Abstract
Transcriptional regulation of secondary wall biosynthesis in Arabidopsis thaliana has been shown to be mediated by a group of secondary wall NAC master switches, including NST1, NST2, SND1 and VND1 to VND7. It has been shown that VND1 to VND7 regulate secondary wall biosynthesis in vessels, NST1 and NST2 function redundantly in anther endothecium, and SND1 and NST1 are required for secondary wall thickening in fibers of stems. However, it is unknown whether NST2 is involved in regulating secondary wall biosynthesis in fibers of stems. In this report, we demonstrated that similar to SND1, NST2 together with NST1 were highly expressed in interfascicular fibers and xylary fibers but not in vessels of stems. Although simultaneous mutations of SND1 and NST1 have been shown to result in a significant impairment of secondary wall thickening in fibers, a small amount of secondary walls was deposited in fibers during the late stage of stem development. In contrast, simultaneous mutations of SND1, NST1 and NST2 led to a complete loss of secondary wall thickening in fibers. These results demonstrate that NST2 together with SND1 and NST1 regulate secondary wall biosynthesis in fibers of stems.
Keywords: Arabidopsis, NAC, NST2, secondary wall, SWN, transcriptional regulation
Introduction
Secondary walls are mainly deposited in tracheary elements and fibers of vascular plants, and they constitute the bulk of plant lignocellulosic biomass targeted for biofuel production.1 Understanding how secondary walls are constructed will provide knowledge foundation for genetic modification of plant biomass tailored for biofuel production. Secondary walls in dicot species are mainly composed of cellulose, xylan and lignin, and their biosynthesis requires coordinate expression of the biosynthetic genes of these secondary wall components. Molecular and genetic analyses have revealed that the coordinate activation of secondary wall biosynthetic genes is mediated by a transcriptional network.2 In this network, the top-level master switches, secondary wall NACs (SWNs), act together with the second-level MYB master switches to turn on the expression of downstream transcription factors and secondary wall biosynthetic genes. SWNs and secondary wall MYB master switches have been functionally characterized in a number of vascular plants and available evidence indicates that their roles in regulating secondary wall biosynthesis are evolutionarily conserved.2-8
In Arabidopsis thaliana, 10 SWNs, including NST1, NST2, SND1 (also called NST3) and VND1 to VND7, have been shown to regulate secondary wall biosynthesis in various secondary wall-forming cell types. VND1 to VND7 are vessel-specific SWNs and their dominant repression causes a loss of secondary wall thickening in vessels.9,10 NST1 and NST2 regulate secondary wall thickening in anther endothecium as their simultaneous mutations lead to a loss of secondary wall thickening in anther endothecium and an anther dehiscence defect.11 SND1 and NST1 have been shown to control secondary wall thickening in interfascicular fibers and xylary fibers of stems12-14 and in the valve endocarp layer and vascular bundles of siliques.15 It is currently unknown whether NST2 plays any role in regulating secondary wall biosynthesis in fibers of stems.
During our study of gene expression of secondary wall NACs in various tissues, we found that NST2 showed an expression profile similar to those of SND1 and NST1, i.e., they are most highly expressed in stems than other organs, which prompted us to investigate a role of NST2 in regulating secondary wall biosynthesis in stems. In this report, we show that NST2 together with NST1 exhibit fiber-specific expression in stems. We further demonstrate that although simultaneous mutations of SND1 and NST1 result in a significant loss of secondary wall thickening in fibers, simultaneous mutations of SND1, NST1 and NST2 are required to achieve a complete loss of secondary wall thickening in fibers. Our results provide genetic evidence demonstrating that NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of stems.
Results and Discussion
The ten Arabidopsis SWNs are phylogenetically grouped into 2 subgroups, SND1, NST1 and NST2 being in one and VND1 to VND7 in another (Fig. 1A). Quantitative PCR analysis showed that similar to SND1 and NST1, NST2 was predominantly expressed in stem internodes near cessation of elongation and non-elongating internodes (Fig. 1B) in which metaxylem and interfascicular fibers undergo massive secondary wall deposition.16 Further expression analysis in microdissected cells of stems revealed that NST2 was highly expressed in both interfascicular fibers and xylem cells with a greater level in interfascicular fibers, a pattern resembling those of SND1 and NST1 (Fig. 1C). These findings raise the possibility that NST2 might be involved in regulating secondary wall biosynthesis in fibers as are SND1 and NST1.
