Abstract
The methylation status of the IGFBP-3 gene is strongly associated with cisplatin sensitivity in patients with non-small cell lung cancer (NSCLC). In this study, we found in vitro evidence that linked the presence of an unmethylated promoter with poor response to radiation. Our data also indicate that radiation might sensitize chemotherapy-resistant cells by reactivating IGFBP-3-expression through promoter demethylation, inactivating the PI3K/AKT pathway. We also explored the IGFBP-3 methylation effect on overall survival (OS) in a population of 40 NSCLC patients who received adjuvant therapy after R0 surgery. Our results indicate that patients harboring an unmethylated promoter could benefit more from a chemotherapy schedule alone than from a multimodality therapy involving radiotherapy and platinum-based treatments, increasing their OS by 2.5 y (p = .03). Our findings discard this epi-marker as a prognostic factor in a patient population without adjuvant therapy, indicating that radiotherapy does not improve survival for patients harboring an unmethylated IGFBP-3 promoter.
Keywords: hypermethylation, IGFIR/AKT, IGFBP-3, NSCLC, radiotherapy
Abbreviations
- ATCC
American Type Culture Collection
- BS
bisulfite sequencing
- CDDP
cisplatin
- ECACC
European Collection of Cell Cultures
- IGFBP-3
insulin-like growth factor binding protein-3
- IR
Ionizing radiation
- NSCLC
non-small cell lung cancer
- OS
overall survival
- qMSP
quantitative methylation specific PCR
Introduction
Non-small cell lung carcinoma (NSCLC) accounts for 1 of every 6 cancer-related deaths worldwide.1 This mortality rate is due to the advanced stage of the disease at diagnosis and its resistance to all therapies. Surgery is the standard treatment in the early stages, and platinum-based adjuvant therapy has been shown to be effective in the advanced stages of the disease.2 Multimodal therapy combining thoracic radiotherapy with chemotherapy after surgery also plays a role in the management of NSCLC,3 primarily for patients at higher risk of local recurrence. However, treatment outcomes vary widely in terms of survival, and increased morbidity is strongly linked with therapy.
The mechanisms of drug resistance in cancer therapy have been widely analyzed, particularly for NSCLC, in which platinum-based therapy has often failed. In fact, we have previously reported that the loss of IGFBP-3 expression by promoter hypermethylation results in reduced tumor cell sensitivity to cisplatin in NSCLC, an effect that is mediated by the activation of the IGF-IR/AKT pathway.4,5 Despite the promising results regarding the usefulness of epigenetic alterations as potential markers in chemotherapy response,6,7 these epi-markers have not been studied extensively in radiotherapy, leading to scarce data concerning epigenetics and radiosensitivity.8 The radioresistance of tumor cells is a less explored and poorly defined field compared with drug resistance, and the role of the IGF-I/IGBP-3 axe on radiosensitivity in cancer is controversial because of the differing results when various tumor types are evaluated.9–11 Furthermore, the relationship between IGFBP-3 promoter hypermethylation and the response to radiotherapy in NSCLC is unknown. Recent studies have reported that radiotherapy induces global DNA hypomethylation,12 which is why, in the present study, we evaluated both, the role of radiotherapy on the biology of IGFBP-3 promoter methylation and its clinical value as a potential tool for deciding on a concomitant radiotherapy after NSCLC surgery.
Methods
Cell lines and radiation-clonogenic cell survival assays
The resistant cell lines H23R and H460R were established previously in our laboratory from the parental H23S and H460S lung cell lines, and together with the cell line H1299 were purchased from the ATCC (Manassas, VA); all were maintained in RPMI supplemented with 10% FBS. The CDDP sensitive and resistant ovarian cancer cell lines 41M and 41MR, hereafter called 41S and 41R respectively, were provided by Dr. L Kelland and were maintained in DMEM supplemented with 10% FBS.5 Each of the paired sensitive and resistant cell lines were irradiated at doses of 0, 2, 4, 6 and 8 Gy using a Cesium-137 irradiator Mark I30 (JL Shepherd and Associates, San Fernando, CA). Immediately following irradiation, the cells were trypsinized, diluted and seeded onto p100 plates. After 14 days, the cells were stained by crystal violet, and colonies with over 50 cells were counted with a ColCount colony counter (Optronix, Oxford, UK). Individual assays were performed in triplicate and repeated at least twice. The survival fraction was calculated as previously described.13 For DNA, RNA and protein extraction, cell lines were cultured at a density of 400,000 cells by 60 mm plate for 72 h after irradiation.
NSCLC clinical samples and data collection
Formalin-fixed, paraffin-embedded (FFPE) surgical specimens were obtained from 40 NSCLC patients who received a chemotherapy schedule based on cisplatin/carboplatin with or without concomitant radiotherapy. Histological slides obtained from each block were reviewed by an expert pathologist (F. Rojo) to confirm diagnosis, and to guarantee at least 90% tumoral content. Follow-up was performed according the criteria used in the Medical Oncology Division from Hospital del Mar, including clinical assessments and thorax CT every 3 months for 2 y and every 6 months thereafter. Clinical, pathological, and radiological data were recorded by an independent observer at the H. del Mar and blinded for statistical analysis. In addition 10 samples obtained from pulmonary biopsies with non-neoplastic lung pathology were used as control tissues. We also included in the study clinical/pathological and IGFBP-3 methylation data from an external group of 36 patients from La Paz University Hospital, as published previously,4,5 who did not receive any therapy after surgery. The results from this group were adjusted by age, gender and stage to establish a control group.
