Abstract
There is growing interest in the fast and robust engineering of protein pH-sensitivity that aims to reduce binding at acidic pH, compared to neutral pH. Here, we describe a novel strategy for the incorporation of pH-sensitive antigen binding functions into antibody variable domains using combinatorial histidine scanning libraries and yeast surface display. The strategy allows simultaneous screening for both, high affinity binding at pH 7.4 and pH-sensitivity, and excludes conventional negative selection steps. As proof of concept, we applied this strategy to incorporate pH-dependent antigen binding into the complementary-determining regions of adalimumab. After 3 consecutive rounds of separate heavy and light chain library screening, pH-sensitive variants could be isolated. Heavy and light chain mutations were combined, resulting in 3 full-length antibody variants that revealed sharp, reversible pH-dependent binding profiles. Dissociation rate constants at pH 6.0 increased 230- to 780-fold, while high affinity binding at pH 7.4 in the sub-nanomolar range was retained. Furthermore, binding to huFcRn and thermal stability were not affected by histidine substitutions. Overall, this study emphasizes a generalizable strategy for engineering pH-switch functions potentially applicable to a variety of antibodies and further proteins-based therapeutics.
Keywords: Yeast surface display, Fab display, antibody engineering, protein engineering, pH-dependent antigen binding, pH-switch, adalimumab
Introduction
In recent years, engineering of pH-sensitive binding into proteins has been of increasing interest due to a large area of potential applications, which range from half-life extension of therapeutic proteins to pH-switchable affinity chromatography materials.1–5 Naturally occurring pH-sensitive protein-protein interactions were shown to fine-tune the regulation of biological functions.6–12 Both the underlying mechanism of pH-sensitivity and the biological outcomes gave guidance for current protein engineering approaches.13 Several studies about ligand-receptor interactions have elucidated how naturally occurring pH-sensitive interactions can modulate the cellular fate of receptors and their cargoes. Following receptor binding of the ligand, the complex becomes internalized and is trafficked through the endosomal pathway. The rescue of the internalized receptor and lysosomal degradation of the ligand can be mediated by the pH-sensitivity of the receptor-ligand interaction. When the complex approaches the late endosomal compartment, a significant difference in pH between the extracellular environment (pH 7.4) and the acidified endosomal compartment (pH ∼6) allows the release of the ligand. As a consequence, released ligands can enter the degradative pathway whereas the free receptor is recycled to the cell surface.14,15 Antibody capture during neonatal Fc receptor (FcRn)-mediated antibody recycling is another well-known example of a pH-switch. The pH-drop in the acidified endosome allows FcRn to capture the antibody, and the FcRn-antibody complex then traffics back to the cell surface where neutral pH induces complex dissociation and release of the antibody to the extracellular space.16-18
As recently reviewed by Igawa and colleagues,19 several studies have proven that incorporation of pH-sensitive antigen binding can result in improved function of engineered antibodies or growth hormones in vivo.1,3,4 Antibodies against interleukin 6 (IL6), interleukin 6-receptor (IL6-R) and against proprotein convertase subtilisin kexin type 9 (PCSK9) were successfully engineered to retain high affinity target-binding at pH 7.4 and to show decreased binding at acidic pH (pH 4.5–6.0). When entering the endosomal pathway, pH-dependent antigen binding allowed dissociation of the antibody-antigen complex in the acidified endosome (∼pH 6.0) and FcRn-mediated recycling of free antibody.1,3,20 The re-use of IL6-R and PCSK9 antibodies resulted in enhanced antigen clearance that may enable less frequent or lower antibody dosing.1,3
For most of the investigated pH-sensitive interactions, pH-dependent binding relies on the presence of ionizable histidines that mediate structural transitions in binding or folding of the interacting protein.7,8,16,18 Alterations of electrostatic interactions that are induced upon histidine protonation at lower pH-values can lead to decreased binding affinity. pH-sensitivity not only depends on the environmental pH, but also on pKa-changes of histidine residues that can occur due to changes of accessibility of polar solvent or spatial proximity to neighboring proton-donating groups.21-24 Murthaugh and colleagues demonstrated that the extent of pH-sensitivity over a modest pH-drop, e.g., from pH 7.4 to pH 6.0, depends on the number of histidines as well as the magnitude of pKa-changes.25
So far, various protein engineering approaches have been described that all aimed to incorporate pH-sensitivity into proteins by using different strategies of histidine substitution. For example, rational design guided by structural modeling of the receptor interaction site of granulocyte colony-stimulating factor (GCSF) allowed for the identification of mutational sites that upon histidine substitution lead to a pH-sensitive variant.4
Apart from rational interface design, histidine scanning was successfully applied by systematically and comprehensively replacing residues with histidine and identifying variants with advantageous substitutions in terms of pH-sensitivity.1-3,26 The assessment of individual positions for histidine substitutions that can mediate pH-sensitivity in proteins is limited by time-consuming and cost-intensive experimental investigation and in addition does not account for synergistic effects. Moreover, guidance by structural information is available for only a few proteins of interest. Although successful studies showed that protein engineering in regard of pH-sensitivity could be addressed by computational design, the complexity of binding or multiple-proton linked events makes it difficult to robustly predict exact binding functionalities.27,28 Screening of combinatorial libraries with suitable high-throughput technologies, e.g., yeast surface display (YSD)29 or phage display, has been proven to be an efficient strategy for engineering pH-sensitive binding in a variety of different protein scaffolds (anti-RNAse A VHH,25 scaffold protein Sso7d,2 Fcab molecules30 and most recently protein A5). YSD combines the advanced eukaryotic expression machinery with the applicability of high-throughput fluorescence activated cell sorting (FACS). Using this technology, a pH-sensitive Sso7d scaffold protein variant and several Fcab variants were isolated from error-prone or parsimonious mutagenesis libraries by multiple alternating screening steps at pH 7.4 and pH 6.0.2,30 Interestingly, selected Sso7d and Fcab variants hardly contained any new histidine substitutions. Therefore it was suggested that exchanges other than histidines contributed to the occurrence of pH-sensitivity. It was assumed that pH-sensitivity is caused by alterations in the microenvironments of existing histidines that were already present in the parental clones.2,30
Here we describe a generic strategy for the engineering of antibody heavy and light chain variable domains in terms of reversible pH-sensitive antigen binding. As proof of concept we generated and screened separate heavy and light chain combinatorial histidine substitution libraries that are based on the VH and VL part of adalimumab, which targets tumor necrosis factor (TNF). Adalimumab is a human IgG antibody that is effective in the treatment of various chronic inflammatory diseases, e.g., rheumatoid arthritis or Crohn's disease.31,32 TNF is known to play an important role in the regulation of pathogenic events.31 VH and VL variants that showed high affinity at pH 7.4 and a significant increase in antigen-release at pH 6.0 were isolated after 3 successive screening rounds. Overall, this study emphasizes the application of yeast display for pH-sensitivity engineering.
