glioma cells proliferation and invasion in vitro and in vivo. These data suggest that miR-302c-3p play a pivotal role in the progression of glioma by targeting MTDH and is a potential inhibitor in glioma treatment.
Keywords: down-regulation, glioma, inhibition, invasion, microRNA, MTDH, proliferation
Abbreviations
- miR-302c-3p
Homo sapiens microRNA-302c-3p
- MTDH
metadherin
- LV-302c-3p
lentivirus expressing vector with miR-302c-3p gene
- LV-NC
lentivirus expressing vector without interest gene
- U87MG-302c-3p
U87MG cell transfected with LV-302c-3p
- U87MG-NC
U87MG cell transfected with LV-NC
- 3′UTR
3′-untranslated region.
Introduction
Human glioma, originating from glia or their precursors within the central nervous system, is the most common form of primary brain tumor in adults, and for highly malignant gliomas (WHO III and IV) there is no successful treatment; patients have an average survival time of approximately 12–15 months after diagnosis.1 Glioma are highly invasive and infiltrate normal brain tissue, and as a result, surgical resection is always incomplete. Treatment for glioma consist of surgical resection combined with radiotherapy plus chemotherapy or target therapy, which has been found to significantly increase survival.2 As yet, the mechanisms that underlie the correlation between high grade and poor prognosis are still unknown, because all grades of glioma are characterized by inappropriate proliferation, invasion of normal brain tissue, and disruption of normal brain functions. So only the molecular mechanism is well-defined can we find out novel therapeutic approaches.
miRNAs are 21 to 23 nucleotides, endogenous small noncoding RNA sequences that negatively regulate gene expressions at a post-translational level by binding to regions of sequence complementarity to 3′ UTR of target mRNAs, resulting in either the degradation or translational inhibition of the mRNA. Many miRNAs have been proved to be proto-oncogenes or tumor suppressors in various human cancers, including in glioma.3-5 Recent studies have revealed that miRNAs, such as miR-7,6,7 miR-124,8-10 miR-128,11 miR-107,12 and miR-181b,13-15 are globally dysregulated in glioma. However, the particular molecular mechanisms through which miRNAs mediate glioma carcinogenesis and metastasis are still largely unknown. So the identification and characterization of the role of novel miRNAs in glioma may provide new insight into understanding the molecular mechanisms of glioma development. Based on Leivonen's study, miR-302c was reduced expressed in their experimental models and others found that it may be involved in hepatocellular carcinoma metastasis, which suggested that miR-302c might play an important role in glioma progression.16-19
In this study, we first detected the expressions of miR-302c-3p in glioma patients' tissues, and a series of in vitro and in vivo studies were then conducted to investigate the mechanisms and impact of miR-302c-3p in glioma. By bioinformatics analysis (RNA22) and experimentally validation, we found metadherin (MTDH) was a direct target of miR-302c-3p. Upregulation of MTDH was also detected in glioma tumors compared with normal brain tissues and is inversely correlated with miR-302c-3p expression. Finally, we find that overexpression miR-302c-3p could dramatically inhibit glioma cells proliferation and invasion both in vitro and in vivo.
Results
miR-302c-3p was down-regulated in human glioma tissues
The expression of miR-302c-3p were examined in both glioma tissues and normal brain tissues by qRT-PCR. As shown in Figure 1A, the expression level of miR-302c-3p was significantly decreased in tumor tissues compared with normal brain tissues (0.39 ± 0.04 vs. 1.06 ± 0.03, P < 0.001). Furthermore, the expression levels of miR-302c-3p in tumors with high grades (WHO III and WHO IV) were significantly lower than those in those with low grades (WHO I and WHO II) (0.27 ± 0.03 vs. 0.66 ± 0.05, P < 0.001), see Figure 1B.
Figure 1.
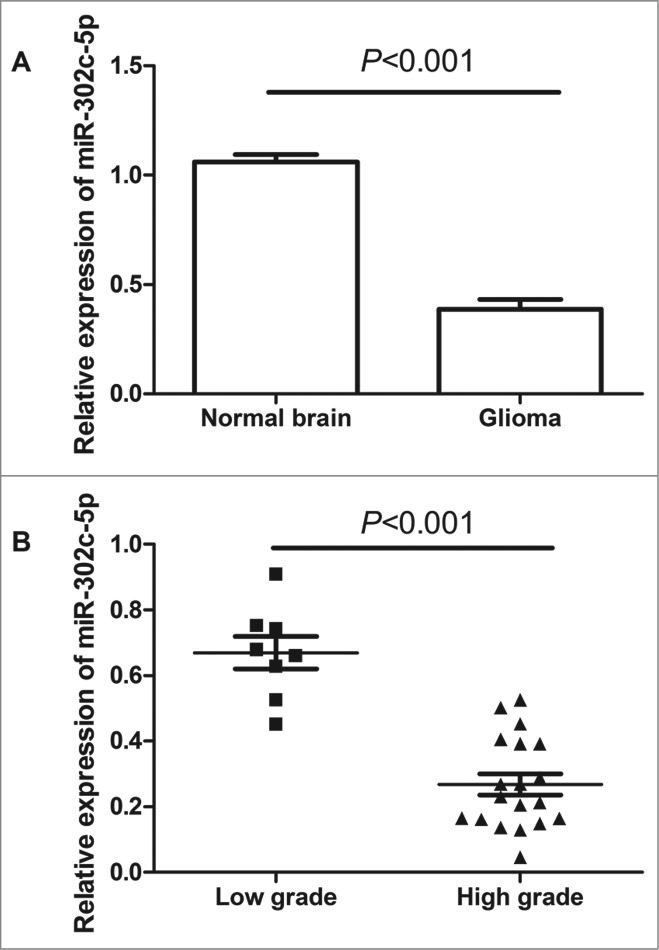
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of miR-302c-3p expression in glioma tissues and normal brain tissues. (A) miR-302c-3p is downregulated in glioma tissues compared with normal brain tissues (P < 0.001). (B) The decreased expression of mi-302c-3p in glioma was positively associated with WHO grade (P < 0.001). Low-grade glioma refers to World Health Organization (WHO) grade I and WHO grade II, high-grade glioma refers to WHO grade III and WHO grade IV.