Western-blot analysis
Whole-cell extracts from the human cancer cell lines and Western-Blot were performed as described.14 Briefly, 20 μg from the 41S and 41R cell lines at 5 IR doses tested were subjected to western blot and the membranes were hybridized with antibodies against AKT (BD Biosciences, NJ, USA), pAKT-Ser473, pERK1/2-thr202/Tyr204 (E10) (Cell Signaling, MA, USA), ERK (C-14) (sc-154), IGFIR, anti-pIGFIR-Tyr1161 (Santa Cruz Biotechnology, Heidelberg, Germany), and anti-α-tubulin (Sigma Aldrich) as a loading control.
DNA and RNA extraction, bisulfite modification, quantitative methylation-specific PCR and qRT-PCR
DNA from human cancer cell lines, 40 paraffin-embedded NSCLC primary specimens, and 10 non-neoplastic lung tissues were isolated and bisulfite modified as described.5 We then measured the IGFBP-3 promoter methylation by qMSP using the following primer/probe set: F:5′-TTTTACGAGGTATATACGAATGC-3′; R:5′-TCTCGAAATAAAATCTCCCTACG-3′; Probe:5′FAM-CCGATATCGAAAAAACT-3′. A primer/probe set for the unmethylated ACTB gene promoter was used as reference.15 Serial dilutions of bisulfite-modified DNA from the SW760 cell line that harbors a methylated promoter for IGFBP-3 were used to construct calibration curves for IGFBP-3 and ACTB genes. PCR reactions were performed in triplicate as described.15
Total RNA from human cancer cell lines was isolated as previously described.16 Reverse transcription and qRT-PCR analysis were performed as described.5 Samples were analyzed in triplicate using the HT7900 Real-Time PCR system (Applied Biosystems, USA), and relative gene expression quantification was calculated according to the comparative threshold cycle method (2−ΔΔCt) using GADPH as an endogenous control gene. Primers and probes for IGFBP-3 and GADPH expression analysis were purchased from Applied Biosystems (IGFBP-3: Hs 00365742_g1) GADPH: Hs03929097_g1).
Infinium humanmethylation27 annotation and TCGA NSCLC data
The Infinium HumanMethylation27 annotation (available at ftp://ftp.illumina.com/Methylation/InfiniumMethylation/HumanMethylation27/) used the National Center for Biotechnology Information (NCBI) relaxed definition of 200 bp length, 50% GC content and 0.6 ObsCpG/ExpCpG for identification of CpG islands in genes in the Consensus Coding Sequence (CCDS) database.17 We first obtained the sequence of the probes from the TCGA Infinium HumanMethylation27 BeadChip annotation in order to determine the position of the probes within the gene IGFBP-3, and interrogate the Infinium probes located within the bona fide CpG island at the IGFBP-3 promoter region. A probe was considered unmethylated if the β-value was ≤0.15, as previously described.18 We correlate the methylation score (raw β-value) of the TCGA NSCLC patients with their clinical-pathological parameters.
Statistics and study approval
Discrete variables (histology, T, N, stage, gender, methylation status at the IGFBP3 promoter and chemotherapy schedule) were compared with the Chi2 test and corrections with Fisher´s exact test were made when needed. DFS was defined as the time from surgery to clinical, radiological or histological evidence of relapse. Statistical significance was defined as P < 0.05.
Survival functions for patients diagnosed with NSCLC with an unmethylated IGFBP3 promoter who were treated with chemotherapy or with chemo-radiotherapy were plotted using Kaplan-Meier methods, and were compared under 3 conditions the log-rank, Breslow and Tarone-Ware methods. For patients without any evidence of survival at the time of analysis, data on OS were censored at the time of the last contact. Statistical analyses were done by use the SPSS software (version 17.0).
Samples were collected following the ethical and confidentiality issues by Dr. Rojo and Dr. De Castro. The present study is approved by the IdiPAZ following rigorous biosecurity and ethical protocols in all procedures, in accordance with the Hospital's Local Ethic Committee.
Results and Discussion
Cell line data and discussion
To investigate the role of IGFBP-3 methylation in radiosensitivity, we first developed radiation-clonogenic cell survival assays with 3 paired CDDP-sensitive and CDDP-resistant cell lines harboring various IGFBP-3 methylation profiles: H23S/R, H460S/R and 41S/R.4 Each of the paired cell lines was irradiated at doses of 0, 2, 4, 6 and 8 Gy. The unmethylated 41S cells showed lower sensitivity to radiotherapy than the H23S cells, which are semimethylated for IGFBP-3, whereas both the CDDP-resistant 41R and H23R cell lines that harbor an IGFBP-3 hypermethylated promoter showed an increased sensitivity to radiotherapy (Figs. 1A, B) compared with their paired sensitive cell lines. These results agree with reported data that show that DNA hypermethylation of the tumor suppressor genes TIMP3, CDH1 and MGMT predicts a better outcome in head and neck cell squamous cell carcinoma and glioblastoma when treated with radiotherapy.19,20 There was no significant change in the radiotherapy sensitivity of the paired H460S/R cell lines, which were used as a negative control experimental group, given we already reported that the resistance to cisplatin treatment in H460R cells is not mediated by changes in IGFBP-3 expression and promoter methylation 4 (Fig. 1C). Our results indicate that those NSCLC cells harboring an IGFBP-3 unmethylated promoter might receive less benefit from radiotherapy-based therapy than those cells with a hypermethylated promoter.
Figure 1.
