Abstract
Hes1 is one mammalian counterpart of the Hairy and Enhancer of split proteins that play a critical role in many physiological processes including cellular differentiation, cell cycle arrest, apoptosis and self-renewal ability. Recent studies have shown that Hes1 functions in the maintenance of cancer stem cells (CSCs), metastasis and antagonizing drug-induced apoptosis. Pathways that are involved in the up-regulation of Hes1 level canonically or non-canonically, such as the Hedgehog, Wnt and hypoxia pathways are frequently aberrant in cancer cells. Here, we summarize the recent data supporting the idea that Hes1 may have an important function in the maintenance of cancer stem cells self-renewal, cancer metastasis, and epithelial–mesenchymal transition (EMT) process induction, as well as chemotherapy resistance, and conclude with the possible mechanisms by which Hes1 functions have their effect, as well as their crosstalk with other carcinogenic signaling pathways.
Keywords: cancer stem cell, chemotherapy resistance, Hes1, metastasis, Notch signaling pathway, non-canonical Notch
Abbreviations
- CSCs
cancer stem cells
- EMT
epithelial–mesenchymal transition
- MASH-1
Mammalian achaete-scute homolog-1
- bHLH
basic helix-loop-helix
- TLE
transducin-like Enhancer of split
- HDACs
histone deacetylases
- dnMAM
dominant-negative mastermind
- Runx2
Runt-related protein 2
- ABC
ATP-binding cassette
- NICD
Notch intracellular domain
- CSL
CBF1/ Suppressor of Hairless / Lag1
- MAML
Mastermind-like protein family
- GSI
γ-secretase inhibitor
Introduction
Since the concept of stem cells has been implicated in cancer, it has taken many decades to increase the understanding of the biological properties of CSCs, which are characterized by a remarkable ability for extensive proliferation, self-renewal, invasion, development of distant metastasis, and drug resistance.1,2 With the efforts to investigate CSCs biology, some of the molecular components that sustain CSC properties are being uncovered. One of these is the Hes1 protein. Notably, the level of Hes1 is a key signature that maintains stem cells, quiescent cells or cancer stem cells in a non-dividing state.3-5 Apart from this, Hes1 has additional cellular functions that have not been previously epitomized. Hes1 also contributes to tumor multidrug resistance as well as metastasis.
In this review, we discuss both the significance of Hes1 function in tumors and CSCs and the mechanisms by which Hes1 contributes to stemness, metastasis and drug resistance. Elucidating the pathways involved in the control of Hes1 expression could provide valuable information for cancer treatment.
Overview of Hes1 Factor
Early described as a transcriptional inhibitor, Hes1, which can antagonize neuronal cell fate regulators, such as Mammalian achaete-scute homolog-1 (MASH-1 in the mouse and HASH-1 in humans), Math, and neurogenin (Ngn), is a member of the basic helix-loop-helix (bHLH) family.6-11 Hes1 factors is identified with 3 conserved domains: the bHLH, Orange and WRPW domains. The bHLH domain of Hes1 is responsible for interaction with lineage-specific bHLH factors; for instance, it can form a functional heterodimer with other bHLH factors, such as E47, Id proteins as well as its homologies hey1/2.12,13-15 Within the bHLH region, Hes1 has a proline residue which exhibits unique binding ability to N box (CACNAG), differing from other bHLH factors which have higher affinity to E box (CANNTG).11 The orange domain is less conserved and confers selection specificity of bHLH heterodimer partner.11,16 Another functional region, the WRPW domain, is characterized with the C-terminal Trp–Arg–Pro–Trp sequence.
There are 2 possible mechanisms that Hes1 promote transcriptional repression. Hes1 forms a non-DNA binding complex with other bHLH factors through its bHLH domain. Second mechanism is active repression. Hes1 binds to N box by forming complexes with co-repressor transducin-like enhancer of split (TLE), homologs of Drosophila Groucho via WRPW motif.17,18 The transcriptional inhibition is mediated by this interaction, and occurs by recruitment of histone deacetylases (HADCs).3, 19 In addition, Hes1 may recruit HADCs using the bHLH domain to repress transcription.12,20
However, recent studies show that Hes1 is more than a repressor. Hes1 amplifies Runx2 expression by cooperating with pRb, and WRPW domain is necessary for Hes1-pRb interacion.42 In addition, Hes1 uses bHLH domain and orange domain to bind with STAT3 and activates STAT3 phosphorylation.58
The role of Hes1 in CSCs maintenance
Hes1 is widely expressed in different tissue and cell types, including neuronal stem cells, embryonic stem cells, quiescent cells, and especially in cells at the precursor stage. Hes1-deficient mice have been shown to exhibit premature differentiation and rigorous defects in brain tissue.6 The overexpression of Hes1 inhibits neuronal and glial differentiation, and the cells most likely maintain their self-renewal ability, if Hes1 is up-regulated.7 Furthermore, the undifferentiated state can be sustained in quiescent cells, endodermal endocrine cells and intestine progenitor cells through Hes1 overexpression.8-10 Hes1 plays an essential role in stem cells in the undifferentiated state due to its repression ability.