Results
Construction of libraries
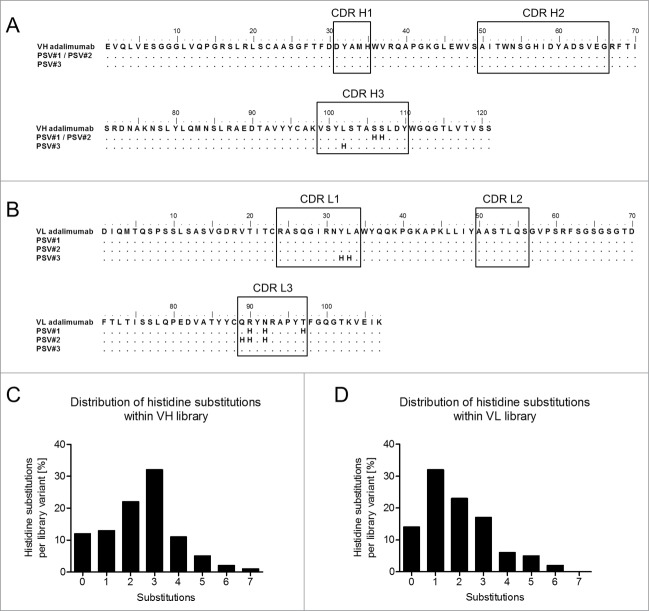
Combinatorial histidine substitution libraries were derived from parental adalimumab sequences by using pre-assembled trinucleotide building blocks during sequence synthesis, where at each selected position the occurrence of a histidine codon was adjusted to 10%. Sampling of 10% histidine codons at each position was chosen to achieve 3 mutations per library variant on average aiming at retained high affinity target binding properties at neutral pH. Two separate libraries were constructed that comprised heavy and light chain variable regions. Mutations were restricted to the complementary-determining regions (CDRs) that included 27 residues of the VL region and 34 residues of the VH region (Fig. 1A and 1B). Sequence analysis of randomly selected variants from both libraries revealed 0–7 and 0–6 histidine substitutions per variant (Fig. 1C and 1D) in addition to the naturally occurring 2 histidines of the VH domain covering all 3 CDRs of VH and VL, respectively. Furthermore, 3 and 1 substitutions per VH or VL regions showed the highest prevalence. Assuming 3 histidine substitutions on average per library member, theoretical diversities were calculated as previously described33 to be approximately 3,000 for the VL library and 10,000 for the VH library. The 2 libraries encoding histidine-doped variable domains were introduced into YSD vectors that allowed for the surface display of adalimumab Fab variants via expression of the heavy chain fused to the Aga2p anchor and coexpression of the light chain.34 Resulting library sizes for the VH library and the VL library of approximately 2 × 106 and 3 × 107 independent transformants were sufficient to cover most of the histidine combinations. Considering the given library sizes, the calculated theoretical numbers of all different combinations of mutational variants carrying 3 histidine substitutions and their prevalence in the libraries, coverages were estimated to be 64-fold and 1700-fold for the VH and VL libraries, respectively.
Figure 1.
Amino acid sequences of adalimumab variable heavy (VH) and light chain (VL) regions and the design of the combinatorial histidine substitution libraries. Amino acid sequence alignments show the VH (A) and VL (B) regions of adalimumab and variants with pH-dependent antigen binding (PSV#1, PSV#2 and PSV#3). CDRs are indicated in black boxes. Distribution of histidines substitutions within CDRs of the VH (C) and VL (D) libraries were calculated from 89 and 80 analyzed sequences, respectively. Genes used for sequencing were also used for the generation of yeast surface display libraries. Mutations with respect to the parental adalimumab sequences are shown with dots indicating identical residues.
Isolation of variants with pH-dependent antigen binding
To discard variants that lost their ability to bind recombinant human (rh)TNF upon histidine substitution, 3 × 106 cells displaying rhTNF binding variants were isolated from each library upon cell staining with labeled rhTNF at pH 7.4 followed by FACS (Fig. S1). Afterwards, a novel selection strategy was applied to isolate pH-sensitive rhTNF-specific variants with high affinity binding at pH 7.4 and reduced affinities at pH 6.0 that relies on the presaturation of antigen binding sites with unlabeled rhTNF at 7.4, followed by re-occupation of antigen binding sites that released the antigen at pH 6.0 with labeled rhTNF (Fig. 2).
Figure 2.
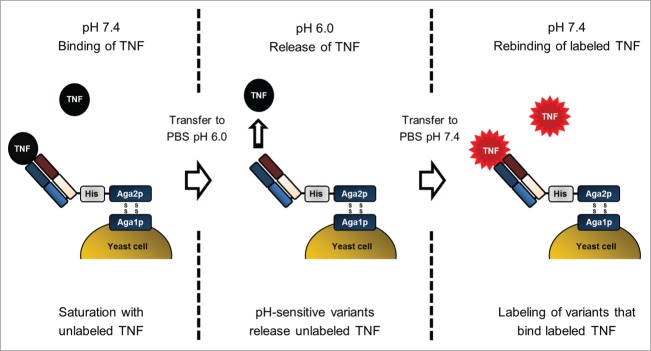
Schematic illustration of the YSD selection strategy for isolation of antibody variants with pH-dependent antigen binding by FACS. The procedure comprises 3 key steps. First, library variants specific for rhTNF were saturated with unlabeled antigen in PBS-1 (pH 7.4). During the second step, pH-sensitive variants release rhTNF after cells were transferred to PBS-2 (pH 6.0). Third, cells were washed and incubated with labeled rhTNF at pH 7.4. Detection of binding signals allows isolation of variants that exhibit reversible pH-dependent antigen binding.
To this end, cells were incubated with unlabeled rhTNF at pH 7.4, which allowed saturation of TNF-specific variants. Afterwards, cells were washed and incubated at pH 6.0, which enabled pH-sensitive variants to release unlabeled rhTNF. As a control, cells were also incubated at pH 7.4 to estimate the fraction of pH-sensitive variants. After incubation at pH 6.0, cells were washed and incubated with labeled rhTNF at pH 7.4. The rebinding of labeled rhTNF allowed discrimination between pH-sensitive variants and variants that were still occupied by unlabeled rhTNF. Three selection rounds were performed to enrich pH-dependent binders exhibiting enhanced binding signals when incubated at pH 6.0 compared to pH 7.4 (Fig. 3). During the third selection round, 2 separate incubation cycles at pH 6.0 were applied (30 min and 10 min). By shortening the incubation time at pH 6.0 from 30 min to 10 min, cell numbers in the gates decreased due to increased off-rate screening stringencies (Fig. 3). In each selection round, approximately 0.5-5 × 105 cells were isolated from both libraries. For validation purposes, the same labeling strategy was also applied to adalimumab wild-type Fab displaying cells, where no pH-dependent binding was observed (Fig. S2). After 3 rounds of screening, single yeast cells were isolated from both screens and subsequently sequenced. From a pool of 38 VH sequence clones, 7 unique variants were identified and 3 were selected (N > 1 ) for further analysis (Fig. S3A). The 98 VL sequences contained 38 unique variants. Of these, 8 individual variants with the highest occurrence (N > 1 ) were chosen (Fig. S3B).
Figure 3.
Screening strategy for enrichment of pH-dependent antibody variants from the yeast displayed (A) VH-library and (B) VL-library using FACS. Yeast cells were incubated with unlabeled rhTNF, transferred to PBS-2 (pH 6.0), followed by double staining with directly or indirectly labeled rhTNF and PE or FITC conjugated anti-Penta-His for simultaneous detection of antigen binding and surface display (upper panels, rounds 1, 2, 3a and 3b). After round 2, library cells were subjected to 2 parallel selections. In round 3, incubation with PBS-2 was done either for 30 min (round 3a) or for 10 min (round 3b). Cells in the sorting gates were isolated, grown and subjected to the next round of selection. Percentages of cells in the gates are indicated. Library displaying cells that served as gating controls were incubated with PBS-1 (pH 7.4) instead of PBS-2 and are shown in the lower panel.
Stability and pH-sensitivity of full length IgG variants
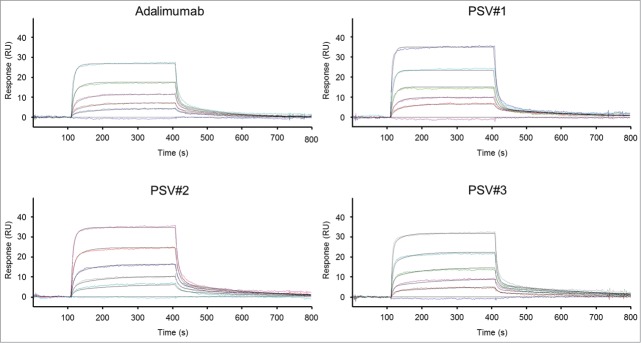
Enriched heavy and light chain variants were subcloned for soluble expression of IgG1 molecules, and all possible 24 combinations of the 3 heavy chain and 8 light chain variants together with 11 variants with parental heavy or light chain pairings were expressed in mammalian cell cultures and profiled for their pH-sensitive binding at pH 7.4 and pH 6.0 via biolayer interferometry (BLI) (data not shown). Three clones (PSV#1, PSV#2 and PSV#3) were selected according to their differently pronounced pH-sensitive binding profiles and subjected to detailed binding kinetic analysis at pH 7.4 and pH 6.0 compared to commercially available adalimumab (Fig. 4). Results indicate single-digit picomolar binding affinity for wild-type adalimumab at pH 7.4, which corresponds to the affinity determined by Kaymakcalan et al.35 (KD: 30.4 pM), considering the KD detection limit of BLI at approximately KD: 100 pM.36 Due to the very slow off-rate of adalimumab, fittings are susceptible to little variances within the blank measurement, and that resulted in an approximately 10-fold enhanced KD due to a 9-fold decreased dissociation rate constant compared to the previously published data. Selected antibody variants PSV#1, PSV#2 and PSV#3 showed approximately 10-, 17- and 24-fold reduced affinities compared to adalimumab, mainly driven by increased off-rates (Fig. 4, left panel; Table 1, Table 2). pH-sensitivity was addressed by measuring dissociation at pH 6.0 after association at pH 7.4 (Fig. 4, right panel; Table 1). All 3 selected variants showed distinct rapid antigen release when dissociation was done at pH 6.0. To determine off-rates of the selected variants, a local partial fitting was applied in which the assumption of complete dissociation within the measured time-frame was made. As adalimumab showed only slow rhTNF release within the measured time-window, accurate Kd-values were determined by using a global fitting (association and dissociation).