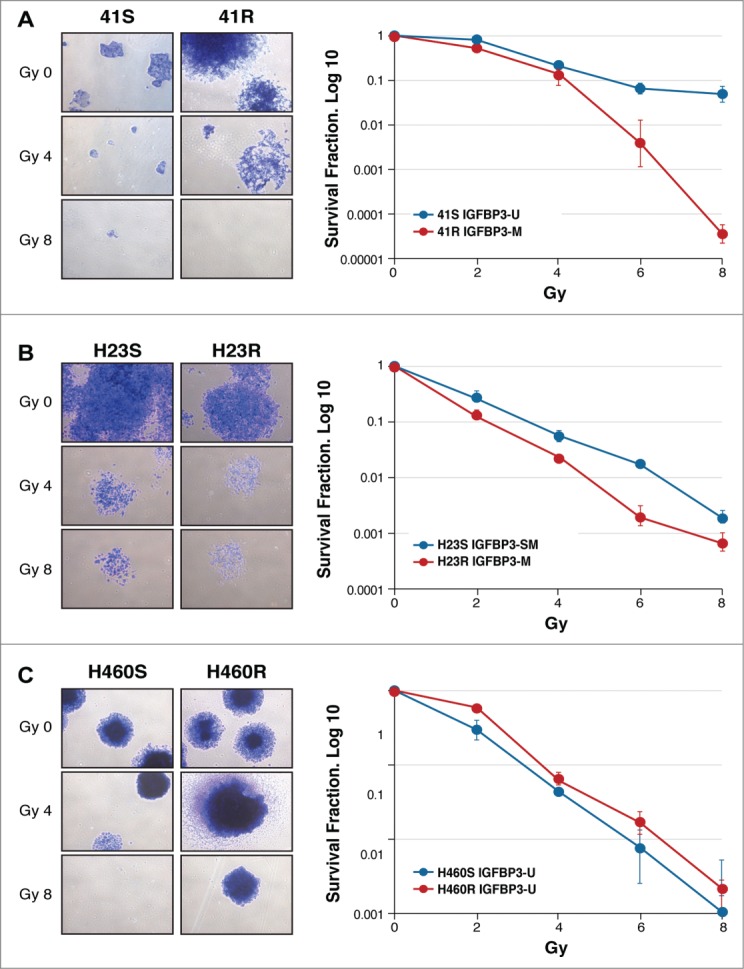
Radiation clonogenic cell survival assays with 3 paired CDDP-sensitive and CDDP-resistant cell lines, 41S/R (A), H23S/R (B) and H460S/R (C). The images are representative of 0, 4 and 8 Gy doses in each paired cell line using a cesium-137 irradiator Mark I30. Individual assays were performed in triplicate and repeated at least twice. The survival fraction (SF) was calculated by the following formula: SF = (number of colonies formed/number of cells seeded) x plating efficiency of the control group, in which plating efficiency was calculated as the ratio between the colonies observed and the number of cells plated. Dose-response clonogenic survival curves were plotted on a log-linear scale.
Radiotherapy is believed to function either by direct ionization or indirectly by DNA interaction of radicals formed by water ionization.21 inducing DNA damage and mitochondrial production of ROS and RNS. The oxidative stress generated results in a complex cellular response, such as the activation of cell signaling, the inhibition of certain proteins and the increased metabolism of chemical compounds in the cells. All these events involve genetic and epigenetic alterations that lead the biological balance toward either death or survival of the treated malignant cells.22 Therefore, to investigate whether the epigenetic regulation of the axis IGFBP-3/IGFIR/AKT is a mechanism that influences radiosensitivity in tumor cells, we studied the changes in the IGFBP-3 expression and promoter methylation levels associated with radiotherapy treatment in the 41S/R and H460S/R paired cell lines. We also used the additional lung cancer cell line H1299, purchased from the American Type Culture Collection (ATCC), because these cells harbor a hypermethylated promoter for the IGFBP-3 gene and present an elevated IC50 (6 ug/ml) to cisplatin.5 We first observed a reactivation of IGFBP-3 expression in a dose-response effect after radiotherapy treatment in the resistant cell line 41R, increasing from 102 (2 Gy) to 107.8 times (at 8 Gy) in comparison with the sensitive cell line 41S treated at the same doses, in which there were no noticeable changes in IGFBP-3 expression (Fig. 2A), probably due to the high basal levels of IGFBP-3 expression in this cell line. As expected, there were no significant changes between the negative control cell lines H460S and H460R (Fig. 2B). These results are in agreement with previous findings that the hydroxyl radicals generated by radiation induce gene expression in mammalian cells.23 The increase in IGFBP-3 expression in the 41R cells is probably mediated by the decrease in IGFBP-3 promoter hypermethylation observed in this cell line after ionizing radiation. IGFBP-3 promoter hypermethylation decreases by approximately 30% at 2 and 4 Gy and 63% at 6 Gy compared with the non radiated cells (0Gy) (Fig. 2C). As expected, we did not find any decrease in the methylation levels in the cell line 41S after radiation therapy, probably because those cells harbor at baseline a completely unmethylated promoter for IGFBP-312 (Fig. 2C).
Figure 2.
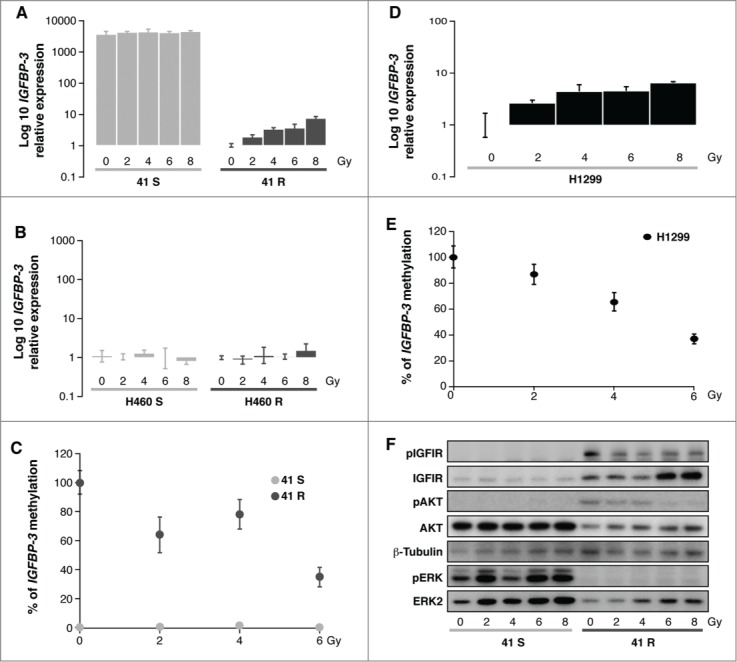
(A, B, D) Quantification of IGFBP-3 expression levels in 41S/R, H460S/R and H1299 cells 72 h after IR treatment using the resistant untreated controls (0Gy) as calibrators. (C, E) Methylation levels of IGFBP-3 in 41S, 41R and H1299 cells 72 h after irradiation.. The calculation of the IGFBP-3 gene to β-actin ratios was based on the fluorescence emission intensity values for both genes at 0, 2, 4 and 6 Gy. The data were normalized to each untreated control, set to 100%, and represent the mean ± standard deviation of at least 3 independent experiments performed in triplicate at each concentration for every cell line analyzed. (F) Activation of the ERK and IGFIR/AKT axes 72 h after radiation in the 41S and 41R cell lines at 5 IR doses.