MTDH is up-regulated in human glioma tissues and its expression correlates with miR-302c-3p expression in glioma
To corroborate the potential importance of MTDH in gliomas, we compared the level of MTDH expression in 27 tumors versus 6 normal brain tissues. The expression of the MTDH accessed by qRT-PCR was significantly increased in gliomas by compared with the normal brain tissues (P = 0.0027, see Fig. 2A). MTDH is overexpressed in 85.2% (23/27) of tumors compared with normal tissues (see Fig. 2A). To further evaluate the correlation between MTDH and miR-302c-3p expression in gliomas, we compared the expression of MTDH and miR-302c-3p in 27 gliomas. Expression of MTDH and miR-302c-3p exhibited a significant inverse correlation (P = 0.003, R2 = 0.4065; Fig. 2B), which significantly suggested that the miR-302c-3p target status of MTDH.
Figure 2.
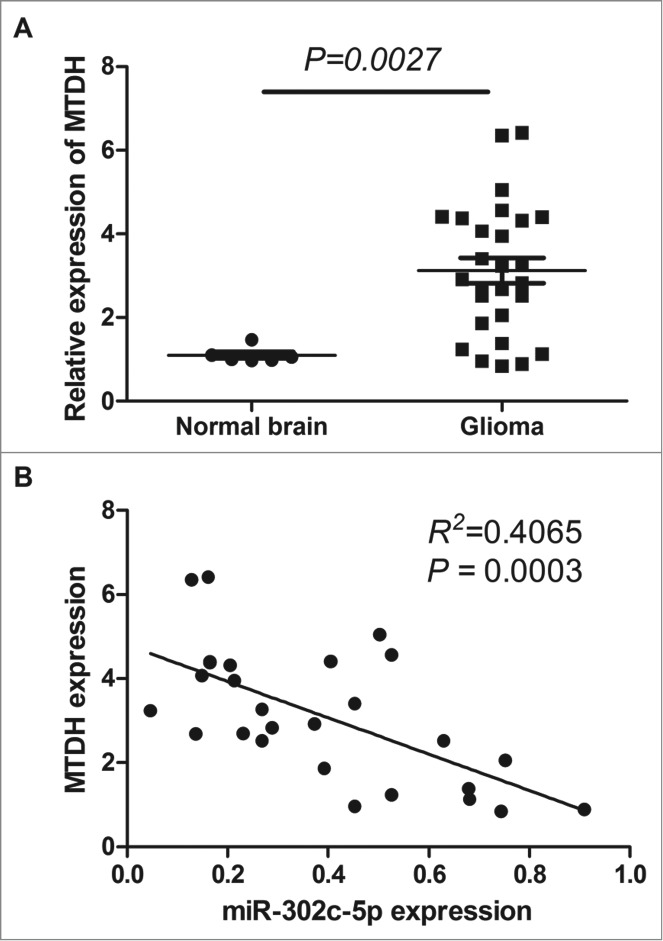
qRT-PCR analysis of MTDH expression in glioma tissues and normal brain tissues. (A) Level of MTDH mRNA is upregulated in glioma tissues compared with normal brain tissues (P = 0.0027). (B) Correlation between MTDH expression and miR-302c-3p expression in glioma tissues (P = 0.003, R2 = 0.4065).
MTDH was a direct target of miR-302c-3p which inhibited its expression via binding to its 3′UTR
On the basis of the prediction software, RNA22 (https://cm.jefferson.edu/rna22v1.0-homo_sapiens/GetInputs.jsp), we find that the 3′UTR of MTDH contains a complementary site (position 2271˜2293) for the seed region of miR-302c-3p, and may be a potential target of miR-302c-3p (Fig. 3A). To validate the miRNA-target interaction and determine whether miR-302c-3p inhibited the expression of MTDH through direct interaction with 3′UTR of MTDH mRNA in the intracellular environment in glioma, Luciferase assay was established. Results showed that miR-302c-3p mimics suppressed the luciferase activities about 69.6% in the wild-type group (P = 0.005, Fig. 3B), while no obvious alteration was detected in the mutant group or the negative control group (Fig. 3B), which indicated that miR-302c-3p could directly bind with MTDH 3′-UTR at the seed sequence.