Figure 4.
BLI sensorgrams of kinetic analyses of rhTNF binding to immobilized adalimumab and pH-dependent binding variants (PSV#1, PSV#2 and PSV#3). Association with rhTNF at indicated concentrations was measured for 300 s at pH 7.4. Dissociation was performed for 1800 s at pH 7.4 (left panel) or at pH 6.0 (right panel). Dissociation times at pH 6.0 varied between 400 s (PSV#1, PSV#2 and PSV#3) and 900 s (adalimumab). Binding curves (colored lines) were fitted with a 1:1 interaction model (red lines). In contrast to the applied global fitting analysis, binding curves of PSV#1, PSV#2 and PSV#3 were fitted using the local partial model when dissociation was carried out at pH 6.0. Representative experiments from triplicate measurements are shown (except: adalimumab at pH 6.0 in duplicates).
Table 1.
Binding kinetics and Tm of adalimumab and pH-dependent variants binding to rhTNF
| Antibody | pH 7.4 |
pH 6kd (× 10−5) | kd ratio pH 6.0 / pH 7.4 | kd ratio (pH 6.0) vs. adalimumab | Tm (C°) pH 7.4 | Tm (C°) pH 6.0 | ||
|---|---|---|---|---|---|---|---|---|
| KD | ka (× 106) | kd (× 10−5) | ||||||
| pM | M−1s−1 | s−1 | s−1 | |||||
| Adalimumab | 4.6 (± 6.3) | 1.32 (± 0.06) | 0.55 (± 0.94) | 4.81 (± 0.86) | 9 | 1 | 69.8 (± 0.2) | 63.4 (± 0.211) |
| PSV#1 | 46.3 (± 60.5) | 0.67 (± 0.61) | 3.26 (± 4.46) | 754 (± 360) | 231 | 157 | 69.7 (± 0.001) | 61.7 (± 0.001) |
| PSV#2 | 77.3 (± 33) | 1.14 (± 0.41) | 9.35 (± 5.75) | 7340 (± 2430) | 785 | 1527 | 69.7 (± 0.002) | 62.9 (± 0.328) |
| PSV#3 | 112 (± 15.3) | 1.93 (± 0.30) | 21.8 (± 5.82) | 11000 (± 2970) | 505 | 2293 | 67.2 (± 0.001) | 63.2 (± 0.001) |
Binding affinity (KD), association rate (ka) and dissociation rate constants (kd) of adalimumab and the selected pH-dependent binding variants, PSV#1, PSV#2 and PSV#3 at pH 7.4. Dissociation rates were also determined at pH 6.0. Mean values and corresponding SD values are shown, except for adalimumab that was measured at pH 6.0 in duplicates. Tm was determined in triplicates in thermal shift assays at pH 7.4 or pH 6.0.
| KD1 (nM) | ka1 (× 105 M−1s−1) | kd1 (× 10−2 s−1) | KD2 (nM) | ka2 (× 105 M−1s−1) | kd2 (× 10−2 s−1) | |
|---|---|---|---|---|---|---|
| Adalimumab | 8.77 (± 3.88) | 9.06 (± 2.60) | 0.73 (± 0.18) | 467 (± 312) | 3.62 (± 1.49) | 14.1 (± 7.35) |
| PSV#1 | 3.14 (± 1.42) | 17.50 (± 7.75) | 0.48 (± 0.11) | 128 (± 25) | 10.4 (± 3.58) | 12.6 (± 6.47) |
| PSV#2 | 6.61 (± 2.11) | 6.16 (± 1.54) | 0.39 (± 0.07) | 123 (± 7.09) | 8.38 (± 0.56) | 10.3 (± 5.04) |
| PSV#3 | 10.2 (± 0.28) | 5.01 (± 2.17) | 0.51 (± 0.21) | 115 (± 7.07) | 13.2 (± 0.85) | 15.2 (± 0.01) |
The results demonstrate that the dissociation rate constants of PSV#1, PSV#2 and PSV#3 at pH 6.0 were increased by 157-, 1527- and 2293-fold compared to adalimumab. Ratios of kd-values determined at pH 6.0 and pH 7.4 (kd at pH 6.0 / kd at pH 7.4) for PSV#1, PSV#2 and PSV#3 revealed a 231-, 785- and 505-fold enhanced release of antigen, whereas adalimumab showed only a ratio by factor 9 (Table 1). In comparison to adalimumab, this indicates that all engineered variants showed a significantly enhanced pH-dependence with only slightly weakened binding affinities at pH 7.4.
To analyze whether the thermal stability of the variants was affected by the histidine substitutions, thermal shift assay measurements were performed at pH 7.4 and pH 6.0 (Table 1; Fig. S4). All variants exhibited high thermal stabilities at physiological pH with the lowest Tm value for PSV#3 (67.2°C) and the highest Tm for adalimumab (69.8°C) indicating only a minor decreases in thermal stability due to the substitution with histidines. In contrast, low thermal stabilities at pH 6.0 were indicated by values that ranged from 61.7°C to 63.4°C for all 3 variants and wild-type adalimumab, indicating that structural modifications that account for pH-sensitivity seem to have only minor effects on the thermal stability.
Reversible pH-dependent antigen binding
To investigate reversible association (pH 7.4) and dissociation (pH 6.0) of rhTNF by pH-sensitive variants PSV#1, PSV#2 and PSV#3, 2 consecutive binding cycles were performed in which antigen association to all 4 immobilized antibodies was carried out at pH 7.4, followed by dissociation at pH 6.0 for 400 s (Fig. 5). In all sensorgrams, the binding curves were aligned to the baselines that were measured at pH 7.4 after every dissociation step. All engineered antibodies showed rapid release of rhTNF during the first dissociation step at pH 6.0. This is in accordance with respective kd-values determined before (Table 1). PSV#2 and PSV#3 showed a complete loss of antigen during dissociation. Therefore, similar binding signals compared to initial binding signals were acquired throughout the second association step.
Figure 5.
Functional analyses of reversible pH-dependent antigen binding of PSV#1, PSV#2, PSV#3 and adalimumab. Two cycles of association to 13 nM rhTNF at pH 7.4 for 300 s and dissociation at pH 6.0 for 400 s were measured. During the third cycle, dissociation was carried out at pH 7.4. Baseline measurements (40 s) at pH 7.4 were done after every dissociation step at pH 6.0. All association steps were aligned to the baseline. Due to slow release of rhTNF at pH 6.0, only little association was measured for adalimumab during the second and third binding cycle.
Due to incomplete antigen dissociation from PSV#1, the binding signals in subsequent association steps were slightly decreased. In contrast to the selected variants, wild type adalimumab showed only very slow rhTNF release at pH 6.0 resulting in almost no observable binding signals in the following steps. The last dissociation step was performed at pH 7.4 to evaluate whether pH-sensitivity was still functional after 2 pH-dependent antigen binding cycles. Different dissociation rates of the antibodies can be qualitatively estimated from the binding curves, which are in accordance with our previous kinetic measurements at pH 7.4 (Table 1) (PSV#3 > PSV#2 > PSV#1 > adalimumab). All together, these findings suggest that pH-dependent antigen binding is not mediated by irreversible structural changes, but rather occurs due to a specific effect of protonated histidine residues.