These results are not specific for the paired cell lines 41S and 41R because the results obtained from the additional cell line H1299 also agree with these observations; there was an increase in the IGFBP-3 mRNA levels that is a dose-response effect after radiotherapy treatment, increasing from 102.6 at 2 Gy to 106.4 times at 8 Gy in the H1299 cells (Fig. 2D). These results are concomitant with a reduction in the methylation levels of the IGFBP-3 promoter after radiation exposure, reaching similar values to that observed in the 41R cells (75% and 63%, respectively) (Fig. 2E). In the 3 experimental groups treated at the higher concentration (8 Gy), we could not obtain a DNA template of sufficient quality to perform the qRT-MSP analysis. Supporting our observation, previous studies have shown that radiotherapy causes global hypomethylation in vitro and in vivo, possibly due to a decreased expression of epigenetic regulators12,24; we did not observe changes in the DNMT3B expression levels between the studied cell lines (data not shown), but in further studies, it would be necessary to get insight into the specific mechanisms responsible for IGFBP-3 demethylation after radiotherapy treatment in cisplatin chemotherapy-resistant cells.
We then analyzed whether the observed changes in IGFBP-3 expression and promoter methylation were linked to modifications affecting the activation of the IGFIR/AKT cellular pathway, which could explain the observed differential sensitivity to ionizing radiation. The results confirmed our previously published results, showing at 0 Gy the phosphorylation of both the IGFIR and AKT proteins in the resistant cell 41R compared with the sensitive cell line 41S at the same dose (Fig. 2F).4 The exposure to ionizing radiation decreased the IGFIR and AKT phosphorylation levels in a dose response manner from 0 Gy to 8 Gy in the 41R cells. These results were concomitant with the dose-response increase in IGFBP-3 expression we observed in the resistant cell line 41R at the same doses. This outcome indicates that the re-expression of IGFBP-3 through promoter demethylation is probably mediating the decrease in the activation of the survival pathway IGF-IR/AKT by sequestering the IGF-I factor, a mechanism that we have already described in those cells.4 The re-silencing of this survival pathway in cisplatin-resistant cells that initially harbored a methylated promoter for IGFBP-3 could result in a gain in sensitivity to radiotherapy treatment. These findings open the door to exploring radiotherapy as an alternative treatment to cisplatin in those tumors that present the hypermethylated promoter of IGFBP-3.
As expected, IR-treatment induced a dose-response increase in the expression of ERK1/2 levels in both sensitive cells and cisplatin-resistant cells. In fact, MAPK signaling can be stimulated by treatment with IR in tumor cells,25,26 probably promoting the activation of the ERBB family receptor, which in turn increases the activity of downstream molecules in the RAS pathway such as RAF-1, MEK 1/2, ERK1/2 and p90rsk.27 The activation of the ERK pathway can either protect or enhance radiation sensitivity, depending on the cell type analyzed.28-30 Our data indicate that this survival pathway is activated by radiotherapy treatment in IGFBP-3 unmethylated 41S cells alone, and although the synthesis of the ERK protein is increased in 41R cells, its activation is inhibited. Therefore, the radioprotection observed in the 41S cells after radiotherapy exposure might be due to the activation of the ERK signaling pathway, as previously reported in the DU145 and A431 human cancer cell lines.31,32 The ERK pathway activation observed in the 41S cells could also be secondarily regulated by the K-RaS/p38 pathway, given it has been proposed that a sublethal dose of radiation can enhance the metastatic potential of cancer cells via the K-Ras pathway.33 These results indicate the possibility of alternative treatments with specific MERK inhibitors such as AZD6244, which enhance the radiation responsiveness of diverse tumor types, including lung and colorectal tumors.30
Primary tumor data and discussion
We next explored the IGFBP-3 methylation effect on overall survival (OS) in a population of 40 patients with NSCLC who received a chemotherapy schedule based on cisplatin or carboplatin with or without concomitant radiotherapy (Figs. 3A, B and Table 1). We also included an external group of 36 patients with NSCLC who did not receive any therapy after surgery, whose results were published previously.4,5 The NSCLC samples were separated into 2 groups based on their IGFBP-3 methylation levels; patients with methylation levels equal to those of the negative control group were considered unmethylated (Fig. 3B). We then analyzed the patients’ responses to radiotherapy and platinum-based treatments in terms of methylation levels. The survival functions were plotted using the Kaplan-Meier estimator and were compared using log-rank under 3 conditions (Fig. 3C). We found a statistically significant association (p = .03) between OS and evidence of IGFBP-3 methylation. Twenty-6 of the 40 patients (65%), harbored an unmethylated promoter and, as expected, approximately, 31% of them underwent combined treatment with IR and Chemotherapy compared with the 69%, who underwent a chemotherapy regimen based on cisplatin or carboplatin without radiotherapy. Our results indicate that patients with an unmethylated IGFBP-3 promoter had an OS of 6.57 y when receiving chemotherapy alone; however, when this group of patients also received radiotherapy, their OS decreased by approximately 2.5 y, confirming our experimental data from human cancer cells. This result could be associated with the initial stage at diagnosis, given that patients with locally advanced stages tend to receive radiotherapy; however, when we analyzed the stages of the patients who received radiotherapy, we found no correlation between stage at diagnosis and radiotherapy (p = .329). There were unfortunately no patients with a regimen of radiotherapy alone; therefore, although we observed a trend toward better survival when patients with a methylated promoter received a combined treatment with chemotherapy and radiotherapy, it was not a statistically significant event (data not shown).