Figure 3.
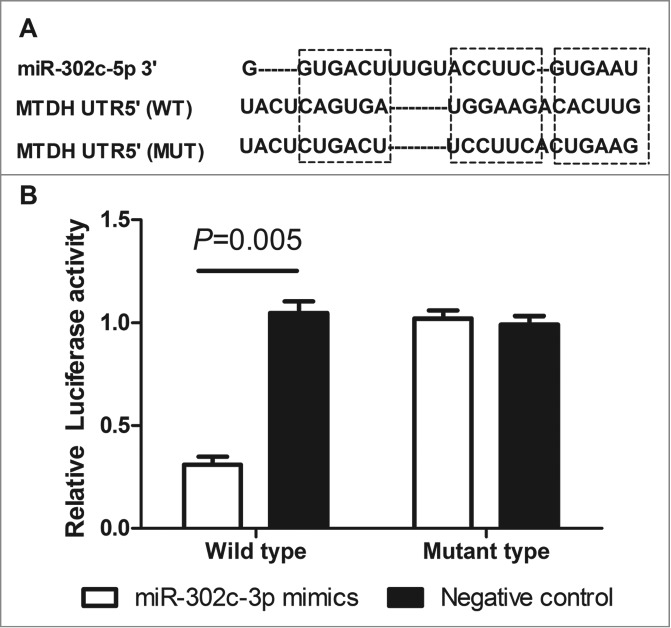
MTDH is a direct functional target of miR-302c-3p. (A) The predicted binding sites of miRNA-302c-3p in the 3′UTR region of MTDH. (B) miR-302c-3p mimics suppressed the luciferase activities in the wild-type group compared with negative control (P = 0.005).
Overexpression of miR-302c-3p inhitibted U87MG proliferation and invasion in vitro
The CCK-8 assays were performed to assess cell proliferation of U87MG cells transfected with LV-302c-3p or LV-NC. A significant reduction in the proliferation rate was observed on the U87MG cells transfected with LV-302c-3p when compared to negative control (Fig. 4A). To explore the effect of overexpression of miR-302c-3p on the invasive capacity of glioma cells, transwell assays were performed. U87MG cells transfected with LV-302c-3p or LV-NC were seeded into the chambers with matrigel, and their invasive potential were determined after 24h culture. The results showed that the invasive capacity of U87MG cells overexpressing miR-302c-3p was reduced by 82.4% compared with the control groups (Fig. 4B, C, D).
Figure 4.
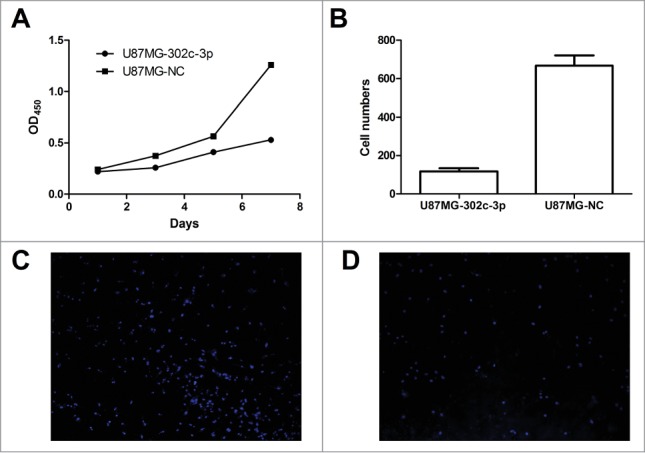
Overexpression of miR-302c-3p results in a dramatic inhibition of glioma cells proliferation and invasion in vitro. (A) CCK-8 assay was used to determine growth rate of U87MG-302c-3p and U87MG-NC cells. (B) Transwell assays were performed to determine invasion ability of U87MG-NC cells (C) and U87MG-302c-3p (D).
miR-302c-3p inhibits glioma cell tumorigenesis in vivo
To further investigate the role of miR-302c-3p inhibition on tumorigenesis in vivo, we constructed a lentivirus vector to mediate the expression of miR-302c-3p and established 2 stable cell lines, which were named U87MG-302c-3p and U87MG-NC. Real-time PCR results showed that U87MG-302c-3p cells expressed 15.4 fold miR-302c-3p than control cells U87MG-NC, indicating the construction was successful (Fig. 5A). Then, we implanted U87MG-302c-3p and U87MG-NC cells into nude mice. As shown in Figure 5B, mice injected with U87MG-NC cells formed larger tumors compared with the U87MG-302c-3p cells (P = 0.004, Fig. 5B, C, D).
Figure 5.