Many tumors, which are composed of a heterogeneous group of cells, have recently been identified as having a small population of cells, also called cancer stem-like cells, which have a high level of stem cell-like properties, including an undifferentiated state, resistance to cell death, self-renewal and tumorigenesis. Considering the tremendous potential that Hes1 has in maintaining self-renewal ability, it was important to investigate the significance of roles these factors play in CSC. Neuroblastoma, originating from the sympathetic nervous system, has embryonic features. The suppression of Hes1 leads to HASH-1 upregulation, which impairs the neuroblastoma undifferentiated state.21 The level of Hes1 in CD133+ glioma, a tumor of the brain, is elevated. Strikingly, after shRNAs against Hes1 are applied, the colony-forming ability of CD133+ glioma is reduced.22 In addition, when the Hes1 level reduces, CD133+ positive cells are depleted.23 Meanwhile, CD133 is identified as a marker of cancer stem cells.24 The mis-expression of Hes1 decreases the tumor sphere-forming capacity and self-renewal ability of oral squamous cell carcinoma cells.25 The same phenomenon can be observed in colon cancer, as well as pancreatic cancer.26,27 The Notch-Hes1 pathway also functions significantly in maintaining breast cancer stem cells.28 During a leukemia blast crisis, patient samples that were tested exhibited elevated levels of Hes1.29 Hes1 may promote tumor cell growth and the self-renewal phenotype under hypoxia conditions or Notch stimulation.30,31 Downstream factors of Hes1, which tend to be differentiating makers, such as Mash1 and NeuroD show dose- and time-dependent response to Hes1 inhibition treatment in cancer progression, confirming the idea that Hes1 maintains the undifferentiated state, possibly through transcriptional repression.21,32,33
Hes1 contributes to cancer metastasis
Tumor metastasis, which occurs in a multistep biological process, wildly spreads cancer cells from a primary tumor to a distant location and indicates a far worse prognosis and increases the risk of death in cancer patients. Importantly, the correlation between Hes1 and tumor metastasis has been revealed through a series of experimental investigation.
The accumulating experimental data indicate that a malignant tumor tending to metastasizes is often accompanied by aberrant Notch signaling and the elevation of Hes1.34-37 Hes1 loss of function via a dominant-negative mastermind (dnMAM) in an osteosarcoma xenograft model, which then results in a significant reduction of lung metastasis neoplasm, while dnMAM has no effect on tumor latency or tumor growth rates.38 Evidence from 56 samples obtained from patients who did well with osteosarcoma and died from metastasis reveals that Hes1 is inversely correlated with survival.37 The metastasis potentiality of Hes1 was further studied in vitro by constructing mutant Hes1 fragments which were then transfected into osteosarcoma cells. The results suggest that Hes1 represses Deltex1 which can block osteosarcoma invasiveness by binding to the Deltex1 promoter.39
There is yet another mechanism by which Hes1 is able to promote tumor metastasis. Hes1 has been implicated in a tumor cell “osteoblast-like” phenotype and tumor bone metastasis. Runt-related protein 2 (Runx2), also known as Cbfa1, has been well documented to be highly expressed in breast tumors and prostate tumors, as well as their secondary tumor sites. Runx2 plays a critical role in tumor bone metastasis, as discussed by Pratap J.40 Interestingly, Hes1 can eliminate the repressive potentiality that the TLE protein exerts on Runx2 by forming a Hes1-Runx2 complex and enhancing the transactivation of Runx2.41 Additionally, Hes1 increases Runx2-DNA binding, stimulates Runx2 transcriptional activity by increasing the stability of Runx2, and associates with pRb synergistically to augment Runx2 transcription.42,43 When examined in prostate tumor bone metastasis, Runx2 DNA-binding activity is enhanced in response to osteogenic induction and is inhibited by Notch-Hes1 inhibition, verifying that Hes1-Runx2 signaling functions critically in tumor bone metastasis.44
Furthermore, EMT process allows the loss of junction with adjacent cells, the polarization of epithelial cells and subsequent acquisition of mesenchymal features. During EMT, cells undergo multiple biologic changes including enhanced motility capacity, invasiveness and resistance to apoptosis has been well established to have an intimate connection with tumor metastasis and also shows its deep relationship to cancer stem cells.45,46 In recent investigations, Hes1 has been found to potentially act as a regulator of EMT, inducing phenotype plasticity.
To initiate EMT, glioma cells lose their phenotype and cell-cell adhesion and become invasive through β-catenin and NF-КB synergistically, a process that is enhanced by Notch1 via AKT signaling.47 Previous studies on Notch and AKT signaling suggest that Notch signaling can stimulate the AKT pathway by suppressing PTEN levels.48,49 Interestingly, Hes1 can effectively abrogate PTEN by binding to its promoter, and when shRNA silence of Hes1 is performed, PTEN transcription shows a moderate increase.48,50 If transfected with Hes1 siRNA, clear cell renal cell carcinoma cells have a significant change in PTEN level and reduced invasiveness.51 In addition, Hes1 is required to promote Snail-dependent EMT in HK-2 cells that are cultured in high glucose conditions.52
Hes1 leads to tumor multidrug resistance
Drug resistance is a major cause of tumor treatment failure and cancer recurrence. Based on the “cancer stem cell” theory, the small population of pluripotent cells develops a range of strategies to promote chemotherapy insensitivity, including ATP-binding cassette (ABC) transporter expression, DNA repair capacity and quiescence.53,54 This may provide a new avenue to target CSCs, as the Hes1 anti-drug effect has been recently discovered.