Interaction of immobilized huFcRn with soluble antibodies
A previous study showed that engineering of the variable fragment regions of antibodies can affect binding affinities to FcRn and thus modulate pharmacokinetics.37 High affinity interaction of FcRn to the Fc-portion in the acidified endosome allows antibody capture and recycling, thereby contributing to long antibody half-life.16 Therefore it was reasonable to investigate binding kinetics of PSV#1, PSV#2, PSV#3 and adalimumab to immobilized huFcRn at pH 6.0 using surface plasmon resonance (SPR). Binding curves were fitted using a heterogeneous binding model as it was previously shown that FcRn immobilization can cause complex binding when FcRn molecules interact with the Fc-portion at one or both Fc-binding sites.38 As a result, 2 sets of calculated kinetic parameters were obtained, including weaker interactions with sub-micromolar affinities and strong binding affinities in the single- to double-digit nanomolar range that represents functional binding to huFcRn. All measurements revealed very similar binding affinities, indicating that histidine substitutions in the CDRs of PSV#1, PSV#2 and PSV#3 have no effect on binding to huFcRn (Fig. 6).
Figure 6.
Surface plasmon resonance analyses of adalimumab and pH-dependent variants binding to immobilized huFcRn at pH 6.0. Binding curves (colored lines) of representative experiments are shown that were simultaneously fitted (black lines) to a heterogeneous binding model. Kinetic parameters are summarized in the table. Mean values of triplicate measurements are shown for each antibody (except for PSV#3 which was measured in duplicates). Corresponding SD values are shown in parentheses.
Discussion
In this study, we describe a protein engineering approach for incorporation of reversible pH-dependent antigen binding into the CDRs of adalimumab using combinatorial histidine substitution libraries and yeast surface display as platform technology. Previously described yeast library screenings that aimed at isolating pH-sensitive variants employed alternating selection steps at different pH values.2,30 Positive selection of high affinity binding variants at pH 7.4 was followed by a negative selection step to select variants that had released their antigens during incubation at acidic pH. However, negative selection steps possess the inherent disadvantage that decreased binding signals may not exclusively be caused by pH-sensitivity. To address this, we applied a selection strategy in which alternating incubation steps at pH 7.4 and pH 6.0 were used to select in one screening round both high affinity binding at pH 7.4 and pH-sensitivity. As a result, the screening process was significantly accelerated and the sorting efficacy increased by empowering accuracy of cell sorting due to exclusive application of positive selections. In addition, gating controls (incubation at pH 7.4 instead of pH 6.0, Figs. 2 and 3) allowed precise quantification of cell fractions in FACS plots that correspond to pH-sensitive binding.
In this study, mutations were restricted to the CDRs. Separate VH and VL histidine substitution libraries were screened in parallel and mutation rates of 2–3 histidines per variant allowed full coverage of all possible combinations of histidine substitutions within achieved library sizes. We anticipated that this approach would address the synergistic interplay of multiple histidine residues that was emphasized by Murthaugh and colleagues in order to enhance pH-sensitivity.25
After 3 rounds of parallel screening for pH-sensitive heavy and light chain variants, the most abundant sequences were subcloned for soluble expression of bivalent full-length IgG. When variants were paired with their corresponding parental chains and immobilized onto biosensors, no significant pH-dependency could be observed using BLI. This phenomenon was similarly observed by Gera et al. and Traxlmayr et al. who suggested that avidity effects can counteract pH-sensitivity.2,30 The loading on biosensors may result in higher surface densities of the IgG compared to yeast surface-displayed monovalent Fab molecules (approximately 5-6 × 105 copies/cell).39 Therefore, avidity effects may occur due to the multivalency of trimeric TNF during BLI experiments that abrogate pH-sensitive binding.
It was shown that only 2 of the 3 protomers contribute to adalimumab Fab binding.40 Therefore, multivalent interactions could be mediated by the unoccupied binding sites interacting with a neighboring IgG molecule on the densely packed biosensor. Similarly, it was shown that pH-sensitive binding was omitted when monovalent interaction partners were immobilized at higher densities on ELISA plates or BLI biosensors and soluble dimeric molecules were applied.2,30 In the first study, immobilization of monomeric pH-sensitive Fc-binding molecules (Sso7d) on ELISA plates and application of soluble dimeric Fc molecules prevented detection of pH-sensitive binding due to multivalent interactions. This hypotheses was strengthened by the restoration of pH-sensitivity, only when monovalent interactions were permitted by immobilization of dimeric Fc on ELISA plates.2 In addition, Traxlmayr and colleagues observed similar effects with Fcabs that showed pronounced pH-sensitivity when measured with soluble monomeric human epidermal growth factor receptor (HER)2 on yeast cells. However, pH-sensitivity of bivalent Fcabs was abrogated when monomeric HER2 at higher concentrations was immobilized on biosensor tips.30 All these data suggest that pH-switch engineering should take into account avidity effects when thinking about final molecule formats and experimental settings during characterization. Consequently, settings in, for example, BLI should consider the final functional application of the engineered moieties. In relation to our separate screenings of heavy and light chain libraries, increased pH-sensitivities upon initial combination of VH and VL libraries would have opposed these counteracting avidity effects; however, this approach would have been more susceptible to limitations of library coverage. Without estimations to which extent avidity effects may counteract pH-sensitivity in engineered adalimumab variants, we therefore focused on using all combinatorial effects within one chain at a time.
Next, we combined different heavy and light chain histidine substitution variants for investigation of pH-sensitive binding of full length IgGs using BLI experiments. Interestingly, most of the combined variants exhibited pronounced pH-sensitive binding. Hence, combinations effectively addressed additive or synergistic effects that acted superior to opposing avidity effects. Three variants (PSV#1, PSV#2 and PSV#3) were selected according to their binding affinities at pH 7.4 and increased off-rates at pH 6.0 and characterized in BLI experiments. All 3 variants carried ‘paired’ histidine substitutions in their heavy or light chains, where 2 histidines are adjacent to each other (Figs. 1A, 1B, and 7). Noticeably, these paired histidine patterns also occurred across many pH-sensitive anti-RNase A VHH- antibodies that were isolated from a combinatorial histidine scanning library using phage display.25 Murtaugh et al. proposed that the close proximity would lower the apparent pKa-values of corresponding histidines by negative cooperativitiy.25 In the human prolactin (hPRL) it was also suggested that clustered histidines contribute to pH-dependent hPRL binding to its receptor.7 Taken together, the data indicate that these double histidines generally may contribute to the mechanism of pH-sensitive protein-protein interaction.
Figure 7.

Comparative analysis of structural models of TNF surface potentials at pH 7.4 (A) or pH 6.0 (B) and analysis of the positioning of residues that were substituted with His in the adalimumab Fab structure (C-E). The electropotential of trimeric TNF at pH 7.4 (A) or pH 6.0 (B) is shown by blue (positive charge density) and red (negative charge density) surface coloring and opposing heavy and light chains are depicted as cartoons. Gray surface coloring indicates neutral/hydrophobic regions. Detailed view of adalimumab Fab–TNF binding interface with TNF surface potential at pH 6.0 (C-E). Side-chains of residues that were replaced with histidines within adalimumab light chain CDRs (green) or heavy chain CDRs (yellow) are shown as balls and sticks (magenta in LC, red in HC) for (C) PSV#1, (D) PSV#2 and (E) PSV#3. The model was generated with published X-ray structures (PDB 1TNF and 3WD5) using YASARA structure and POVRay and the electropotential was displayed with 60 kJ/mol. Within the light chain, Asp-1 and Ile-2 residues were not displayed to allow better insights into the binding interface.
Residues that have been replaced with histidines were analyzed in structural models of the adalimumab Fab in complex with trimeric TNF. Heavy and light chain CDRs (CDR H1-H3 and CDR L1-L3) form loops that are buried in deep binding pockets of 2 adjacent monomers (Fig. 7).40 In addition, the electrostatic potential of the TNF surface was analyzed (Fig. 7A and 7B). In contrast to the surface potential at pH 7.4, a broadly positively charged surface of TNF at pH 6.0 indicates that repulsion between local positive charge densities on adalimumab variants and the positively charged TNF surface may contribute to increased complex destabilization (Fig. 7B).