Figure 3.
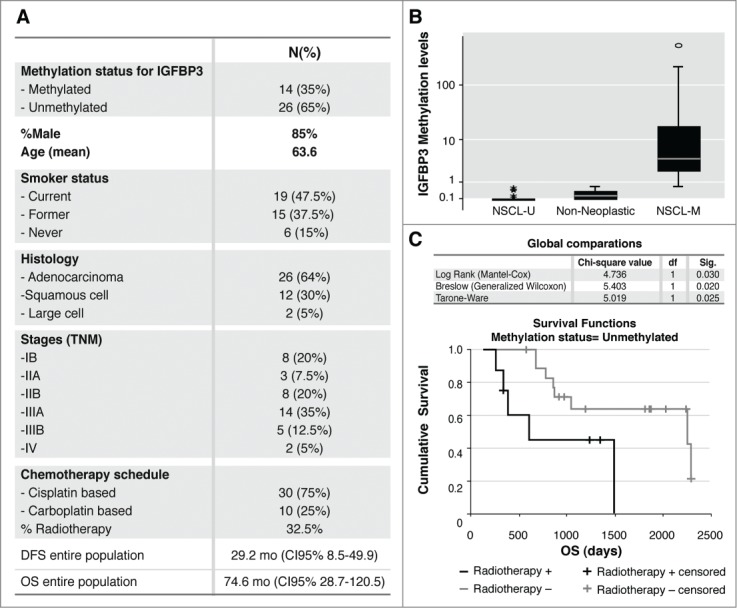
(A) Clinicopathological parameters of the entire population: Methylation status for the IGFBP-3 promoter, gender, age, smoking status, histology, stages, chemo-radiotherapy schemes, disease-free survival (DFS) and overall survival (OS). (B) Box plot for the IGFBP-3/ACTB ratios determined by quantitative methylation-specific PCR (qMSP), in DNA from 40 paraffin-embedded tumors and 10 non-neoplastic lung tissue samples, the obtained ratios were multiplied by 1000 for easier tabulation, as described.15 The values designated as 0.1 and 0.01 are zero values, which cannot be plotted correctly on a log scale. NSCLC-M: the samples considered methylated for IGFBP-3, with higher promoter methylation levels than the controls; NSCLC-U: the samples considered negative, with methylation levels less than or equal to those of the negative control group. (C) Kaplan-Meier. Comparison between IGFBP-3 methylation status (unmethylated) and cumulative survival (days) in 40 patients diagnosed with NSCLC who were treated with chemotherapy (radiotherapy-) or with chemo-radiotherapy (radiotherapy+).
Table 1.
Clinicopathological parameters recorded from 40 NSCLC patients. Adeno, Adenocarcinoma; SCCA, Squamous Cell Carcinoma; M, Methylated; U, Unmethylated; 1, Cisplatin-Vinorelbine; 2, Carboplatin-Vinorelbine; 3, Cisplatin-Gemcitabine; 4, Carboplatin-Paclitaxel
| Patient | Methylation levels (IGFBP3/ACTB)*1000 | Methylation status | Age, y | Sex | Histology | Stage (TNM) | Chemotherapy | Radiotherapy | Start of Chemotherapy | End of Chemotherapy | Last contact | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 496.148 | M | 58 | Female | Adeno | IIIB | 4 | Yes | 2002-06-20 | 2002-08-08 | 2003-07-19 | Exitus |
| 2 | 210.679 | M | 70 | Female | SCCA | IIIA | 1 | No | 2009-01-20 | 2009-04-14 | 2012-01-27 | Alive |
| 3 | 62.698 | M | 70 | Female | SCCA | IB | 1 | No | 2005-08-25 | 2005-09-22 | 2006-07-25 | Exitus |
| 4 | 18.000 | M | 65 | Female | SCCA | IB | 1 | No | 2009-05-26 | 2009-07-14 | 2012-05-08 | Alive |
| 5 | 15.546 | M | 61 | Female | Adeno | IV | 1 | Yes | 2009-02-26 | 2009-05-07 | 2012-05-17 | Alive |
| 6 | 7.550 | M | 73 | Male | Adeno | IIIA | 1 | Yes | 2007-09-17 | 2007-09-25 | 2012-02-09 | Alive |
| 7 | 4.377 | M | 70 | Female | SCCA | IB | 1 | No | 2006-05-11 | 2006-07-27 | 2006-10-05 | Exitus |
| 8 | 3.845 | M | 56 | Female | Adeno | IIIA | 2 | No | 2006-06-29 | 2006-09-07 | 2011-11-08 | Alive |
| 9 | 3.100 | M | 63 | Female | Adeno | IIIA | 1 | Yes | 2009-06-29 | 2009-09-07 | 2012-04-16 | Alive |
| 10 | 2.593 | M | 63 | Female | SCCA | IV | 1 | No | 2005-07-28 | 2005-08-26 | 2005-10-01 | Exitus |
| 11 | 2.088 | M | 68 | Female | Adeno | IIA | 1 | No | 2006-11-16 | 2007-01-25 | 2012-04-18 | Alive |
| 12 | 1.915 | M | 54 | Female | Adeno | IIB | 1 | No | 2009-12-14 | 2010-02-22 | 2012-05-31 | Alive |
| 13 | 1.507 | M | 56 | Female | Adeno | IIB | 1 | No | 2008-12-23 | 2009-03-12 | 2012-02-02 | Alive |
| 14 | 0.670 | M | 72 | Female | Adeno | IIIA | 2 | Yes | 2010-10-14 | 2010-12-02 | 2011-03-01 | Exitus |
| 15 | 0.555 | U | 63 | Female | Adeno | IIB | 1 | Yes | 2009-06-08 | 2009-08-17 | 2012-06-04 | Alive |