The overexpression of Hes1 antagonizes a pro-apoptosis state of myeloma cells.55 Correspondingly, the Hes1 level is observed to be higher in chemically resistant, as well as cyclophosphamide-treated hepatocellular carcinoma.56 There are several latent mechanisms by which cancer cells are able to promote transformations that increase drug resistance. Such instances include high level of Hes1 expression and enhancing STAT3 phosphorylation.57 On the other hand, the modulation of STAT3 phosphorylation is affected by Hes1 binding to STAT3 directly and then recruiting JAK and Src protein effectively.58,59
Tumor dormancy appears to be the underlying mechanism for tumor resistance to chemotherapy. Hes1 correlates with dormancy, and this is mediated, in part, by retaining the tumor in a quiescent stage that can re-enter the cell cycle.3,60-62 Indeed, it should be noted that cancer cell dormancy resulting from cell cycle arrest has been observed in solid tumors, while the quiescent stage is reached by several pathways, in addition to cell cycle arrest.63 Experiments in vitro have shown the involvement of Notch receptor, Hes1, and p21 activities in the drug resistance of quiescent tumors. Notch1 activates the p21-induced cell-cycle-arrest tumor model, which is identified as a drug-resistant phenotype.64,65 However, sustained activation of Notch-p21 contributes to an irreversible senescent phenotype.8,66 In contrast, cells that express full-length Hes1 genes resume proliferation, after a p21-induced senescent phenotype, as Hes1 transcriptionally represses p21 in a bHLH domain-dependent manner and Hes1 keeps quiescent cells from the differentiation state mentioned previously.8,67 Based on this, we may conclude that the quiescence-based dormancy relies on the balance between Hes1 and p21.
As seen below, we conclude that Hes1 plays an extraordinarily important role in tumor progression (Fig. 1).
Figure 1.
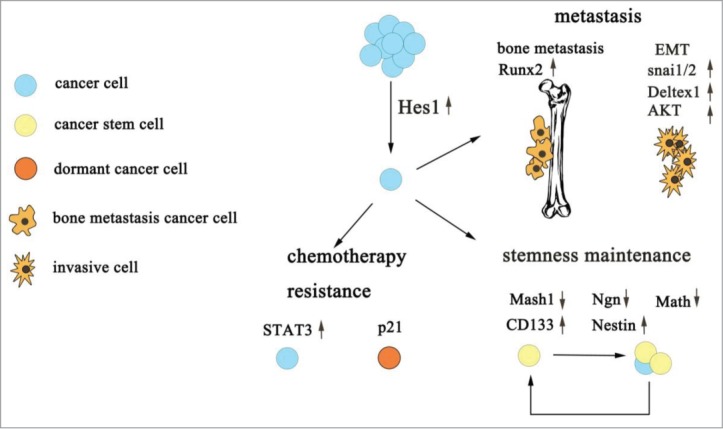
The role of Hes1 in tumor progression. Hes1 contributes to CSCs maintenance, cancer chemotherapy resistance and tumor metastasis mainly through its downstream protein.
Control of Hes1 expression
The mechanism that leads to an induction of Hes1 cancer cells could possibly represent a promising therapeutic target. In this section, we have only highlighted some signaling pathways that have been implicated in the induction of Hes1. We will discuss what is currently known about the mechanism by which they up regulate Hes1 (Fig. 2).
Figure 2.
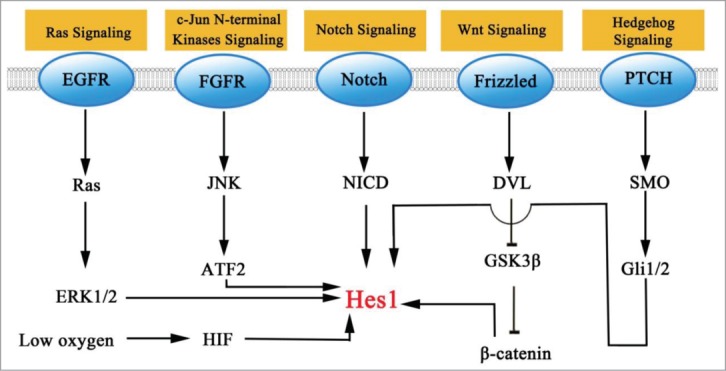
Signaling pathways that modulate the level of Hes1.