The positioning of residues that were substituted in PSV#1, PSV#2 and PSV#3 was analyzed in the adalimumab-TNF complex (Fig. 7C–E). The Kabat numbering scheme was used in all of the following descriptions.41 The positioning of substituted residues suggests that intramolecular changes in the antibodies may also occur that affect pH-sensitivity. In PSV#1 and PSV#2, protonation at S100.bH and S100.cH, may place positive charges in close spatial proximity to protonated histidines of the light chains of PSV#1 (R90H, N92H, T97H) and PSV#2 (Q89H, R90H, N92H) that might induce an aberrant paratope conformation (Fig. 7C and 7D). However, these potential structural changes at pH 6.0 cannot be resolved by differences in the thermal stability measurements (Table 1; Fig. S4). The unique mutational pattern of PSV#3 comprises fewer substitutions (L98H in CDR-H3 and Y32H, L33H in CDR-L3) than PSV#1 and PSV#2. The structural model of the adalimumab Fab-TNF complex indicates that the side chains of Leu-98 and Tyr-32 that are mutated in PSV#3 project deep into binding pockets on TNF (Fig. 7E). Consequently intermolecular repulsion forces between PSV#3 and TNF might contribute to the fastest antigen release at pH 6.0 compared to PSV#1 and PSV#2. In comparison to the enhanced TNF release of the pH-sensitive variants at pH 6, adalimumab showed only a moderate 9-fold increased dissociation rate constant. Interestingly, wild-type adalimumab includes 2 histidine residues in the heavy chain CDRs at position 35 of the CDR-H1 and position 56 of the CDR-H2, the latter contributing to binding interface interactions. In addition, the adalimumab epitope on TNF includes histidine residue at position 73. If these residues were protonated at acidic pH, the resulting complex destabilization could explain the moderately increased dissociation rate. Overall, de-protonated histidines seemed to be very well tolerated, which resulted in retained high affinity binding even though 3 to 5 binding interface positions were mutated.
It is important to note that PSV#1-#3 have 10–24-fold reduced binding affinity at pH7.4 compared to adalimumab, which could abolish the dosage reduction effect of the pH-dependent binding. Carefully designed animals models that mimic human pharmacokinetics of TNF and antibodies are required to answer this question aimed at correlating physicochemical properties and in vivo pharmacokinetics.
To achieve additional cycles of target-binding, reversible antigen binding is critical. For this reason, it was shown that all pH-sensitive antibodies PSV#1, PSV#2 and PSV#3 can reversibly bind to TNF after the release of the antigen was performed at pH 6.0 (Fig. 5).
How fast the antibodies release their antigens also affects the fraction of free antibody that becomes recycled as the duration is limited at which the antibody-antigen complexes stay in acidic endosomal environment. This was also indicated by Devanaboyina and colleagues,20 who showed a correlation between the degrees of pH-sensitivity in anti-IL6 antibodies with the lysosomal localization of antigen. However, the detailed spatiotemporal resolution of subcellular IgG trafficking remains under investigation. Nevertheless, it was proposed that the half-time of IgG trafficking could be estimated due to the overlapping intracellular pathways between the transferrin-receptor and FcRn.42 Trafficking of transferrin-receptors revealed for the endosomal IgG transit a half-life of approximately 450 s.42-44 Notably, PSV#1, PSV#2 and PSV#3 were able to release almost all of the previously bound antigen at pH 6.0 within 400 s (Fig. 5), indicating that most likely all of the antibodies may be recycled in the unbound state when applied in vivo. Furthermore, all 3 variants maintained their high affinity binding to huFcRn at pH 6.0, which may be essential for enhanced antigen clearance in vivo (Fig. 6). An important next step could be the evaluation of the proposed functionalities of PSV#1–3 in PK rodent models.
This study demonstrates that efficient incorporation of pH-sensitive antigen-binding into antibodies can be performed using combinatorial histidine scanning libraries and YSD. To our knowledge, it is the first time that antibody Fab histidine scanning libraries were screened with YSD that comprehensively considered combinatorial effects of multiple histidine substitutions. Furthermore, the screening strategy described here allows simultaneous screening for both high affinity binding and pH-sensitivity and excludes conventional negative selection steps that are used to select for pH-sensitivity. This strategy could be potentially applied to a variety of antibodies and other protein-based therapeutics and may significantly accelerate the engineering of their pH-sensitive binding functionalities.
Materials and Methods
Yeast strain and media
S. cerevisiae EBY100 strain (Invitrogen). YPD medium was composed of 20 g/L dextrose (EMD Millipore), 20 g/L peptone (EMD Millipore), 10 g/L yeast extract (BD), 10 mL/L penstrep (Gibco). Yeast transformants were grown in liquid cultures using minimal SD-base (Clontech) supplemented with 8.56 g/L NaH2PO4, 5.4 g/L Na2HPO4 (both EMD Millipore) and 10 mL/L Penstrep (Gibco), as well as dropout supplements containing all essential reagents except tryptophan or leucin (Clontech). For induction of expression, cells were cultured in minimal SD Gal/Raf (SG) base, completed with dropout supplements and 8.56 g/L NaH2PO4, 5.4 g/L Na2HPO4, 10 mL/L Penstrep (Gibco), as well as 11% w/v polyethylene glycol 8000 (Euroclone). Agar plates were prepared using minimal SD Agar base supplemented with respective dropout mixes.
Construction of combinatorial histidine substitution libraries
YSD vectors that enable covalent cell surface display of heterodimeric antibody Fab fragments were derivatives of pYD1 vectors (Yeast Display Vector Kit Version D, Catalog no. V835–01, Invitrogen). Vectors carrying adalimumab variable heavy and light chain regions genes, which were codon optimized for expression in mammalian cells (pYD-VH-CH1 and pYD-VL-CL), were used. For selection in E. coli, both vectors contained ampicillin marker genes. Selection of transformed yeast cells was enabled through the heavy chain vector encoded TRP1-auxotrophic marker gene or the LEU2 marker gene for selection of cells that carry the corresponding light chain vector. Gene expression was under the control of the galactose-inducible Gal1-promoter. pYD-VH-CH1 encoded for the AGA2 signal peptide, a variable heavy chain region of adalimumab fused to the CH1 IgG1 region followed by AGA2 gene, thereby allowing covalent surface display of the following N-terminal fusion protein: Xpress-(G4S)-VH-CH1-(G4S)-His-(G4S)3-Aga2p. pYD-VL-CL encoded a αMFpp8 leader sequence for soluble light chain secretion45 followed by the VL region of adalimumab and the constant kappa region (IMGT reference: IGKC*01). Preparation of electrocompetent yeast cells and transformation of heavy or light chain containing DNA plasmids was performed according to Suga and Hatakeyama.46 Cell suspension was mixed with 1–2 μg plasmid DNA, transferred to a chilled cuvette (0.2 cm Gene Pulse, Bio-Rad), pulsed at 1.5 kV, 25 μF, 200 Ω using Gene Pulser Xcell (Bio-Rad), and then plated on minimal selective agar-plates. Combinatorial histidine substitution libraries were synthesized by GeneArt® (Life Technologies). Synthesis of the variable regions of the parental adalimumab Fab fragment allowed combinatorial histidine substitutions within all 3 CDRs of the VH and VL regions (CDR H1-H3 and CDR L1-L3). Gene libraries were delivered in pMK plasmids. Library amino acid substitution patterns within CDRs were analyzed by colony sequencing from plates with pMK transformed E.coli.