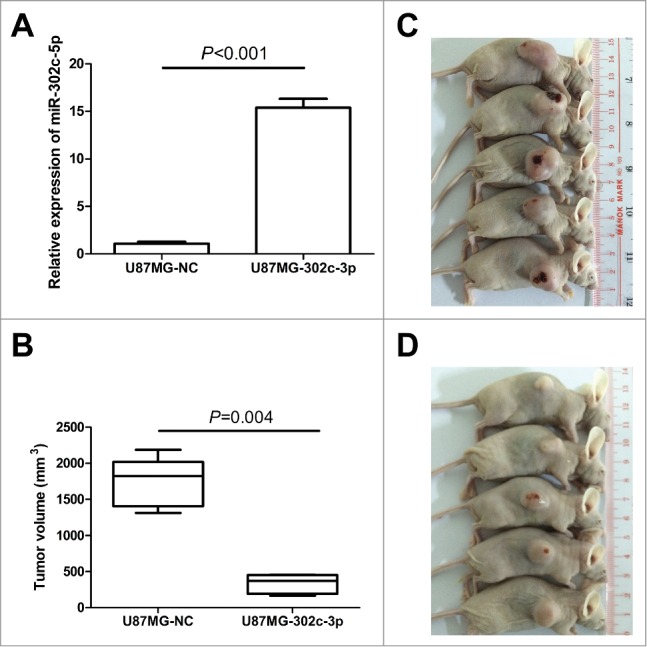
Exogenetic expression of miR-302c-3p suppresses glioma cell tumorigenesis in vivo. (A) Establishment of the stable cell lines overexpressing miR-302c-3p. (B) Quantification of the tumor volume of U87MG-NC group (C) and U87MG-302c-3p group (D).
miR-302c-3p inhibits glioma cell tumorigenesis in the brain
To further investigate the role of miR-302c-3p inhibition on tumorigenesis in the brain, C6 were transfected with LV-302c-3p and then injected into the brain. Cells transfected with LV-NC were used as a control. After 21th day post-injection, MR image analysis and histopathological analysis were performed. Results showed that the right brain injected with C6-NC cells formed much larger tumors compared with the C6–302c-3p cells (Fig. 6A, B, C, D).
Figure 6.
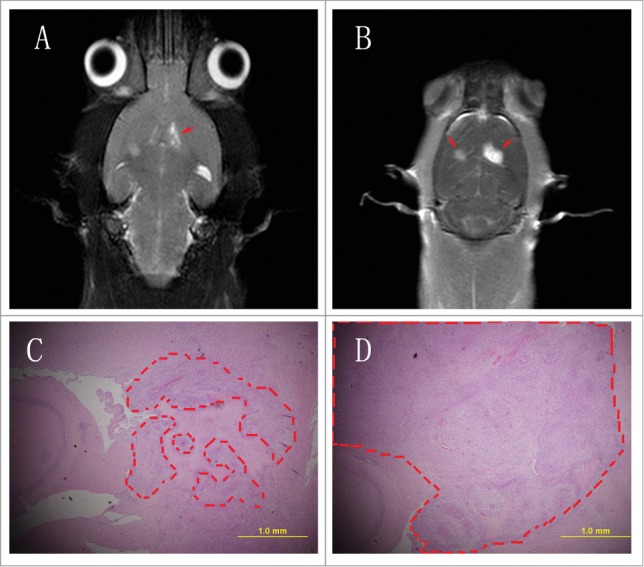
Exogenetic expression of miR-302c-3p suppresses glioma cell tumorigenesis in rat brain. (A) T2 weight image. The left side is C6–302c-3p cells. The right side is C6-NC cells. Red arrows indicated the tumors. (B) Enhanced T2 weighted images. The left side is C6–302c-3p cells. The right side is C6-NC cells. Red arrows indicated the tumors. (C) C6–302c-3p group (C) and C6-NC group (D). Dashed box indicated tumors (original magnification 40×).
Discussion
Gliomas are the most frequent primary tumors of the brain, which are highly invasive and infiltrate normal brain tissue, and as a result, surgical resection is always incomplete treatment.20 Despite efforts in developing multimodal treatments that have been tremendous, the clinical prognosis of glioma patients remains unfavorable.21 It is of great significance to investigate the underlining molecular mechanisms in human gliomas. Recently, more and more evidences have showed that deregulated miRNAs participate in glioma carcinogenesis. In our study, we found that MTDH is a direct target of miR-302c-3p. Overexpression of miR-302c-3p can reduce glioma cell growth through targeting MTDH. Meanwhile, we found that exogenous miR-302c-3p expression can significantly abrogate the invasion of glioma cells. In addition, in vivo tumorigenesis experiments confirmed that stable overexpression of miR-302c-3p inhibited the tumorigenesis ability of glioma cells.
miRNAs, a series of special regulators which could interact with multiple target genes simultaneously, influenced different downstream pathways.22 Recent studies have demonstrated the aberrant expression of various miRNAs in gliomas, such as miR-7, miR-124, miR-128, miR-107, and miR-181b. In this study, we proved for the first time that the expression of miR-302c-3p in glioma tissues was significantly lower than those in normal brain tissues. Furthermore, the decreased expression of mi-302c-3p in glioma was positively associated with high WHO grade. Overexpression of miR-302c-3p can reduce glioma cell growth and invasion both in vitro and in vivo.