Figure 1.
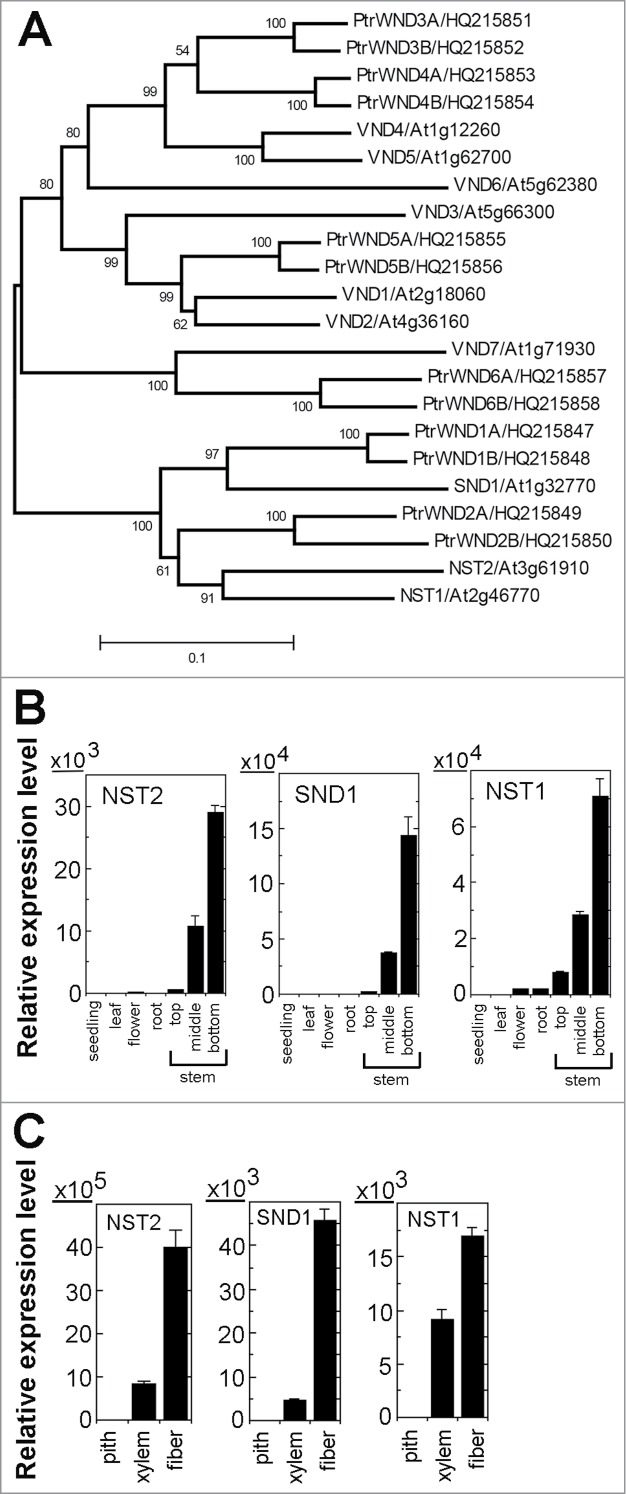
Expression analysis of NST2 in comparison with SND1 and NST1 in various organs and cell types. (A) Phylogenetic relationship of secondary wall NAC master switches from Arabidopsis and Populus trichocarpa (Ptr). The phylogenetic tree was constructed with the neighbor-joining algorithm using the neighbor-joining method in MEGA5.2.19 Bootstrap values are shown in percentages at the nodes. The 0.1 scale denotes 10% change. (B) Quantitative PCR analysis showing the predominant expression of NST2, SND1 and NST1 in stems. The expression of each gene in seedlings was set to 1. (C) Quantitative PCR analysis showing the expression of NST2, SND1 and NST1 in both interfascicular fibers and xylem cells isolated from Arabidopsis inflorescence stems. The expression of each gene in pith cells was set to 1. Error bars denote SE of 3 biological replicates.