Canonical Notch signaling pathway
The Notch signaling pathway in mammals consists of 5 transmembrane ligands (Delta-like 1, 3 and 4 and Jagged 1 and 2) and 4 membrane bound receptors (Notch 1, 2, 3 and 4). Following ligand binding, the Notch receptor undergoes the second cleavage mediated by ADAM/TACE, and the third proteolytic cleavage catalyzed by γ-secretase and releases the Notch intracellular domain (NICD), which translocates to the nucleus and then associates with DNA-binding factors CSL (CBF1/ Suppressor of Hairless / Lag1) and the Mastermind-like protein family (MAML), activating Hes family transcription.6-11
The expression of Hes1 induced by Notch can be detected more than embryo neural stem cells, intestinal crypt progenitor cells, pancreatic stem cells, bone marrow mesenchymal progenitors and quiescent skeletal muscle stem cells.9,10,68-71 Treatment with γ-secretase inhibitor (GSI) results in the suppression of Hes1 and the differentiating state of those stem cells, indicating the role of the Notch pathway in Hes1 expression, as well as the essential status of the Notch-Hes1 pathway in facilitating cell fate decisions and regulating differentiation during development.
Indeed, the upregulation of Hes1 is involved in maintaining stemness in mammals and reflects elevated Notch signaling pathway activity, which may contribute to the growth of those tumors. Aberrant Notch-Hes1 axis is presented in cancer stem cells, such as glioblastoma, breast cancer, pancreatic cancer, and osteosarcoma.27,36,72,73 The inhibition of the Notch signaling pathway can reverse cancer cell survival and induce apoptosis. In glioma, the up-regulation of Notch enhances Nestin and CD133 expression.23,74 After Notch is blocked by GSI, we observe a population of cancer stem cell depletion.23 Aberrant Notch-Hes1 pathway is observed in invasive cells which undergo EMT process.48-51 In dormant cancer cells, Notch-Hes1 activation can be detected.64,65 Above all, the Notch-Hes1 signaling pathway may play a functional role in self-renewal, CSCs maintenance and tumor progression.
Furthermore, crosstalk has been implicated between canonical Notch, Wnt, and hypoxia signaling. β-catenin, a downstream factor of Wnt signaling, increases Hes1 expression by binding with NICD and stabilizes the CSL-NICD complex by recruiting p300 and P/CAF.75-77 HIF-1 is required to promote interactions with NICD and potentiate the stabilization of the CSL-NICD complex under hypoxia conditions, while induction of Hes1 protects progenitor cells, as well as tumor stem cells, against differentiation.78-81
Non-canonical Notch signaling pathway
Notch signaling pathway can exert its biological function independently of its ligands, receptors or CSL in vertebrates. Early evidence for non-canonical Notch has been implicated in normal tissue as well as tumor.82,83 The downstream Notch target gene Hes1 is modulated in a Notch independent manner as well.
Hes1 upregulation mediated by Hedgehog signaling promotes stem/glial cell undifferentiated state in ventricular zone and is not influenced by CSL deletion.84 Hes1 inhibition which is conducted by cyclopamine incubation indicates that Hes1 expression involves canonical Hedgehog signaling, because cyclopamine blocks Hedgehog downstream factor Smo function.85 In the retina progenitor cells, Hedgehog signaling activates Hes1 expression in the presence of GSI, as Hedgehog downstream factor Gli2 directly binds to Hes1 promoter and accelerate Hes1 level without activating CSL.86 The requirement of Gli2 in Hes1 regulation is also observed in telencephalic neuroepithelial stem cells.87 Gli1 also shows its regulation on Hes1. In neural stem cells, overexpression of Gli1 is accompanied with Hes1 upregulation.88 In addition, the Hedgehog-Hes1 pathway can be detected in tumor cells as well. To initiate tumor or maintain undifferentiated properties, Hes1 induction mediated by Hedgehog pathway can be detected during tumorigenesis.89 Interestingly, Hes1 inhibits Hedgehog signaling by binding to the first intron of Gli1 in brain, prostate, lung, ovarian, skin, and hematopoetic cell cancers.90 This may suggests a potential chemotherapy resistance mechanism that tumor survives long term Notch-Hes1 inhibition with Gli1 rescuing expression.
Furthermore, other signaling pathways have involved in non-canonical Notch pathway. FGF2 activates c-Jun N-terminal kinase (JNK) in neural progenitors, and the activated JNK further phosphorylates ATF2. Phospho-ATF2 binds to the Hes1 promoter to enhance Hes1 expression and high level of Hes1 maintains a neural progenitor cells pool.91 In the TGFα-stimulates neuroblastoma, Hes1 upregulation follows ERK1/2 phosphorylation without NICD activation and low level of HASH-1 suppressed by Hes1 contributes to maintain neuroblastoma embryonic non-dividing states and proliferative capacity.92
Conclusion
Several therapeutic avenues to reverse aberrant Notch and non-canonical Notch have been applied in researches. Since Hes1 lies at the crossroads of multiple signaling pathways, co-inhibition of these pathways might represent a new strategy for cancer therapy. For instance, when treated with anti-hedgehog and anti-Notch compounds in vitro, glioma cells show apoptosis phenotype, CSCs depletion and sensitivity to temozolomide.90,93 As discussed earlier, the third cleavage mediated by γ-secretase is a crucial step for Notch activation and is targeted by GSI inhibitors which have been evaluated in phase 1 clinical trials, such as MK-0752, PF-03084014, RO-4929097, reviewed by Takebe N.94 With the rigorous estimation in preclinical models, it is safe to say that GSI inhibitors do show its anti-proliferation, anti-CSC and pro-apoptotic effects on tumor. However, GSI inhibits Notch target genes without selection which causes a rapid differentiation of intestinal progenitor cells into goblet cells.95 This may be the primary cause of gastrointestinal toxicities associated with GSI. Aiming at Hes1 may results in fewer side effects because many other Notch target genes will be unaffected.