The regulatory mechanism of miR-302c was a complex framework and sometimes organ specific. For instance, Rosa et al. revealed that the miR-302 family can influence the differentiation of human embryonic stem cells; other studies have indicated that miR-302c directly targets the estrogen receptor in human breast cancer, and it has also been reported to be dysregulated in biliary tract cancer and thyroid cancer. Zhu's study showed that miR-302c in endothelial cells (ECs) may suppress EC-mediated tumor growth through inhibiting metadherin expression.18 To identify the putative miR-302c-3p target genes, we applied combined in prediction software analysis and luciferase reporter assays. We demonstrated that MTDH is a direct target of miR-302c-3p, with the evidence that overexpression of miR-302c-3p led to reduced luciferase activity with MTDH 3′UTR. In our study, we found that TWIST2 is a direct target of miR-302c-3p.
Metadherin (MTDH), also known as astrocyte elevated gene-1 (AEG-1) and lysine-rich CEACAM1 coisolated (LYRIC), is a multifunctional oncogene that is overexpressed in a variety of human cancers.23,24 As a target of Ras, MTDH activates multiple oncogenic signaling pathways including phosphoinositide-3 kinase (PI3K)–Akt, mitogen-activated protein kinase (MAPK), Wnt, and NF-kB involved in regulation of proliferation, invasion, chemoresistance, angiogenesis, and metastasis.25-28 In particular, MTDH showed higher expression, compared with that in normal brain tissues, in tumors of the CNS, such as neuroblastoma, glioblastoma multiforme, and oligodendroglioma.29-31 Here, we found that MTDH was overexpressed in glioma tissues compared with normal brain tissues. And the expression of MTDH and miR-302c-3p in glioma tissues is inversely correlated. These results inferred that up-regulation of miR-302c-3p abrogates the tumorigenesis ability of glioma cells. And we speculate that miR-302c-3p may function as a tumor inhibitor through the inhibition of glioma proliferation and invasion by targeting MTDH.
The potential clinical application of miR-302c-3p in CRC has never been reported before. In this study, we extended our analyses by assaying miR-302c-3p expression in specimens from 27 human glioma patients. We have identified that miR-302c-3p is downregulated in glioma. miR-302c-3p suppressed glioma cells proliferation and invasion, at least in part via inhibition of oncogene MTDH. And miR-302c-3p may be a candidate tumor suppressive miRNA of glioma and might be a new prognostic biomarker for glioma patients.
Materials and Methods
Patients and tissue samples
Tissue samples were obtained after informed consent from the Department of Neurosurgery, Yuquan Hospital, Tsinghua University (Beijing, China) and Department of Neurosurgery, Shanxi Dayi hospital, Shanxi Medical University (Taiyuan, China). Among them there were 6 normal brain tissue samples collected from patients undergoing internal decompression surgery following brain trauma (3 female, 3 male patients; median age of 39.5, with a range of 29–62 y old). A total of 16 males and 11 females (1.45:1) were enrolled in this research, and the median age was 42.1 y (range, 4–75 years). Eight of the 27 gliomas were classified as low-grade (3 WHO I, 5 WHO II), and 19 gliomas were classified as high-grade (7 WHO III, 12 WHO IV) gliomas. None of the patients had received chemotherapy or radiotherapy preoperatively. Resected tissue samples were immediately divided into two parts: one part was used for histopathological diagnosis by two pathologists according to the 2007 WHO classification, and the other part was stored in liquid nitrogen until used for RNA extraction. All the protocols were reviewed by the Joint Ethics Committee of the Central South University Health Authority and performed following national guidelines.
Cell culture
Human glioma cell line U87MG, C6 and human embryonic kidney cell line 293T were obtained from Cell Library of China Infrastructure of Cell Line Resources. U87MG and C6 was cultured in MEM medium (12492–013, Gibco) supplemented with 10% fetal bovine serum (FBS, 10099–141, Gibco). 293T was cultured in DMEM medium (12430–054, Gibco) supplemented with 10% FBS. All the cells were maintained at 37°C in a humidified incubator containing 5% carbon dioxide.
RNA isolation and quantitative real-time RT-PCR (qRT-PCR)
Total RNA from cells and tissues was isolated using TRIzol reagent (15596–018, Invitrogen) and was eluted in 50 μL nuclease free water. The details for qRT-PCR were conducted as previously described. Briefly speaking, miRNAs and mRNAs were converted to cDNAs using PrimeScript miRNA cDNA Synthesis Kit (D350A, Takara) and the SuperScript® III First-Strand Synthesis System (18080–051, Invitrogen) respectively. SYBR green real-time PCR was performed to compare the expressing levels of miRNAs or mRNAs using TransStart Top Green qPCR SuperMix (AQ131–03, TransGen) by Mx3000P PCR thermal cycler. U6 snRNA and GAPDH were selected for normalization to the expressing levels of miRNAs and mRNAs respectively. Relative quantitations (RQs) of miRNAs and mRNAs were calculated using the 2−ΔΔCt method. The sequences of RT-PCR primers are listed in Table 1.