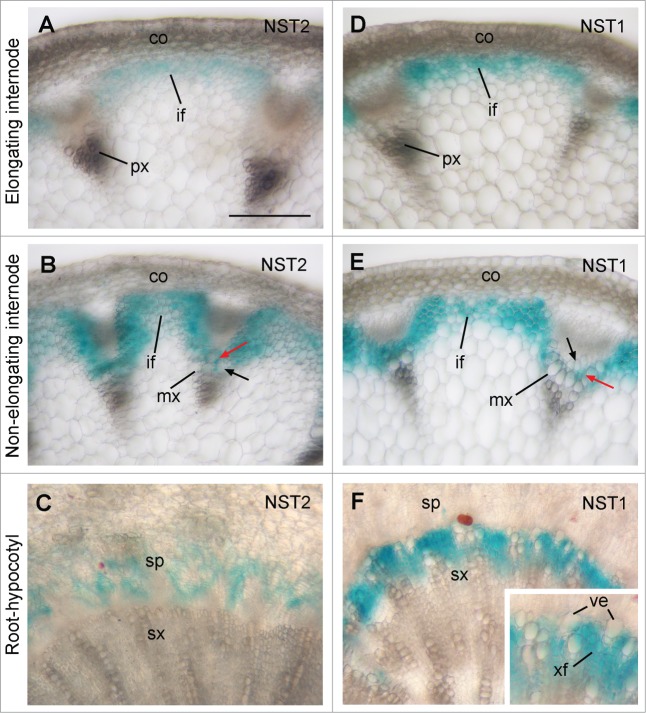
A previous expression analysis using NST2 promoter-GUS reporter gene showed little GUS signal in stems,11 indicating that the promoter sequence used may not contain all the cis-elements required for the proper expression of the endogenous NST2 gene since some cis-elements for certain genes may be located intragenically.17 To further ascertain the cell-type specific expression pattern of NST2 observed by quantitative PCR analyses, we employed the entire NST2 gene sequence, including a 3-kb 5′ upstream sequence, the entire exon and intron region, and a 2-kb 3′ downstream sequence, to perform GUS reporter gene expression analysis. Examination of GUS expression in transgenic plants showed that the GUS staining was only present in interfascicular fibers but not in vessels of the protoxylem in elongating internodes of stems (Fig. 2A). In non-elongating internodes where secondary wall deposition is evident in interfascicular fibers and in vessels and xylary fibers of the metaxylem, the GUS staining was prominent in interfascicular fibers and xylary fibers but not in vessels (Fig. 2B). These results demonstrate that NST2 is expressed in both interfascicular fibers and xylary fibers in stems, implicating its possible role in regulating secondary wall biosynthesis in these cell types. In the root-hypocotyl region where extensive secondary growth occurrs, the GUS staining was seen in the secondary phloem but not in the secondary xylem (Fig. 2C). Since fiber cells are present in the secondary phloem of the root-hypocotyl region, it is possible that NST2 is involved in regulating secondary wall biosynthesis in phloem fibers in the root-hypocotyl region.
Figure 2.
Fiber-specific expression of NST2 and NST1 in Arabidopsis stems and root-hypocotyl region. The NST2 and NST1 genes were fused with the GUS reporter gene to generate the expression constructs, which were transformed into Arabidopsis plants. The first generation of transgenic plants was used for examination of GUS activity (shown as blue). (A and D) Cross sections of elongating internodes showing specific expression of NST2-GUS (A) and NST1-GUS (D) in developing interfascicular fibers. (B and E) Cross sections of non-elongating internodes showing specific expression of NST2-GUS (B) and NST1-GUS (E) in interfascicular fibers and xylary fibers (red arrows) but not in vessels (black arrows). (C and F) Cross sections of the root-hypocotyl region showing expression of NST2-GUS in the secondary phloem (C) and NST1-GUS in the secondary xylem (F). The inset in (F) is an enlarged portion of the secondary xylem showing GUS staining in xylary fibers but not in vessels. co, cortex; if, interfascicular fiber; mx, metaxylem; px, protoxylem; sx, secondary xylem; ve, vessel; xf, xylary fiber. Bar in (A) = 145 μm for (A) to (F).