As we have discussed the fundamental role of Hes1 molecule in tumor cells and the strategies used by tumor cells to maintain stemness, metastasize and resist chemotherapy, targeting Hes1 may represent a potential therapeutic possibility to target cancer. Therefore, significant attention should be paid to clinical development to target Hes1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No.31201028).
Reference
- 1. Fessler E, Dijkgraaf FE, De Sousa E, Melo F, Medema JP. Cancer stem cell dynamics in tumor progression and metastasis: Is the microenvironment to blame?. Cancer Lett 2013; 341(1):97-104; PMID:23089245; http://dx.doi.org/ 10.1016/j.canlet.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 2. Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells—what challenges do they pose?. Nat Rev Drug Disc 2014; 13(7):497-512; PMID:24981363; http://dx.doi.org/ 10.1038/nrd4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med 2010; 16(1):17-26; PMID:20022559; http://dx.doi.org/ 10.1016/j.molmed.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tian C, Zheng G, Cao Z, Li Q, Ju Z, Wang J, Yuan W, Cheng T. Hes1 mediates the different responses of hematopoietic stem and progenitor cells to T cell leukemic environment. Cell Cycle 2013; 12(2):322-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun H, Ghaffari S, Taneja R. bHLH-Orange transcription factors in development and cancer. Translational Oncogenomics 2007; 2:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res 2005; 306(2):343-8; PMID:15925590; http://dx.doi.org/ 10.1016/j.yexcr.2005.03.015 [DOI] [PubMed] [Google Scholar]
- 7. Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J 1994; 13(8):1799; PMID:7909512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 2008; 321(5892):1095-100; PMID:18719287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet 2000; 24(1):36-44; PMID:10615124 [DOI] [PubMed] [Google Scholar]
- 10. Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature 2005; 435(7044):964-8; PMID:15959516 [DOI] [PubMed] [Google Scholar]
- 11. Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 2007; 134(7):1243-51. [DOI] [PubMed] [Google Scholar]
- 12. Fischer A, Gessler M. Delta–Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res 2007; 35(14):4583-96; PMID:17586813; http://dx.doi.org/ 10.1093/nar/gkm477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirata H, Ohtsuka T, Bessho Y, Kageyama R. Generation of structurally and functionally distinct factors from the basic helix-loop-helix gene Hes3 by alternative first exons. J Biol Chem 2000; 275(25):19083-9; PMID:10858455 [DOI] [PubMed] [Google Scholar]
- 14. Jögi A, Persson P, Grynfeld A, Påhlman S, Axelson H. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J Biol Chem 2002; 277(11):9118-26; PMID:11756408 [DOI] [PubMed] [Google Scholar]
- 15. Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 2003; 194(3):237-55. [DOI] [PubMed] [Google Scholar]
- 16. Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 2001; 20(58):8342-57. [DOI] [PubMed] [Google Scholar]
- 17. Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 1994; 79(5):805-15; PMID:8001118 [DOI] [PubMed] [Google Scholar]
- 18. Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol 1996; 16(6):2670-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao J, Lai E, Stifani S. The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol Cell Biol 2001; 21(6):1962-72; PMID:11238932; http://dx.doi.org/ 10.1128/MCB.21.6.1962-1972.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1-and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun 2003; 301(1):250-7; PMID:12535671; http://dx.doi.org/ 10.1016/S0006-291X(02)03020-6 [DOI] [PubMed] [Google Scholar]
- 21. Grynfeld A, Påhlman S, Axelson H. Induced neuroblastoma cell differentiation, associated with transient HES-1 activity and reduced HASH-1 expression, is inhibited by Notch1. Int J Cancer 2000; 88(3):401-10. [PubMed] [Google Scholar]
- 22. Fang KM, Lin TC, Chan TC, Ma SZ, Tzou BC, Chang WR, Liu JJ, Chiou SH, Yang CS, Tzeng SF. Enhanced cell growth and tumorigenicity of rat glioma cells by stable expression of human CD133 through multiple molecular actions. Glia 2013; 61(9):1402-17; PMID:23832679 [DOI] [PubMed] [Google Scholar]