Table 1.
Primers for qRT-PCR
| Gene name | Primer sequence (5′ to 3′) | |
|---|---|---|
| miR-302c-3p | RT primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCACTG |
| Forward | GCGTGCTTCCATGTTTCAGTGG | |
| Reverse | CAGTGCAGGGTCCGAGGTAT | |
| MTDH | Forward | TGCAGCCGAGGAATAAAGGA |
| Reverse | CTGTGCATAAGATCCAAGGAATTG | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward | TGCACCACCAACTGCTTAGC |
| Reverse | GGCATGGACTGTGGTCATGAG | |
Vector construction and dual-luciferase reporter assay
For luciferase assays, the potential miR-302c-3p binding site in the MTDH 3′UTR was predicted at position 2271–2293 by RNA22 (https://cm.jefferson.edu/rna22v1.0-homo_sapiens/GetInputs.jsp). The wild-type and mutant 3′UTR of MTDH containing the seed sequence were synthesized and cloned into the pMIR-Report vector (AM5795, Invitrogen). The wild-type 3′UTR of MTDH was mutated following the principle of complementary base pairing to disrupt the miR-302c-3p binding sites. U87MG cells were transfected with miR-302c-3p mimics (40 nmol) for 24 hours and then the cells were co-transfected with wild-type or mutated 3′UTR of MTDH using Lipofectamine 2000 (11668–500, Invitrogen). Forty-eight hours later, cell lysates were incubated with dual-luciferase reagents (E1483, Promega) and the luciferase activities were determined by the luciferase assay kit (E2810, Promega). Pre-miR™ miRNA Precursor Negative Control (AM17110, Ambion®) was used as a negative control.
Lentivirus packaging and stable cell line establishment
The stem-loop sequence miR-302c (MI0000773) was synthesized from Human Genome DNA with primer pair hsa-mir-302c-sence (5′-CCTTTGCTTTAACATGGGGGTACCTGCTGTGTGAAACAAAAGTAAGTGCTTCCATGTTTCAGTGGAGGTTTTTTGT-3′) and hsa-mir-302c-antisence (5′-CTAGACAAAAAACCTCCACTGAAACATGGAAGCACTTACTTTTGTTTCACACAGCAGGTACCCCCATGTTAAAGCAAAGG-3′). The generated sequence was then cloned into cloned into the XbaI and SmaI site of the lentiviral vector H1 (provided by Professor Qin Shen) to generate a miR-302c-3p stable expressing vector (pH1-miR-302c-3p). Lentivirus expressing vector with miR-302c-3p gene (LV-302c-3p) was produced by co-transfecting with 3 plasmids (pH1-miR-302c-3p, pSVS and pCMV) into 293 T cells. 293 T cells were cultured in DMEM with 5% FBS in a 37°C incubator with 5% CO2. Forty-eight hours after transfection, the supernatant was harvested, filtered and cleared by centrifugation at 500 × g for 10 min. The lentivirus expressing vector without interest gene was selected as negative control (LV-NC). The final obtained virus was re-suspended in phosphate-buffered saline (PBS) and then titrated. The final concentration of the virus suspension was adjusted to 5 × 108 TU/ml. LV-302c-3p or LV-NC was transfected into U87MG, named U87MG-302c-3p and U87MG-NC respectively, and the cells would express EGFP stably 72 h after transfection. LV-302c-3p or LV-NC transfected into C6 were named C6–302c-3p and C6-NC, respectively.
Cell proliferation and invasion assays
To assess the proliferation of U87MG transduced by LV-302c-3p or LV-NC, cells were determined by using CCK-8 (CK04–3000T, DOJINDO). Briefly, 100 μL 1 × 104 cell/ml cell suspensions were initially seeded onto each well in the 96-well plate. After 1, 3, 5, 7 days, 10 μL CCK-8 was added to each well and incubated at 37°C for 4 h. Then, 100 μL of solution was aspirated from each well and poured in a 96-well plate. The absorbance in each well at 450 nm was measured with an ELISA reader.
For cell invasion assay, a specialized Chamber with BD BioCoat Matrigel (354480, BD Biosciences) was used according to the manufacturer's instructions. Briefly, cells (5 × 104 cells/mL) were loaded into chamber inserts containing an 8-μm pore size membrane with a thin layer matrigel matrix. Cells migrating to the lower surface of the membrane during 24 or 48 h were fixed with 100% methanol and non-invading cells on the upper surface of the membrane were removed. The membranes, with migrated cells, were then stained with DAPI (D9564, Sigma), scanned, and digital images were obtained with a microscope. The number of invasive cells was then determined for 5 independent fields under a microscope. The mean of triplicate assays for each experimental condition was used for analysis.
In vivo assays
Twenty female athymic BABL/c nude mice (4–6 weeks old) were obtained from Beijing Vital River Laboratories (Beijing, China), and randomly divided into 2 groups: U87MG-302c-3p injected group and U87MG-NC injected group. 200 μL cells were injected into the subcutaneous tissue at a density of 1 × 107 cells/ml. Tumor diameters were measured and tumor volume = (length × width2)/2. All the mice were sacrificed on the 28th day post-injection. All animal experiments were carried out in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80–23) and approved by the Beijing Administration Committee of Experimental Animals.