| 16 | 0.514 | U | 65 | Female | Large Cell | IIIB | 4 | Yes | 2006-01-02 | 2006-03-06 | 2006-04-18 | Exitus |
| 17 | 0.511 | U | 72 | Female | Large Cell | IIIA | 3 | Yes | 2003-01-30 | 2003-03-26 | 2004-04-24 | Exitus |
| 18 | 0.439 | U | 59 | Female | Large Cell | IIIA | 1 | No | 2005-11-24 | 2006-01-19 | 2012-01-18 | Alive |
| 19 | 0.169 | U | 79 | Female | Adeno | IIIB | 2 | Yes | 2008-04-11 | 2008-05-30 | 2011-09-16 | Alive |
| 20 | 0.077 | U | 69 | Female | Adeno | IIIA | 1 | No | 2010-03-23 | 2010-06-08 | 2011-08-21 | Exitus |
| 21 | 0.001 | U | 70 | Male | Adeno | IB | 1 | No | 2006-06-01 | 2006-07-20 | 2011-05-09 | Alive |
| 22 | 0.001 | U | 56 | Female | SCCA | IIIA | 1 | No | 2006-03-09 | 2006-05-22 | 2007-12-29 | Exitus |
| 23 | 0 | U | 72 | Female | SCCA | IB | 3 | No | 2006-01-19 | 2006-03-30 | 2007-12-13 | Exitus |
| 24 | 0 | U | 65 | Female | Adeno | IIA | 3 | No | 2007-01-26 | 2007-02-02 | 2012-06-14 | Alive |
| 25 | 0 | U | 67 | Female | SCCA | IIIB | 2 | No | 2009-04-16 | 2009-06-30 | 2012-03-30 | Alive |
| 26 | 0 | U | 75 | Male | Adeno | II | 2 | No | 2006-05-18 | 2006-07-27 | 2012-05-16 | Alive |
| 27 | 0 | U | 47 | Male | Adeno | IB | 1 | No | 2007-07-20 | 2007-09-07 | 2012-06-14 | Alive |
| 28 | 0 | U | 52 | Male | Adeno | IIB | 1 | No | 2007-08-06 | 2007-10-08 | 2012-04-20 | Alive |
| 29 | 0 | U | 61 | Female | Adeno | IIIA | 1 | No | 2007-12-03 | 2008-02-18 | 2009-02-12 | Alive |
| 30 | 0 | U | 73 | Female | Adeno | IIIA | 2 | Yes | 2009-10-01 | 2009-11-19 | 2010-03-23 | Alive |
| 31 | 0 | U | 55 | Female | Adeno | IIB | 1 | No | 2010-01-21 | 2010-04-06 | 2012-03-15 | Alive |
| 32 | 0 | U | 64 | Female | SCCA | IIB | 1 | No | 2010-01-22 | 2010-04-01 | 2012-05-21 | Alive |
| 33 | 0 | U | 69 | Male | Adeno | IIIA | 4 | Yes | 2002-06-13 | 2002-08-16 | 2006-05-17 | Exitus |
| 34 | 0 | U | 52 | Female | Adeno | IB | 3 | No | 2002-06-28 | 2002-08-26 | 2008-06-22 | Exitus |
| 35 | 0 | U | 61 | Female | SCCA | IIB | 3 | No | 2004-10-07 | 2004-12-23 | 2007-04-08 | Exitus |
| 36 | 0 | U | 50 | Female | SCCA | IB | 1 | No | 2008-03-17 | 2008-05-26 | 2010-03-06 | Exitus |
| 37 | 0 | U | 67 | Female | SCCA | IIB | 1 | Yes | 2009-05-28 | 2009-08-31 | 2009-11-23 | Exitus |
| 38 | 0 | U | 69 | Female | Adeno | IIIA | 1 | No | 2010-03-23 | 2010-06-08 | 2011-08-21 | Exitus |
| 39 | 0 | U | 57 | Female | Adeno | IIIB | 3 | No | 1999-05-07 | 1999-07-21 | 2005-05-17 | Exitus |
| 40 | 0 | U | 57 | Female | Adeno | IIIA | 4 | Yes | 2002-04-12 | 2002-05-23 | 2002-11-29 | Exitus |
We also interrogated the methylation status of IGFBP-3 in silico using The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/). We found that most probes hybridized within the area located from −600 to −450 bp from the TSS, which is a hot spot for methylation at the CpG island located in the IGFBP-3 promoter.5 When we examined the raw β-value (probe methylation), the histology, the survival factors and the chemotherapy schedule in the TCGA dataset of 149 patients with NSCLC, (32 adenocarcinoma and 117 squamous cell carcinoma), we found that in the absence of methylation patients live longer when receiving chemotherapy as a unique treatment, whereas concomitant treatment with radiotherapy decreases the survival by half (p = .034).