- 23. Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al. NOTCH Pathway Blockade Depletes CD133-Positive Glioblastoma Cells and Inhibits Growth of Tumor Neurospheres and Xenografts. Stem Cells 2010; 28(1):5-16; PMID:19904829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng JX, Liu BL, Zhang X. How powerful is CD133 as a cancer stem cell marker in brain tumors?. Cancer Treat Rev 2009; 35(5):403-8; PMID:19369008; http://dx.doi.org/ 10.1016/j.ctrv.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 25. Lee SH, Hong HS, Liu ZX, Kim RH, Kang MK, Park NH , Shin KH. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Commun 2012; 424(1):58-64; PMID:22728043; http://dx.doi.org/ 10.1016/j.bbrc.2012.06.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao F, Zhang YQ, Wang SC, Liu Y, Zheng L, Yang J, Huang W, Ye Y, Luo W, Xiao D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Rep 2014; 4:3963; PMID:24492635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abel EV, Kim EJ, Wu J, Hynes M, Bednar F, Proctor E, Wang L, Dziubinski ML, Simeone DM. The Notch pathway is important in maintaining the cancer stem cell population in pancreatic cancer. PloS One 2014; 9(3): e91983; PMID:24647545; http://dx.doi.org/ 10.1371/journal.pone.0091983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong NKY, Fuller M, Sung S, Wong F, Karsan A. Heterogeneity of breast cancer stem cells as evidenced with Notch-dependent and Notch-independent populations. Cancer Med 2012; 1(2):105-13; PMID:23342261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K, Kumano K , Harada Y, Harada H, Kitaura J, et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 2010; 115(14):2872-81; PMID:19861684 [DOI] [PubMed] [Google Scholar]
- 30. Chen J, Imanaka N, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 2010; 102(2):351-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danza G, Di Serio C, Rosati F, Lonetto G, Sturli N, Kacer D, Pennella A, Ventimiglia G, Barucci R, Piscazzi A, et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol Cancer Res 2012; 10(2):230-8; PMID:22172337; http://dx.doi.org/ 10.1158/1541-7786.MCR-11-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu YY, Zheng MH, Cheng G, Li L, Liang L, Gao F, Wei YN, Fu LA, Han H. Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer 2011; 11(1):82; PMID:21342503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ball DW. Achaete–scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett 2004; 204(2):159-69. [DOI] [PubMed] [Google Scholar]
- 34. Zayzafoon M, Abdulkadir SA, McDonald JM. The role of Notch signaling in the osteomimetic properties of prostate cancer bone metastases. Proc Am Assoc Cancer Res 2004; 2004(1):789. [DOI] [PubMed] [Google Scholar]
- 35. Mu X, Isaac C, Greco N, Huard J, Weiss K. Notch signaling is associated with ALDH activity and an aggressive metastatic phenotype in murine osteosarcoma cells. Front Oncol 2013; 3:143; PMID:23805413; http://dx.doi.org/ 10.3389/fonc.2013.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dailey DD, Anfinsen KP, Pfaff LE, Ehrhart EJ, Charles JB, Bønsdorff TB, Thamm DH, Powers BE, Jonasdottir TJ, Duval DL. HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC Vet Res 2013; 9(1):130; PMID:23816051; http://dx.doi.org/ 10.1186/1746-6148-9-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hughes DPM. How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Pediatric and Adolescent Osteosarcoma. USA: Springer, 2010:479-96. [DOI] [PubMed] [Google Scholar]
- 38. Zhang P, Yang Y, Zweidler-McKay PA, Hughes DP. Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin Cancer Res 2008; 14(10):2962-9; PMID:18483362; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-1992 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DP. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene 2010; 29(20):2916-26; PMID:20208568; http://dx.doi.org/ 10.1038/onc.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metast Rev 2006; 25(4):589-600; PMID:17165130; http://dx.doi.org/ 10.1007/s10555-006-9032-0 [DOI] [PubMed] [Google Scholar]
- 41. McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani . The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem 2000; 275(1):530-8; PMID:10617648 [DOI] [PubMed] [Google Scholar]
- 42. Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW. HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Mineral Res 2006; 21(6):921-33; PMID:16753023; http://dx.doi.org/ 10.1359/jbmr.060303 [DOI] [PubMed] [Google Scholar]
- 43. Suh JH, Lee HW, Lee JW, Kim JB. Hes1 stimulates transcriptional activity of Runx2 by increasing protein stabilization during osteoblast differentiation. Biochem Biophys Res Commun 2008; 367(1):97-102; PMID:18162173; http://dx.doi.org/ 10.1016/j.bbrc.2007.12.100 [DOI] [PubMed] [Google Scholar]
- 44. Zayzafoon M, Abdulkadir SA, McDonald JM. Notch signaling and ERK activation are important for the osteomimetic properties of prostate cancer bone metastatic cell lines. J Biol Chem 2004; 279(5):3662-70. [DOI] [PubMed] [Google Scholar]