Gap repair cloning by homologous recombination in yeast was used to generate YSD libraries.47 For this, VH and VL regions were amplified from library genes with the primer combinations Gap_VH_F/ Gap_VH_R and Gap_VL_F/Gap_VL_R, respectively. HPLC purified primers were obtained from MWG biotech and sequences for all primers are listed in Table S1. PCR was performed using Phusion® High-Fidelity DNA polymerase (NEB) and conditions were: 98°C for 30 s, 10 cycles of 98°C for 15 s, 66.3°C (Gap_VH_F/ Gap_VH_R) or 63.4°C (Gap_VL_F/Gap_VL_R) for 15 s and 72°C for 15 s, followed by 15 cycles with 98°C for 15 s and 72°C for 30 s and final elongation for 2 min at 72°C. PCR products were purified using the Wizard® SV Gel and PCR Clean-Up system (Promega). Acceptor vectors, pYD-VH-CH1 and pYD-VL-CL were linearized with StuI/Pstl-HF and Acal/Alel restriction enzymes, respectively. Linearized vectors were isolated by sucrose gradient ultracentrifugation. EBY100 cells were first transformed with either pYD-VH-CH1 or pYD-VL-CL, and then used to generate 2 separate libraries via gap repair cloning according to the protocol of Benatuil and colleagues.47 The first library was generated by electroporation of cells containing parental pYD-VH-CH1 with amplified light chain library and linearized pYD-VL-CL vector. The second library was created by electroporation of cells containing parental pYD-VL-CL with amplified heavy chain library and linearized pYD-VH-CH1 vector. Transformed cells were used for serial dilution plating to calculate the library size.
Screening of libraries
Transformed yeast cells were grown overnight in a shaking incubator at 30°C, 250 rpm in SD-Trp/-Leu medium and then transferred into SG-Trp/-Leu medium with a cell density of 107 cells/mL for yeast cell surface display. Following cell washing and labeling steps were performed with DPBS (Gibco, without CaCl2, MgCl2) that was adjusted to either pH 7.4 (PBS-1) or pH 6.0 (PBS-2) by using 1 M HCl or 1 M NaCl (both EMD Millipore). Cell staining was done on ice with 107 cells/20 μL. rhTNF (R&D systems) was biotinylated using EZ-Link™Sulfo-NHS-Biotin (Thermo Scientific) or directly fluorescence-labeled using DyLight650 NHS-Ester (Thermo Scientific). Unbound NHS-biotin or fluorophores were removed using desalting spin columns (Thermo Scientific) equilibrated with DPBS. To estimate binding saturation concentrations, labeled rhTNF species were titrated on yeast cells displaying adalimumab Fab fragments. Selections were performed using a MoFlo Legacy cell sorter (Beckman Coulter) and Summit 5.3 software. After cultivation for 48 h in SG-Trp/-Leu medium at 20°C, 225 rpm, cells for initial selection of antigen-binding variants within VH and VL libraries were collected. Cells were washed and stained for 30 min with 84 nM TNF-biotin in PBS-1, followed by a washing step with PBS-1 and cell staining using streptavidin R-phycoerythrin conjugates (SAPE, Life Technologies, 1:10 diluted) in PBS-1 for detection of target binding. Cells were washed, resuspended in 500 μL PBS-1 and subjected to FACS.
Another labeling strategy was applied when pH-dependent antigen binding was addressed.
A pH-shift by incubation in PBS-2 was performed, which allowed the pH-sensitive variants to release unlabeled rhTNF and to re-capture labeled rhTNF when staining was done afterwards in PBS-1. Simultaneous antigen labeling and detection of surface display allowed 2-dimensional screenings. In round 1 and 3, cells were saturated with 167 nM unlabeled rhTNF for 30 min in PBS-1, then washed and subjected to 500 μL PBS-2. After 30 min incubation time, cells were harvested and simultaneously stained with 84 nM rhTNF-biotin and anti-Penta-His AlexaFluor647 conjugate (Qiagen, diluted 1:10) in PBS-1 for detection of surface presentation followed by a final wash step before cells were resuspended in 500 μL PBS-1 and sorted. During the second screening round, staining was done with 960 nM DyLight650 conjugated rhTNF and anti-Penta-His FITC conjugate (LSBio, diluted 1:4). During sorting round 3, cells were incubated in PBS-2 for 30 min (round 3a) and 10 min (round 3b). In round 2 and 3, additional gating controls were prepared in which PBS-1 instead of PBS-2 was used during cell staining. After 3 rounds of FACS, single cells were plated on SD-Trp/-Leu agar plates and colonies were transferred in SD-Trp/-Leu. Plasmid DNA was isolated from yeast cells cultures using the RPM yeast plasmid isolation kit (MP Biomedicals) followed by PCR amplification of VH and VL regions with vector specific primer combinations (FW_AGA2/RV_CH1 and FW_app8/RV_CL) and sequencing (MWG Biotech) of the PCR products.
Cloning, expression and purification of antibodies
Subcloning of VH /VL regions from pYD vectors into pTT5 vectors was done using the intracellular machinery in E. coli Top10 that allows homologous recombination of linear plasmid DNA with PCR amplified insert through overlapping DNA regions.48
Variable regions were amplified with primers adding flanking sites that were identical to target vector regions (primer pairs: VH_Luc_f1/ VH_Luc_r1 and VK_Luc_f1/ VK_Luc_r1). Incorporation of PCR fragments into linearized pTT5 mammalian expression vectors (Expresso CMV based system, Lucigen) was achieved after transformation of One Shot® TOP10 chemically competent E. coli cells (Invitrogen).
Cloned constructs were sequence verified and expressed in Expi293 cells after Expifectamin mediated transient transfection following the manufacturer´s instructions (Life Technologies). Media was harvested and secreted antibodies were purified after filtration of supernatant through a Stericup filter device (EMD Millipore) using 5 mL HiTrap MabSelect SuRe columns and ÄKTA Explorer 100 (both GE Healthcare Life Sciences). For size exclusion chromatography (SEC) on ÄKTA Explorer 100, proteins were loaded on HiLoad Superdex 200 pg 26/60 columns (GE Healthcare) using DPBS running buffer. Selected fractions from SEC were pooled, concentrated with 50 kDa Amicon Ultra filter units and filtered through 0.22 μM Millex® Syringe Filter Units (both EMD Millipore). Finally, protein concentrations were determined by UV A280 spectroscopy and antibody purity was determined by gel electrophoresis with 12% NuPAGE BisTris gels and MOPS buffer (Invitrogen), as well as analytical SE-HPLC by using TSKgel SuperSW3000 columns (Sigma-Aldrich).
Biolayer interferometry analysis
BLI-based measurements addressing the pH-sensitive antigen binding were obtained applying the Octet RED system (ForteBio) at 25°C, orbital sensor agitation at 1,000 rpm in 200 μL volume. First, anti-human Fc (AHC) biosensors (ForteBio) were pre-wet for 10 min in DPBS followed by capture of antibodies (10 μg/mL in DPBS) for 120 s. After sensor blocking for 100 s (PBS pH 7.4 supplemented with 1% BSA, albumin Fraction V and 0.1% Tween-20 (both EMD Millipore) and 10 μg/mL biocytin (ThermoScientific) sensors were rinsed in kinetics buffer (KB; DPBS pH 7.4, 0.1% BSA, albumin fraction V and 0.02% Tween®-20) that served as background buffer for 200 s. Initial titration of rhTNF concentrations for BLI experiments revealed heterogeneous binding in the binding curves of the PSV#1, PSV#2 and PSV#3 antibodies at higher antigen concentrations. To overcome these effects, low rhTNF concentrations were used (0.26–2 nM). Association with freshly prepared rhTNF (0.26–2 nM) was monitored for 300 s followed by the dissociation step at pH 7.4 for 1800 s. Alternatively, dissociation was measured at pH 6.0 (KB, adjusted with 1 M HCL) for 400 s for engineered antibodies or for 900 s for the parental antibody. Data from the 0 nM concentration were subtracted from all binding curves, followed by fitting of the sensorgrams using a 1:1 Langmuir binding model (Analysis Software version 8.0). Binding curves of engineered antibodies that were obtained during dissociation at pH 6.0 were analyzed using local partial fitting, whereas curves of parental antibody were analyzed by global fitting. One additional experiment addressed the ability of immobilized antibodies (on AHC biosensors) to reversibly bind rhTNF at pH 7.4, after dissociation at pH 6.0. Subsequent to antibody loading (120 s), sensors were blocked (100 s) and rinsed (100 s), followed by 2 cycles of association with 13 nM rhTNF (pH 7.4, 200 s) and dissociation at pH 6.0 (400 s). During the third cycle, dissociation was carried out at pH 7.4. Baseline measurements (40 s) at pH 7.4 were done after every dissociation step at pH 6.0.