Finally, the identification of a predictor for therapy could reflect biological changes in cancer cells that are independent of any type of therapy used. To evaluate this possibility, we tested IGFBP-3 methylation status in a cohort of patients diagnosed with early-stage NSCLC who underwent an R0 resection without any adjuvant therapy. In this regard, there was no statistical significance (p = .09) in OS according to the methylation status (data not shown). In summary, our results indicate that the unmethylated IGFBP-3 promoter is associated with resistance to radiotherapy in NSCLC. Specifically, the differences in survival suggest that patients harboring an unmethylated IGFBP-3 promoter would not benefit from adding radiotherapy to adjuvant chemotherapy.
The limitations of this study are associated with the small number of patients analyzed, due mainly to different treatment arms, which limited the number of patients in each group; however, our findings are promising given there is currently no DNA methylation marker or marker panel that can predict radiotherapy response.34 Future prospective multicentric studies including additional and larger NSCLC cohorts need to be performed in future. Nevertheless, IGFBP-3 methylation status is worth considering prior to using radiation therapy after surgery for patients with NSCLC.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We gratefully acknowledge Javier Perez for the artwork and J. Siegfried for English grammar corrections.
Funding
Supported by FIS (ISCIII), PI12/00386, PI12/01463, PI11/00949; PI11/00537; PTA2012/7141-I, all supported by FEDER funds. IIC was supported by the “Miguel Servet” program (CP 08/000689).
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/ 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002; 346:92-8; PMID:11784875; http://dx.doi.org/ 10.1056/NEJMoa011954 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida EJ, Chen H, Torres MA, Curran WJ, Liu T. Spectrophotometer and ultrasound evaluation of late toxicity following breast-cancer radiotherapy. Med Phys 2011; 38:5747-55; PMID:21992389; http://dx.doi.org/ 10.1118/1.3633942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cortes-Sempere M, de Miguel MP, Pernia O, Rodriguez C, de Castro Carpeno J, Nistal M, Conde E, Lopez-Rios F, Belda-Iniesta C, Perona R, et al. IGFBP-3 methylation-derived deficiency mediates the resistance to cisplatin through the activation of the IGFIRAkt pathway in non-small cell lung cancer. Oncogene 2013; 32:1274-83; PMID:22543588; http://dx.doi.org/ 10.1038/onc.2012.146 [DOI] [PubMed] [Google Scholar]
- 5.Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, Machado-Pinilla R, Rodriguez-Fanjul V, Manguan-Garcia C, Cejas P, Lopez-Rios F, Paz-Ares L, de CastroCarpeno J, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010; 29:1681-90; PMID:20023704; http://dx.doi.org/ 10.1038/onc.2009.454 [DOI] [PubMed] [Google Scholar]
- 6. Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000; 343:1350-4; PMID:11070098; http://dx.doi.org/ 10.1056/NEJM200011093431901 [DOI] [PubMed] [Google Scholar]
- 7.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012; 13:679-92; PMID:22945394; http://dx.doi.org/ 10.1038/nrg3270 [DOI] [PubMed] [Google Scholar]
- 8.Roossink F, de Jong S, Wisman GB, van der Zee AG, Schuuring E. DNA hypermethylation biomarkers to predict response to cisplatin treatment, radiotherapy or chemoradiation: the present state of art. Cell Oncol (Dordr) 2012; 35:231-41; PMID:22836879; http://dx.doi.org/ 10.1007/s13402-012-0091-7 [DOI] [PubMed] [Google Scholar]
- 9.Isohashi F, Endo H, Mukai M, Inoue T, Inoue M. Insulin-like growth factor stimulation increases radiosensitivity of a pancreatic cancer cell line through endoplasmic reticulum stress under hypoxic conditions. Cancer Sci 2008; 99:2395-401; PMID:19018773; http://dx.doi.org/ 10.1111/j.1349-7006.2008.00970.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, He L, Xi M, Cai M, Shen J, Li Q, Liao Y, Qian D, Feng Z, Zeng Y, et al. Nimotuzumab promotes radiosensitivity of EGFR-overexpression esophageal squamous cell carcinoma cells by upregulating IGFBP-3. J Transl Med 2012; 10:249; PMID:23232108; http://dx.doi.org/ 10.1186/1479-5876-10-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshino K, Motoyama S, Koyota S, Shibuya K, Usami S, Maruyama K, Saito H, Minamiya Y, Sugiyama T, Ogawa J. IGFBP3 and BAG1 enhance radiation-induced apoptosis in squamous esophageal cancer cells. Biochem Biophys Res Commun 2011; 404:1070-5; PMID:21195059; http://dx.doi.org/ 10.1016/j.bbrc.2010.12.115 [DOI] [PubMed] [Google Scholar]
- 12.Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics 2013; 8:839-48; PMID:23880508; http://dx.doi.org/ 10.4161/epi.25498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skvortsova I, Skvortsov S, Stasyk T, Raju U, Popper BA, Schiestl B, von Guggenberg E, Neher A, Bonn GK, Huber LA, et al. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics 2008; 8:4521-33; PMID:18821526; http://dx.doi.org/ 10.1002/pmic.200800113 [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Perez I, Murguia JR, Perona R. Cisplatin induces a persistent activation of JNK that is related to cell death. Oncogene 1998; 16:533-40; PMID:9484843; http://dx.doi.org/ 10.1038/sj.onc.1201578 [DOI] [PubMed] [Google Scholar]
- 15.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, Westra WH, Schoenberg M, Zahurak M, Goodman SN, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst 2006; 98:996-1004; PMID: 16849682; http://dx.doi.org/ 10.1093/jnci/djj265 [DOI] [PubMed] [Google Scholar]