- 45. Geiger TR, Peeper DS. Metastasis mechanisms. Biochimica et Biophysica Acta (BBA)-Rev Cancer 2009; 1796(2):293-308; PMID:19683560; http://dx.doi.org/ 10.1016/j.bbcan.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 46. Kong D, Li Y, Wang Z, Sarkar FH. Cancer stem cells and epithelial-to-mesenchymal transition (EMT)-phenotypic cells: are they cousins or twins? Cancers 2011; 3(1):716-29; PMID:21643534; http://dx.doi.org/ 10.3390/cancers30100716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang X, Chen T, Zhang J, Mao Q, Li S, Xiong W, Qiu Y, Xie Q, Ge J. Notch1 promotes glioma cell migration and invasion by stimulating β-catenin and NF-КB signaling via AKT activation. Cancer Sci 2012; 103(2):181-90; PMID:22093097; http://dx.doi.org/ 10.1111/j.1349-7006.2011.02154.x [DOI] [PubMed] [Google Scholar]
- 48. Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13(10):1203-10; PMID:17873882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu ZJ, Xiao M, Balint K, Soma A, Pinnix CC, Capobianco AJ, Velazquez OC, Herlyn M. Inhibition of endothelial cell proliferation by Notch1 signaling is mediated by repressing MAPK and PI3K/Akt pathways and requires MAML1. FASEB J 2006; 20(7):1009-11; PMID:16571776 [DOI] [PubMed] [Google Scholar]
- 50. Palomero T, Dominguez M, Ferrando AA. The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle-Landes Biosci- 2008; 7(8):965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu S, Ma X, Ai Q, Huang Q, Shi T, Zhu M, Wang B, Zhang X. NOTCH1 functions as an oncogene by regulating the PTEN/PI3K/AKT pathway in clear cell renal cell carcinoma. Urologic Oncology: Seminars and Original Investigations. Elsevier 2013; 31(6):938-48; PMID:21993533 [DOI] [PubMed] [Google Scholar]
- 52. Sumual S, Saad S, Tang O, Yong R, McGinn S, Chen XM, Pollock CA. Differential regulation of Snail by hypoxia and hyperglycemia in human proximal tubule cells. Int J Biochem Cell Biol 2010; 42(10):1689-97; PMID:20620220; http://dx.doi.org/ 10.1016/j.biocel.2010.06.023 [DOI] [PubMed] [Google Scholar]
- 53. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011:313-9; PMID:21386835; http://dx.doi.org/ 10.1038/nm.2304 [DOI] [PubMed] [Google Scholar]
- 54. Holohan C, Van Schaeybroeck S, Longley D B, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13(10):714-26; PMID:24060863; http://dx.doi.org/ 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- 55. Nefedova Y, Sullivan DM, Bolick SC, Dalton WS, Gabrilovich DI. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood 2008; 111(4):2220-9; PMID:18039953 [DOI] [PubMed] [Google Scholar]
- 56. Marfels C, Hoehn M, Wagner E, Günther M. Characterization of in vivo chemoresistant human hepatocellular carcinoma cells with transendothelial differentiation capacities. BMC cancer 2013; 13(1):176; PMID:23547746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gu F, Ma Y, Zhang Z, Zhao J, Kobayashi H, Zhang L, Fu L. Expression of Stat3 and Notch1 is associated with cisplatin resistance in head and neck squamous cell carcinoma. Oncol Rep 2010; 23(3):671-6; PMID:20127005 [DOI] [PubMed] [Google Scholar]
- 58. Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat Cell Biol 2004; 6(6):547-54; PMID:15156153; http://dx.doi.org/ 10.1038/ncb1138 [DOI] [PubMed] [Google Scholar]
- 59. Lee JH, Suk J, Park J, Kim SB, Kwak SS, Kim JW, Lee CH, Byun B, Ahn JK, Joe CO. Notch signal activates hypoxia pathway through HES1-dependent SRC/signal transducers and activators of transcription 3 pathway. Mol Cancer Res 2009; 7(10):1663-71; PMID:19808903; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0191 [DOI] [PubMed] [Google Scholar]
- 60. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011:313-9; PMID:21386835; http://dx.doi.org/ 10.1038/nm.2304 [DOI] [PubMed] [Google Scholar]
- 61. Almog N. Molecular mechanisms underlying tumor dormancy. Cancer Lett 2010; 294(2):139-46; PMID:20363069; http://dx.doi.org/ 10.1016/j.canlet.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 62. Ruppender NS, Morrissey C, Lange PH, Vessella RL. Dormancy in solid tumors: implications for prostate cancer. Cancer Metast Rev 2013; 32(3-4):501-9; PMID:23612741; http://dx.doi.org/ 10.1007/s10555-013-9422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kusumbe AP, Bapat SA. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res 2009; 69(24):9245-53. [DOI] [PubMed] [Google Scholar]
- 64. Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood 2004; 103(9):3503-10; PMID:14670925 [DOI] [PubMed] [Google Scholar]
- 65. Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, Ball DW. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001; 61(7):3200-5; PMID:11306509 [PubMed] [Google Scholar]