Surface plasmon resonance analysis
SPR was used to determine antibody-huFcRn binding using a Biacore 3000 (GE Healthcare) at 25°C using running and sample buffer (50 mM NaPO4, 150 mM NaCl, 0.05% P20, pH 6.0). First, huFcRn was immobilized (100 to 150 RU) to a CM5 sensor chip via amine coupling by using EDC/NHS chemistry according to the manufactures protocol. Kinetic measurements were performed by injecting serial antibody dilutions (5 concentrations: 6.25 nM −100 nM) with a flow rate of 30 μl/min for 300 s across all flow cells, followed by measurement of dissociation for 300 s. Regeneration of the surface was done by sequential injection of 2 buffers for 30 s and 30 μl/min (buffer A: 50 mM NaPO4, 150 mM NaCl, 0.05% P20, pH 7.3 and buffer B: 0.1 M Tris-HCl, 150 mM NaCl, 0.05% P20, pH 8.0). All sensorgrams were referenced by subtracting values of a blank flow cell from each binding curve. Kinetic constants were calculated with a predefined heterogeneous ligand model (BIAcore Evaluation Software).
Thermal shift assay
Protein stability measurements were run in triplicates using a StepOnePlus Real-Time PCR System (Life Technologies) according to previous descriptions.49 All samples were prepared in PBS, pH 7.4 or pH 6.0 with SYPRO Orange (Invitrogen) and 0.9 molar antibody solutions. SYPRO Orange was diluted 1:167 for measurements at pH 7.4 or 1:125 for measurements at pH 6.0. Melting curves were measured from 25°C to 99 °C with 1°C / 60 s. Data analysis was performed with the Protein Thermal Shift™ Software version 1.0.
In silico modeling and calculations
Models of the adalimumab Fab-TNF complex were derived from the structure alignment of trimeric TNF (PDB 1TNF) and the adalimumab Fab fragment in complex with monomeric TNF (PDB 3WD5) using YASARA structure50,51 and the MUSTANG algorithm. All in silico experiments were performed applying the YASARA 2 force field and a simulation cell with the distance of 5 Å around all atoms. The complex structure was energy minimized using vacuo at 298.16 K and 0.9 M NaCl (aq) at pH 7.4 and pH 6.0, respectively. The electrostatic potential of the surfaces of trimeric TNF were displayed with 60 kJ/mol for both pH values. All graphics were prepared with POVRay.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful for the support and advice from a number of scientists at EMD Serono and Merck Serono, in particular Vanita Sood, Simon Krah, Carolin Sellmann and Birgit Piater. In addition, we thank Dirk Müller-Pompalla, Moritz Classen, Stephan Keller and Alexander Müller for the purification of the pH-sensitive antibody variants and Jessica Dawson for valuable comments on the manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Chaparro-Riggers J, Liang H, DeVay RM, Bai L, Sutton JE, Chen W, Geng T, Lindquist K, Casas MG, Boustany LM, et al. . Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J Biol Chem 2012; 287:11090-7; PMID:22294692; http://dx.doi.org/ 10.1074/jbc.M111.319764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gera N, Hill AB, White DP, Carbonell RG, Rao BM. Design of pH sensitive binding proteins from the hyperthermophilic Sso7d scaffold. PLoS One 2012; 7:e48928; PMID:23145025; http://dx.doi.org/ 10.1371/journal.pone.0048928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, et al. . Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol 2010; 28:1203-7; PMID:20953198; http://dx.doi.org/ 10.1038/nbt.1691 [DOI] [PubMed] [Google Scholar]
- 4. Sarkar CA, Lowenhaupt K, Horan T, Boone TC, Tidor B, Lauffenburger DA. Rational cytokine design for increased lifetime and enhanced potency using pH-activated "histidine switching.” Nat Biotechnol 2002; 20:908-13; PMID:12161759; http://dx.doi.org/ 10.1038/nbt725 [DOI] [PubMed] [Google Scholar]
- 5. Tsukamoto M, Watanabe H, Ooishi A, Honda S. Engineered protein A ligands, derived from a histidine-scanning library, facilitate the affinity purification of IgG under mild acidic conditions. J Biol Eng 2014; 8:15; PMID:25057290; http://dx.doi.org/ 10.1186/1754-1611-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. French AR, Tadaki DK, Niyogi SK, Lauffenburger DA. Intracellular trafficking of epidermal growth factor family ligands is directly influenced by the pH sensitivity of the receptor/ligand interaction. J Biol Chem 1995; 270:4334-40; PMID:7876195; http://dx.doi.org/ 10.1074/jbc.270.9.4334 [DOI] [PubMed] [Google Scholar]
- 7. Kulkarni MV, Tettamanzi MC, Murphy JW, Keeler C, Myszka DG, Chayen NE, Lolis EJ, Hodsdon ME. Two independent histidines, one in human prolactin and one in its receptor, are critical for pH-dependent receptor recognition and activation. J Biol Chem 2010; 285:38524-33; PMID:20889499; http://dx.doi.org/ 10.1074/jbc.M110.172072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maeda K, Kato Y, Sugiyama Y. pH-dependent receptor/ligand dissociation as a determining factor for intracellular sorting of ligands for epidermal growth factor receptors in rat hepatocytes. J Control Release 2002; 82:71-82; PMID:12106978; http://dx.doi.org/ 10.1016/S0168-3659(02)00126-8 [DOI] [PubMed] [Google Scholar]
- 9. Lauffenburger DA, Fallon EM, Haugh JM. Scratching the (cell) surface: cytokine engineering for improved ligand/receptor trafficking dynamics. Chem Biol 1998; 5:R257-63; PMID:9818145; http://dx.doi.org/ 10.1016/S1074-5521(98)90110-7 [DOI] [PubMed] [Google Scholar]
- 10. Ebner R, Derynck R. Epidermal growth factor and transforming growth factor-alpha: differential intracellular routing and processing of ligand-receptor complexes. Cell Regul 1991; 2:599-612; PMID:1777504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waterman H, Sabanai I, Geiger B, Yarden Y. Alternative intracellular routing of ErbB receptors may determine signaling potency. J Biol Chem 1998; 273:13819-27; PMID:9593726; http://dx.doi.org/ 10.1074/jbc.273.22.13819 [DOI] [PubMed] [Google Scholar]
- 12. Cama A, Sierra ML, Kadowaki T, Kadowaki H, Quon MJ, Rudiger HW, Dreyer M, Taylor SI. Two mutant alleles of the insulin receptor gene in a family with a genetic form of insulin resistance: a 10 base pair deletion in exon 1 and a mutation substituting serine for asparagine-462. Hum Genet 1995; 95:174-82; PMID:7860063; http://dx.doi.org/ 10.1007/BF00209397 [DOI] [PubMed] [Google Scholar]
- 13. Fallon EM, Liparoto SF, Lee KJ, Ciardelli TL, Lauffenburger DA. Increased endosomal sorting of ligand to recycling enhances potency of an interleukin-2 analog. J Biol Chem 2000; 275:6790-7; PMID:10702236; http://dx.doi.org/ 10.1074/jbc.275.10.6790 [DOI] [PubMed] [Google Scholar]
- 14. Keeler C, Jablonski EM, Albert YB, Taylor BD, Myszka DG, Clevenger CV, Hodsdon ME. The kinetics of binding human prolactin, but not growth hormone, to the prolactin receptor vary over a physiologic pH range. Biochemistry 2007; 46:2398-410; PMID:17279774; http://dx.doi.org/ 10.1021/bi061958v [DOI] [PubMed] [Google Scholar]
- 15. Yamamoto T, Chen HC, Guigard E, Kay CM, Ryan RO. Molecular studies of pH-dependent ligand interactions with the low-density lipoprotein receptor. Biochemistry 2008; 47:11647-52; PMID:18847225; http://dx.doi.org/ 10.1021/bi801117t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol 2007; 7:715-25; PMID:17703228; http://dx.doi.org/ 10.1038/nri2155 [DOI] [PubMed] [Google Scholar]
- 17. Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature 1989; 337:184-7; PMID:2911353; http://dx.doi.org/ 10.1038/337184a0 [DOI] [PubMed] [Google Scholar]