NST1 was previously reported to be expressed in both fibers and protoxylem vessels in stems based on the NST1 promoter-GUS reporter gene expression analysis, which led to the suggestion that NST1 regulates secondary wall thickening in both fibers and vessels.11,14 However, simultaneous mutations of SND1 and NST1 only affected secondary wall thickening in fiber cells but not that in vessels. Therefore, it is important to discern whether NST1 is expressed in both fibers and vessels. GUS reporter gene expression analysis using the entire NST1 gene sequence, including a 3-kb 5′ upstream sequence, the entire exon and intron region, and a 2-kb 3′ downstream sequence, revealed that NST1 was specifically expressed in interfascicular fibers and xylary fibers but not in vessels in stems (Fig. 2D and E). In the root-hypocotyl region, NST1 expression was prominent in developing xylary fibers but not in vessels in the secondary xylem (Fig. 2F). These results demonstrate that similar to SND1 [12] and NST2 (Fig. 2A and B), NST1 is specifically expressed in fiber cells in stems, which is consistent with the observation that the snd1 nst1 double mutant only showed defective secondary wall thickening in fibers. It also indicates that like that of NST2, the promoter sequence of NST1 used in the previous studies [11,14] does not contain all the cis-elements required for its proper expression. It’d be interesting to find out what cis-elements are responsible for fiber-specific expression of NST1 and NST2.
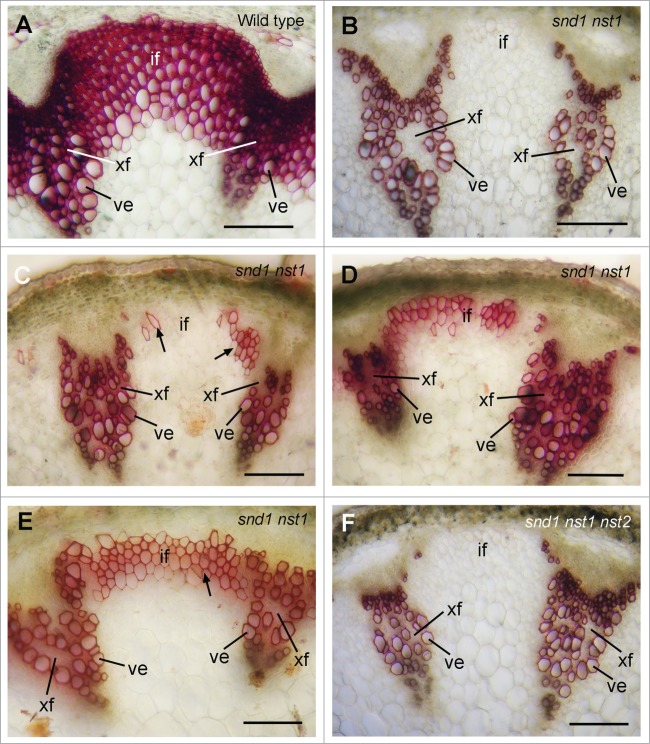
We next investigated a role of NST2 in regulating secondary wall biosynthesis in fibers of stems. Single mutation of SND1, NST1 or NST3 does not cause apparent defects in secondary wall thickening.11,13,14 The snd1 nst1 double mutant had a pendent stem phenotype (Fig. 3) due to a loss of secondary wall thickening in both interfascicular fibers and xylary fibers in stems.13,14 Closer examination of stems revealed that although secondary wall thickening was not seen in interfascicular fibers and xylary fibers of 7-week-old snd1 nst1 double mutant (Fig. 4B),13,14 various degrees of secondary wall thickening was observed in interfascicular fibers of 9-week-old snd1 nst1 mutant plants (Figs. 4C–E). It was noted that patches of interfascicular fiber cells had secondary walls albeit with a much lesser degree of thickening compared with the massive secondary wall thickening in the fibers of wild-type stems (Fig. 4A). This finding suggests that another SWN may play a redundant role in regulating the secondary wall thickening of interfascicular fibers. To find out whether NST2, which is highly expressed in fiber cells as shown in Figure 2, plays such a role, we created snd1 nst1 nst2 triple mutant. The mutant plants exhibited the same pendent stem phenotype as the snd1 nst1 double mutant (Fig. 3A). Examination of stems of 9-week-old snd1 nst1 nst2 triple mutant revealed no secondary wall thickening in interfascicular fibers and xylary fibers (Fig. 4F), indicating that NST2 is responsible for the partial deposition of secondary walls in the interfascicular fibers of 9-week-old snd1 nst1 double mutant. In addition, it was noticed that although the snd1 nst1 double mutant was as fertile as the wild type, the triple mutant was sterile (Fig. 3B) due to anther indehiscence caused by the mutations of NST1 and NST2.11
Figure 3.