Stereotactic surgeries
Ten rats were used in this experiment. Animals anesthetized by intraperitoneal injection of 5% chloral hydrate (350 mg/kg) and placed in the stereotaxic instrument. C6–302c-3p suspensions 5 μl (1 × 107 cells/ml) was delivered at 0.4 μl/min at the following stereotaxic coordinates: 0 mm posterior and 3.5 mm left-lateral to the bregma and 5.5 mm ventral to the skull surface. The needle was withdrawn to 1.3 mm after injection of 1.5 μl and then injection was continued. The needle was left in situ for an additional 5 min after completing the injection, and then withdrawn slowly. C6-NC suspensions, as a negative control, was injected into the contralateral position of the same rats.
MR image analysis
All rats experienced high resolution MR scanning preoperatively on the 21th day post-injection. The MR images were acquired using a 3.0 tesla clinical unit (Philips Achieva 3.0T TX). The rats were anesthetized with intraperitoneal injection of 5% chloral hydrate (350 mg/kg). After coronal T2 weighted images had been obtained, enhanced T2 weighted images were acquired by an intravenous injection of dimeglumine gadopentetate (Magnevist, Bayer Schering Parma, Germany) at a dose of 500 μl/250 g body weight via caudal vein.
Histopathological analysis
Rats were sacrificed for histopathological examination following MR image analysis. After anesthetization with intraperitoneal injection of 5% chloral hydrate solution, each rat's thorax was rapidly opened and the right atrium was incised. Normal saline followed by 4% paraformaldehyde was pumped into the rat's circulation through a cannula inserted into the left ventricle. When the perfusate flowing from the right atrium was clear, brain tissues were dissected and placed in 10% methanal. After two-day fixation, the brain was sectioned in 5-μm slices for haematoxylin-eosin staining. The images were examined with light microscopy and digitally recorded.
Statistical analysis
All computations were carried out using the software of GraphPad Prism 5 (GraphPad Software Inc., CA, USA). Data were expressed as mean ± SD. The differential expression of miR-302c-3p and MTDH between glioma tissues and normal brain tissues was evaluated by independent sample t test.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by Shanxi Scholarship Council of China (2011–102) and National Natural Science Foundation of China (81472817).
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008; 359: 492-507; PMID:18669428; http://dx.doi.org/ 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, et al.. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A 2005; 102:5814-9; PMID:15827123; http://dx.doi.org/ 10.1073/pnas.0402870102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 2011; 11:644-56; PMID:21822212; http://dx.doi.org/ 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- 4.Petrocca F, Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. J Clin Oncol 2011; 29:747-54; PMID:21079135; http://dx.doi.org/ 10.1200/JCO.2009.27.6287 [DOI] [PubMed] [Google Scholar]
- 5.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci 2010; 101:2309-15; PMID:20726859; http://dx.doi.org/ 10.1111/j.1349-7006.2010.01683.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Li G, Su Z, Jiang Z, Chen L, Wang J, Yu S, Liu Z. Poly(amido amine) is an ideal carrier of miR-7 for enhancing gene silencing effects on the EGFR pathway in U251 glioma cells. Oncol Rep 2013; 29:1387-94; PMID:23404538 [DOI] [PubMed] [Google Scholar]
- 7.Lu ZJ, Liu SY, Yao YQ, Zhou YJ, Zhang S, Dai L, Tian HW, Zhou Y, Deng HX, Yang JL, et al.. The effect of miR-7 on behavior and global protein expression in glioma cell lines. Electrophoresis 2011; 32:3612-20; PMID:22120825; http://dx.doi.org/ 10.1002/elps.201100230 [DOI] [PubMed] [Google Scholar]
- 8.Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang C, Liu X, Wang X, Li H, Kang C, et al.. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol 2014; 16:1341-53; PMID:24861879; http://dx.doi.org/ 10.1093/neuonc/nou084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li A, Lin X, Tan X, Yin B, Han W, Zhao J, Yuan J, Qiang B, Peng X. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett 2013; 587:2455-60; PMID:23792158; http://dx.doi.org/ 10.1016/j.febslet.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 10.Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, Yang Y, McEnery K, Jethwa K, Gjyshi O, et al.. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res 2013; 73:3913-26; PMID:23636127; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch DH, Barres BA, et al.. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 2012; 31:1884-95; PMID:21874051; http://dx.doi.org/ 10.1038/onc.2011.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y, Wei Y, Wang J, Ao Q, Gong K, Zuo H. Decreased expression of microRNA-107 predicts poorer prognosis in glioma. Tumour Biol 2015; PMID:25596705; http://dx.doi.org/ 10.1007/s13277-015-3086-y [DOI] [PubMed] [Google Scholar]