- 18. Tesar DB, Bjorkman PJ. An intracellular traffic jam: Fc receptor-mediated transport of immunoglobulin G. Curr Opin Struct Biol 2010; 20:226-33; PMID:20171874; http://dx.doi.org/ 10.1016/j.sbi.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Igawa T, Mimoto F, Hattori K. pH-dependent antigen-binding antibodies as a novel therapeutic modality. Biochim Biophys Acta 2014; 11:1943-50; http://dx.doi.org/ 10.1016/j.bbapap.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 20. Devanaboyina SC, Lynch SM, Ober RJ, Ram S, Kim D, Puig-Canto A, Breen S, Kasturirangan S, Fowler S, Peng L, et al. . The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. MAbs 2013; 5:851-9; PMID:24492341; http://dx.doi.org/ 10.4161/mabs.26389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem 1970; 24:1-95; PMID:4912353; http://dx.doi.org/ 10.1016/S0065-3233(08)60241-7 [DOI] [PubMed] [Google Scholar]
- 22. Warwicker J. A theoretical study of the acidification of the rhinovirus capsid. FEBS Lett 1989; 257:403-7; PMID:2555222; http://dx.doi.org/ 10.1016/0014-5793(89)81582-0 [DOI] [PubMed] [Google Scholar]
- 23. Warwicker J. Model for the differential stabilities of rhinovirus and poliovirus to mild acidic pH, based on electrostatics calculations. J Mol Biol 1992; 223:247-57; PMID:1309885; http://dx.doi.org/ 10.1016/0022-2836(92)90729-4 [DOI] [PubMed] [Google Scholar]
- 24. Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The Role of histidine residues in low-pH-mediated viral membrane fusion. Structure 2006; 14:1481-7; PMID:17027497; http://dx.doi.org/ 10.1016/j.str.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 25. Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR. A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches. Protein Sci 2011; 20:1619-31; PMID:21766385; http://dx.doi.org/ 10.1002/pro.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito W, Sakato N, Fujio H, Yutani K, Arata Y, Kurosawa Y. The His-probe method: effects of histidine residues introduced into the complementarity-determining regions of antibodies on antigen-antibody interactions at different pH values. FEBS Lett 1992; 309:85-8; PMID:1511750; http://dx.doi.org/ 10.1016/0014-5793(92)80745-3 [DOI] [PubMed] [Google Scholar]
- 27. Strauch EM, Fleishman SJ, Baker D. Computational design of a pH-sensitive IgG binding protein. Proc Natl Acad Sci USA 2014; 111:675-80; PMID:24381156; http://dx.doi.org/ 10.1073/pnas.1313605111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spassov VZ, Yan L. pH-selective mutagenesis of protein-protein interfaces: in silico design of therapeutic antibodies with prolonged half-life. Proteins 2013; 81:704-14; PMID:23239118; http://dx.doi.org/ 10.1002/prot.24230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doerner A, Rhiel L, Zielonka S, Kolmar H. Therapeutic antibody engineering by high efficiency cell screening. FEBS Lett 2014; 588:278-87; PMID:24291259; http://dx.doi.org/ 10.1016/j.febslet.2013.11.025 [DOI] [PubMed] [Google Scholar]
- 30. Traxlmayr MW, Lobner E, Hasenhindl C, Stadlmayr G, Oostenbrink C, Ruker F, Obinger C. Construction of pH-sensitive Her2-binding IgG1-Fc by directed evolution. Biotechnol J 2014; 9:1013-22; PMID:24964247; http://dx.doi.org/ 10.1002/biot.201300483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tracey D, Klareskog L, Sasso EH, Salfeld JG, Tak PP. Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol Ther 2008; 117:244-79; PMID:18155297; http://dx.doi.org/ 10.1016/j.pharmthera.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 32. den BA, van de Putte L, Rau R, Schattenkirchner M, Van RP, Sander O, Binder C, Fenner H, Bankmann Y, Velagapudi R, et al. . A single dose, placebo controlled study of the fully human anti-tumor necrosis factor-alpha antibody adalimumab (D2E7) in patients with rheumatoid arthritis. J.Rheumatol 2002; 29:2288-98; PMID:12415583 [PubMed] [Google Scholar]
- 33. Arnold FH. Directed evolution: creating biocatalysts for the future. Chem Eng Sci 1996; 51:5091-102; http://dx.doi.org/ 10.1016/S0009-2509(96)00288-6 [DOI] [Google Scholar]
- 34. Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553-7; PMID:9181578; http://dx.doi.org/ 10.1038/nbt0697-553 [DOI] [PubMed] [Google Scholar]
- 35. Kaymakcalan Z, Sakorafas P, Bose S, Scesney S, Xiong L, Hanzatian DK, Salfeld J, Sasso EH. Comparisons of affinities, avidities, and complement activation of adalimumab, infliximab, and etanercept in binding to soluble and membrane tumor necrosis factor. Clin Immunol 2009; 131:308-16; PMID:19188093; http://dx.doi.org/ 10.1016/j.clim.2009.01.002 [DOI] [PubMed] [Google Scholar]
- 36. Estep P, Reid F, Nauman C, Liu Y, Sun T, Sun J, Xu Y. High throughput solution-based measurement of antibody-antigen affinity and epitope binning. MAbs 2013; 5:270-8; PMID:23575269; http://dx.doi.org/ 10.4161/mabs.23049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang W, Lu P, Fang Y, Hamuro L, Pittman T, Carr B, Hochman J, Prueksaritanont T. Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab Dispos 2011; 39:1469-77; PMID:21610128; http://dx.doi.org/ 10.1124/dmd.111.039453 [DOI] [PubMed] [Google Scholar]
- 38. Martin WL, Bjorkman PJ. Characterization of the 2:1 complex between the class I MHC-related Fc receptor and its Fc ligand in solution. Biochemistry 1999; 38:12639-47; PMID:10504233; http://dx.doi.org/ 10.1021/bi9913505 [DOI] [PubMed] [Google Scholar]
- 39. Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screen 2008; 11:127-34; PMID:18336206; http://dx.doi.org/ 10.2174/138620708783744516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu S, Liang S, Guo H, Zhang D, Li H, Wang X, Yang W, Qian W, Hou S, Wang H, et al. . Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view. J Biol Chem 2013; 288:27059-67; PMID:23943614; http://dx.doi.org/ 10.1074/jbc.M113.491530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson G, Wu TT. Kabat Database and its applications: future directions. Nucleic Acids Res 2001; 29:205-6; PMID:11125092; http://dx.doi.org/ 10.1093/nar/29.1.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gurbaxani B, Dostalek M, Gardner I. Are endosomal trafficking parameters better targets for improving mAb pharmacokinetics than FcRn binding affinity? Mol Immunol 2013; 56:660-74; PMID:23917469; http://dx.doi.org/ 10.1016/j.molimm.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 43. Hopkins CR, Trowbridge IS. Internalization and processing of transferrin and the transferrin receptor in human carcinoma A431 cells. J Cell Biol 1983; 97:508-21; PMID:6309862; http://dx.doi.org/ 10.1083/jcb.97.2.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J Immunol 2004; 172:2021-9; PMID:14764666; http://dx.doi.org/ 10.4049/jimmunol.172.4.2021 [DOI] [PubMed] [Google Scholar]
- 45. Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng 2009; 103:1192-201; PMID:19459139; http://dx.doi.org/ 10.1002/bit.22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suga M, Hatakeyama T. High-efficiency electroporation by freezing intact yeast cells with addition of calcium. Curr Genet 2003; 43:206-11; PMID:12684838 [DOI] [PubMed] [Google Scholar]
- 47. Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel 2010; 23:155-9; PMID:20130105; http://dx.doi.org/ 10.1093/protein/gzq002 [DOI] [PubMed] [Google Scholar]
- 48. Bubeck P, Winkler M, Bautsch W. Rapid cloning by homologous recombination in vivo. Nucleic Acids Res 1993; 21:3601-2; PMID:8346047; http://dx.doi.org/ 10.1093/nar/21.15.3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem 2004; 332:153-9; PMID:15301960; http://dx.doi.org/ 10.1016/j.ab.2004.04.031 [DOI] [PubMed] [Google Scholar]
- 50. Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. . A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 2003; 24:1999-2012; PMID:14531054; http://dx.doi.org/ 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- 51. Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 2004; 57:678-83; PMID:15390263; http://dx.doi.org/ 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.