- 16.Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 2006; 66:5021-8; PMID:16707423; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-3365 [DOI] [PubMed] [Google Scholar]
- 17.Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics 2009; 1:177-200; PMID:22122642; http://dx.doi.org/ 10.2217/epi.09.14 [DOI] [PubMed] [Google Scholar]
- 18. Ibragimova I, Maradeo M, Dulaimi E, Cairns P. Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2, KDM6A and other chromatin-modifying genes is absent or rare in clear cell RCC. Epigenetics 2013; 8:486-93; PMID:23644518; http://dx.doi.org/ 10.4161/epi.24552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Schutter H, Geeraerts H, Verbeken E, Nuyts S. Promoter methylation of TIMP3 and CDH1 predicts better outcome in head and neck squamous cell carcinoma treated by radiotherapy only. Oncol Rep 2009; 21:507-13; PMID:19148529 [PubMed] [Google Scholar]
- 20.Niyazi M, Schnell O, Suchorska B, Schwarz SB, Ganswindt U, Geisler J, Bartenstein P, Kreth FW, Tonn JC, Eigenbrod S, et al. FET-PET assessed recurrence pattern after radio-chemotherapy in newly diagnosed patients with glioblastoma is influenced by MGMT methylation status. Radiother Oncol 2012; 104:78-82; PMID:22673727; http://dx.doi.org/ 10.1016/j.radonc.2012.04.022 [DOI] [PubMed] [Google Scholar]
- 21. Moeller BJ, Richardson RA, Dewhirst MW. Hypoxia and radiotherapy: opportunities for improved outcomes in cancer treatment. Cancer Metastasis Rev 2007; 26:241-8; PMID:17440683; http://dx.doi.org/ 10.1007/s10555-007-9056-0 [DOI] [PubMed] [Google Scholar]
- 22.Campos AC, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D’Almeida V, Correa M, Jasiulionis MG. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 2007; 9:1111-21; PMID:18084618; http://dx.doi.org/ 10.1593/neo.07712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan CL, Wu Z, Eastman A, Bresnick E. Irradiation-induced expression of O6-methylguanine-DNA methyltransferase in mammalian cells. Cancer Res 1992; 52:1804-9; PMID:1372530 [PubMed] [Google Scholar]
- 24.Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun 2004; 325:39-47; PMID:15522198; http://dx.doi.org/ 10.1016/j.bbrc.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 25.Carter S, Auer KL, Reardon DB, Birrer M, Fisher PB, Valerie K, Schmidt-Ullrich R, Mikkelsen R, Dent P. Inhibition of the mitogen activated protein (MAP) kinase cascade potentiates cell killing by low dose ionizing radiation in A431 human squamous carcinoma cells. Oncogene 1998; 16:2787-96; PMID:9652746; http://dx.doi.org/ 10.1038/sj.onc.1201802 [DOI] [PubMed] [Google Scholar]
- 26.Hagan M, Wang L, Hanley JR, Park JS, Dent P. Ionizing radiation-induced mitogen-activated protein (MAP) kinase activation in DU145 prostate carcinoma cells: MAP kinase inhibition enhances radiation-induced cell killing and G2M-phase arrest. Radiat Res 2000; 153:371-83; PMID:10760996; http://dx.doi.org/ 10.1667/0033-7587(2000)153%5b0371:IRIMAP%5d2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 27.Valerie K, Yacoub A, Hagan MP, Curiel DT, Fisher PB, Grant S, Dent P. Radiation-induced cell signaling: inside-out and outside-in. Mol Cancer Ther 2007; 6:789-801; PMID:17363476; http://dx.doi.org/ 10.1158/1535-7163.MCT-06-0596 [DOI] [PubMed] [Google Scholar]
- 28.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK pathways in radiation responses. Oncogene 2003; 22:5885-96; PMID:12947395; http://dx.doi.org/ 10.1038/sj.onc.1206701 [DOI] [PubMed] [Google Scholar]
- 29.Che J, Lu YW, Sun KK, Feng C, Dong AJ, Jiao Y. Overexpression of TOB1 confers radioprotection to bronchial epithelial cells through the MAPKERK pathway. Oncol Rep 2013; 30:637-42; PMID:23756562 [DOI] [PubMed] [Google Scholar]
- 30.Shannon AM, Telfer BA, Smith PD, Babur M, Logie A, Wilkinson RW, Debray C, Stratford IJ, Williams KJ, Wedge SR. The mitogen-activated proteinextracellular signal-regulated kinase kinase 12 inhibitor AZD6244 (ARRY-142886) enhances the radiation responsiveness of lung and colorectal tumor xenografts. Clin Cancer Res 2009; 15:6619-29; PMID:19843666; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-2958 [DOI] [PubMed] [Google Scholar]
- 31.Grant S, Qiao L, Dent P. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci 2002; 7:d376-89; PMID:11815285; http://dx.doi.org/ 10.2741/grant [DOI] [PubMed] [Google Scholar]
- 32.Qiao L, Yacoub A, McKinstry R, Park JS, Caron R, Fisher PB, Hagan MP, Grant S, Dent P. Pharmocologic inhibitors of the mitogen activated protein kinase cascade have the potential to interact with ionizing radiation exposure to induce cell death in carcinoma cells by multiple mechanisms. Cancer Biol Ther 2002; 1:168-76; PMID:12170777; http://dx.doi.org/ 10.4161/cbt.64 [DOI] [PubMed] [Google Scholar]
- 33.Su WH, Chuang PC, Huang EY, Yang KD. Radiation-induced increase in cell migration and metastatic potential of cervical cancer cells operates via the K-Ras pathway. Am J Pathol 2012; 180:862-71; PMID:22138581; http://dx.doi.org/ 10.1016/j.ajpath.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 34.Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG, Starmans MH, Boutros PC, Vooijs M, van Engeland M, et al. Epigenetics in radiotherapy: Where are we heading? Radiother Oncol 2014; 111:168-77; PMID:24861629; http://dx.doi.org/ 10.1016/j.radonc.2014.05.001 [DOI] [PubMed] [Google Scholar]