Plant morphology of the snd1 nst1 double mutant and the snd1 nst1 nst2 triple mutant. Seven-week-old (A) and 9-week-old (B) snd1 nst1 double mutant and snd1 nst1 nst2 triple mutant showing pendent stem phenotype compared with the wild type (WT). The inset in (B) shows the sterile inflorescence of the snd1 nst1 nst2 triple mutant compared with the wild type and the snd1 nst1 double mutant.
Figure 4.
Complete loss of secondary wall deposition in fibers of the snd1 nst1 nst2 triple mutant. Cross sections of stems were stained for lignin (shown as red) with phloroglucinol-HCl. (A) Nine-week-old wild type showing thick lignified walls in interfascicular fibers, xylary fibers and vessels. (B) Seven-week-old snd1 nst1 double mutant showing an absence of lignified walls in interfascicular fibers and xylary fibers. (C–E) Nine-week-old snd1 nst1 double mutant showing various degrees of lignified walls in interfascicular fibers. (F) Nine-week-old snd1 nst1 nst2 triple mutant showing a complete absence of lignified walls in interfascicular fibers and xylary fibers. if, interfascicular fiber; ve, vessel; xf, xylary fiber. Bars = 115 μm.
In summary, we have demonstrated that like SND1, both NST1 and NST2 are specifically expressed in interfascicular fibers and xylary fibers in stems and that simultaneous mutations of all these 3 SWNs are required for a complete loss of secondary wall thickening in fibers. Our findings provide genetic evidence indicating that NST2 together with SND1 and NST1 are involved in regulating secondary wall biosynthesis in fibers. The fact that only a small amount of secondary walls was deposited in the fibers of the snd1 nst1 double mutant during the late stage of stem development indicates that NST2 plays a minor role in the regulation of secondary wall biosynthesis in fibers. However, expression of NST2 driven by the SND1 promoter could rescue the secondary wall defects of the snd1 nst1 double mutant,18 indicating that the endogenous expression level of NST2 is a primary determinant of its functional significance in regulating secondary wall biosynthesis in fibers. It is now apparent that regulation of secondary wall biosynthesis in Arabidopsis inflorescence stems involves all 10 SWNs, vessel-specific VND1 to VND7 and fiber-specific SND1, NST1 and NST2. Their roles in regulating secondary wall biosynthesis are largely interchangeable since it has been demonstrated that expression of any of the 10 SWNs driven by the SND1 promoter is capable of rescuing the secondary wall defects of the snd1 nst1 double mutant.10,18
Materials and Methods
Plant growth conditions
Plants were grown in a greenhouse with supplemental lights under 14-h-light/10-h-dark cycles. Common garden potting soil was used for growing plants with biweekly application of plant fertilizers.
Quantitative PCR analysis
Total RNA was isolated from different organs and cell types with a Qiagen RNA isolation kit (Qiagen). The seedlings used were 5 d old. Leaves and roots were from 6-week-old plants. Stems from 6-week-old plants were divided into top, middle and bottom parts, which represent the rapidly elongating internodes, internodes near cessation of elongation and non-elongating internodes, respectively. Different cell types, including interfascicular fibers, xylem and pith cells, were isolated from stems of 6-week-old plants using PALM microlaser system (PALM Microlaser Technologies). Total RNAs were first converted into first strand cDNAs, which served as templates for real-time quantitative PCR analysis with the QuantiTect SYBR Green PCR kit (Clontech). The PCR primers for SND1 were 5′-actccaagcaaactcgatttctct-3′ and 5′-tacagataaatgaagaagtgggtc-3′; those for NST1 were 5′-gcttaacggacccacatcatattc-3′ and 5′-ttatccactaccattcgacacgtg-3′; those for NST2 were 5′-tcaacaactgccacgtcagcaaag-3′ and 5′-ttatccactaccgttcaacaagtg-3′. The relative expression level was calculated by normalizing the PCR threshold cycle number of each gene with that of the EF1α reference gene. The data were the average of 3 biological replicates.
GUS reporter gene analysis
The NST1 or NST2 gene containing a 3-kb 5′ upstream sequence, the entire coding region, and a 2-kb 3′ downstream sequence was used for gene expression analysis with the GUS reporter gene. The GUS gene was inserted in frame right before the stop codon of these genes, and then cloned into pBI101 (Clontech) to create the GUS reporter constructs. The constructs were transformed into wild-type Arabidopsis (ecotype Columbia) plants by the agrobacterium-mediated transformation to generate the GUS reporter transgenic plants. Inflorescence stems from 6-week-old transgenic plants were examined for GUS activity as described previously [12].