- 13.Sun YC, Wang J, Guo CC, Sai K, Wang J, Chen FR, Yang QY, Chen YS, Wang J, To TS, et al.. MiR-181b sensitizes glioma cells to teniposide by targeting MDM2. BMC Cancer 2014; 14:611; PMID:25151861; http://dx.doi.org/ 10.1186/1471-2407-14-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Sai K, Chen FR, Chen ZP. miR-181b modulates glioma cell sensitivity to temozolomide by targeting MEK1. Cancer Chemother Pharmacol 2013; 72:147-58; PMID:23645289; http://dx.doi.org/ 10.1007/s00280-013-2180-3 [DOI] [PubMed] [Google Scholar]
- 15.Li P, Lu X, Wang Y, Sun L, Qian C, Yan W, Liu N, You Y, Fu Z. MiR-181b suppresses proliferation of and reduces chemoresistance to temozolomide in U87 glioma stem cells. J Biomed Res 2010; 24:436-43; PMID:23554660; http://dx.doi.org/ 10.1016/S1674-8301(10)60058-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min D, Lv XB, Wang X, Zhang B, Meng W, Yu F, Hu H. Downregulation of miR-302c and miR-520c by 1,25(OH)2D3 treatment enhances the susceptibility of tumour cells to natural killer cell-mediated cytotoxicity. Br J Cancer 2013; 109:723-30; PMID:23820258; http://dx.doi.org/ 10.1038/bjc.2013.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res 2010; 12; PMID:20346098; http://dx.doi.org/ 10.1186/bcr2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu K, Pan Q, Jia LQ, Dai Z, Ke AW, Zeng HY, Tang ZY, Fan J, Zhou J. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep 2014; 4:5524; PMID:25027009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, et al.. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ 2012; 19:232-44; PMID:21720384; http://dx.doi.org/ 10.1038/cdd.2011.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froklage FE, Oosterbaan LJ, Sizoo EM, de Groot M, Bosma I, Sanchez E, Douw L, Heimans JJ, Reijneveld JC, Lagerwaard FJ, et al.. Central neurotoxicity of standard treatment in patients with newly-diagnosed high-grade glioma: a prospective longitudinal study. J Neurooncol 2014; 116:387-94; PMID:24264531; http://dx.doi.org/ 10.1007/s11060-013-1310-4 [DOI] [PubMed] [Google Scholar]
- 21.Khan UA, Bhavsar A, Asif H, Karabatsou K, Leggate JR, Sofat A, Kamaly-Asl ID. Treatment by specialist surgical neurooncologists improves survival times for patients with malignant glioma. J Neurosurg 2015; 122:297-302; PMID:25415070; http://dx.doi.org/ 10.3171/2014.10.JNS132057 [DOI] [PubMed] [Google Scholar]
- 22.He H, Hao SJ, Yao L, Yang F, Di Y, Li J, Jiang YJ, Jin C, Fu DL. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol Ther 2014; 15:1333-9; PMID:25010661; http://dx.doi.org/ 10.4161/cbt.29706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Li LP. Progress of cancer research on astrocyte elevated gene-1/Metadherin (Review). Oncol Lett 2014; 8:493-501; PMID:25009642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SG, Kim K, Kegelman TP, Dash R, Das SK, Choi JK, Emdad L, Howlett EL, Jeon HY, Su ZZ, et al.. Oncogene AEG-1 promotes glioma-induced neurodegeneration by increasing glutamate excitotoxicity. Cancer research 2011; 71:6514-23; PMID:21852380; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci U S A 2006; 103:17390-5; PMID:17088530; http://dx.doi.org/ 10.1073/pnas.0608386103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF, Fisher PB. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene 2008; 27:1114-21; PMID:17704808; http://dx.doi.org/ 10.1038/sj.onc.1210713 [DOI] [PubMed] [Google Scholar]
- 27.Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche H, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci U S A 2009; 106:21300-5; PMID:19940250; http://dx.doi.org/ 10.1073/pnas.0910936106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo BK, Chen D, Su ZZ, Gredler R, Yoo J, Shah K, Fisher PB, Sarkar D. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res 2010; 70:3249-58; PMID:20388796; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emdad L, Sarkar D, Lee SG, Su ZZ, Yoo BK, Dash R, Yacoub A, Fuller CE, Shah K, Dent P, et al.. Astrocyte elevated gene-1: a novel target for human glioma therapy. Mol Cancer Ther 2010; 9:79-88; PMID:20053777; http://dx.doi.org/ 10.1158/1535-7163.MCT-09-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noch E, Bookland M, Khalili K. Astrocyte-elevated gene-1 (AEG-1) induction by hypoxia and glucose deprivation in glioblastoma. Cancer Biol Ther 2014; 11:32-9; PMID:21084864; http://dx.doi.org/ 10.4161/cbt.11.1.13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumann BC, Dorsey JF. Astrocyte-elevated gene-1 (AEG-1): Glioblastoma's helping hand during times of hypoxia and glucose deprivation? Cancer Biol Ther 2014; 11:40-2; PMID:21150314; http://dx.doi.org/ 10.4161/cbt.11.1.14139 [DOI] [PubMed] [Google Scholar]