Generation of snd1 nst1 nst2 triple mutant
The T-DNA insertion lines of SND1 (SALK_015495), NST1 (SALK_120377) and NST2 (SALK_022022) were crossed to generate the snd1 nst1 nst2 triple mutant. The homozygous T-DNA insertion mutants were identified by PCR analysis. The bottom internodes of inflorescence stems of 10 plants for each genotype were stained for lignin with phloroglucinol-HCl.
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for genes used in this study are SND1 (At1g32770), NST1 (At2g46770), NST2 (At3g61910).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The T-DNA insertion mutants of SND1 (SALK_015495), NST1 (SALK_120377) and NST2 (SALK_022022) were obtained from Arabidopsis Biological Resource Center.
Funding
This work was funded by the US Department of Agriculture National Institute of Food and Agriculture [AFRI Plant Biology program (#2010–65116–20468)] and the National Science Foundation (ISO-1051900).
References
- 1. Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol 2009; 60:165-82; PMID:19014348; http://dx.doi.org/ 10.1146/annurev.arplant.043008.092125 [DOI] [PubMed] [Google Scholar]
- 2. Zhong R, Lee C, Ye Z-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 2010; 15:625-31; PMID:20833576; http://dx.doi.org/ 10.1016/j.tplants.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 3. Zhong R, McCarthy RL, Lee C, Ye Z-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 2011; 157:1452-68; PMID:21908685; http://dx.doi.org/ 10.1104/pp.111.181354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohtani M, Nishikubo N, Xu B, Yamaguchi M, Mitsuda N, Goué N, Shi F, Ohme-Takagi M, Demura T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J 2011; 67:499-512; PMID:21649762; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04614.x [DOI] [PubMed] [Google Scholar]
- 5. Duval I, Lachance D, Giguère I, Bomal C, Morency MJ, Pelletier G, Boyle B, MacKay JJ, Séguin A. Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J Exp Bot 2014; 65:2319-33; PMID:24713992; http://dx.doi.org/ 10.1093/jxb/eru116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye Z-H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 2011; 52:1856-71; PMID:21908441; http://dx.doi.org/ 10.1093/pcp/pcr123 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida K, Sakamoto S, Kawai T, Kobayashi Y, Sato K, Ichinose Y, Yaoi K, Akiyoshi-Endo M, Sato H, Takamizo T, et al. Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front Plant Sci 2013; 4:383; PMID:24098302; http://dx.doi.org/ 10.3389/fpls.2013.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I, Sampedro J, Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J Exp Bot 2013; 64:1333-43; PMID:23386682; http://dx.doi.org/ 10.1093/jxb/ers394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 2005; 19:1855-60; PMID:16103214; http://dx.doi.org/ 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou J, Zhong R, Ye Z-H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 2014; 9:e105726; PMID:25148240; http://dx.doi.org/ 10.1371/journal.pone.0105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell 2005; 17:2993-3006; PMID:16214898; http://dx.doi.org/ 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhong R, Demura T, Ye Z-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006; 18:3158-70; PMID:17114348; http://dx.doi.org/ 10.1105/tpc.106.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong R, Richardson EA, Ye Z-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007; 225:1603-11; PMID:17333250; http://dx.doi.org/ 10.1007/s00425-007-0498-y [DOI] [PubMed] [Google Scholar]
- 14. Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007; 19:270-80; PMID:17237351; http://dx.doi.org/ 10.1105/tpc.106.047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 2008; 56:768-78; PMID:18657234; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03633.x [DOI] [PubMed] [Google Scholar]
- 16. Ye Z-H, Freshour G, Hahn MG, Burk DH, Zhong R. Vascular development in Arabidopsis. Int Rev Cytol 2002; 220:225-56; PMID:12224550; http://dx.doi.org/ 10.1016/S0074-7696(02)20007-8 [DOI] [PubMed] [Google Scholar]
- 17. Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 1997; 9:355-65; PMID:9090880; http://dx.doi.org/ 10.1105/tpc.9.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong R, Lee C, Ye Z-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 2010; 3:1087-103; PMID:20935069; http://dx.doi.org/ 10.1093/mp/ssq062 [DOI] [PubMed] [Google Scholar]
- 19. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-39; PMID:21546353; http://dx.doi.org/ 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]