- 66. Cui H, Kong Y, Xu M, Zhang H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer Res 2013; 73(11):3451-9; PMID:23610446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Castella P, Sawai S, Nakao K, Wagner JA, Caudy M. HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression. Mol Cell Biol 2000; 20(16):6170-83; PMID:10913198; http://dx.doi.org/ 10.1128/MCB.20.16.6170-6183.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J 1999; 18(8):2196-07; PMID:10205173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rooman I, De Medts N, Baeyens L, Lardon J, De Breuck S, Heimberg H, Bouwens L. Expression of the Notch signaling pathway and effect on exocrine cell proliferation in adult rat pancreas. Am J Pathol 2006; 169(4):1206-14; PMID:17003479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 2008; 14(3):306-14; PMID:18297083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 2012; 30(2):243-52; PMID:22069237 [DOI] [PubMed] [Google Scholar]
- 72. Chen J, Kesari S, Rooney C, Strack PR, Chen J, Shen H, Wu L, Griffin JD. Inhibition of notch signaling blocks growth of glioblastoma cell lines and tumor neurospheres. Genes & Cancer 2010; 1(8):822-35; PMID:21127729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, Golde TE, Miele L, Foreman KE. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res 2010; 30(10):3853-67; PMID:21036696 [PubMed] [Google Scholar]
- 74. Shih AH, Holland EC. Notch signaling enhances nestin expression in gliomas. Neoplasia (New York, NY) 2006; 8(12):1072; PMID:17217625; http://dx.doi.org/ 10.1593/neo.06526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Espinosa L, Inglés-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3β down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem 2003; 278(34):32227-35; PMID:12794074 [DOI] [PubMed] [Google Scholar]
- 76. Jin YH, Kim H, Ki H, Yang I, Yang N, Lee KY, Kim N, Park HS, Kim K. Beta-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochimica et Biophysica Acta (BBA)-Mol Cell Res 2009; 1793(2):290-9; PMID:19000719; http://dx.doi.org/ 10.1016/j.bbamcr.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 77. Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized β-catenin functions through TCF/LEF proteins and the Notch/RBP-JК complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol 2008; 28(24):7427-41; PMID:18852283; http://dx.doi.org/ 10.1128/MCB.01962-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 2005; 9(5):617-28; PMID:16256737 [DOI] [PubMed] [Google Scholar]
- 79. Danza G, Di Serio C, Rosati F, Lonetto G, Sturli N, Kacer D, Pennella A, Ventimiglia G, Barucci R, Piscazzi A, et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol Cancer Res 2012; 10(2):230-8; PMID:22172337; http://dx.doi.org/ 10.1158/1541-7786.MCR-11-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Qiang L, Wu T, Zhang HW, Lu N, Hu R, Wang YJ, Zhao L, Chen FH, Wang XT, You QD, et al. HIF-1α is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating Notch signaling pathway. Cell Death Differ 2012; 19(2):284-94; PMID:21818118; http://dx.doi.org/ 10.1038/cdd.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pistollato F, Rampazzo E, Persano L, Abbadi S, Frasson C, Denaro L, D'Avella D, Panchision DM, Della Puppa A, Scienza R, et al. Interaction of hypoxia-inducible factor-1α and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells 2010; 28(11):1918-29; PMID:20827750; http://dx.doi.org/ 10.1002/stem.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Veeraraghavalu K, Subbaiah VK, Srivastava S, Chakrabarti O, Syal R, Krishna S. Complementation of human papillomavirus type 16 E6 and E7 by Jagged1-specific Notch1-phosphatidylinositol 3-kinase signaling involves pleiotropic oncogenic functions independent of CBF1; Su (H); Lag-1 activation. J Virol 2005; 79(12):7889-98; PMID:15919944; http://dx.doi.org/ 10.1128/JVI.79.12.7889-7898.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nofziger D, Miyamoto A, Lyons KM, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 1999; 126(8):1689-702; PMID:10079231 [DOI] [PubMed] [Google Scholar]
- 84. Dave RK, Ellis T, Toumpas MC, Robson JP, Julian E, Adolphe C, Bartlett PF, Cooper HM, Reynolds BA, Wainwright BJ. Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PloS One 2011; 6(2): e14680; PMID:21379383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene 2007; 27(10):1489-500; PMID:17873912 [DOI] [PubMed] [Google Scholar]
- 86. Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol 2009; 184(1):101-12; PMID:19124651; http://dx.doi.org/ 10.1083/jcb.200805155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Takanaga H, Tsuchida-Straeten N, Nishide K, Watanabe A, Aburatani H, Kondo T. Gli2 is a novel regulator of sox2 expression in telencephalic neuroepithelial cells. Stem Cells 2009; 27(1):165-74; PMID:18927476; http://dx.doi.org/ 10.1634/stemcells.2008-0580 [DOI] [PubMed] [Google Scholar]
- 88. Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J 2009; 28(6):663-76; PMID:19214186; http://dx.doi.org/ 10.1038/emboj.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Julian E, Dave RK, Robson JP, Hallahan AR, Wainwright BJ. Canonical Notch signaling is not required for the growth of Hedgehog pathway-induced medulloblastoma. Oncogene 2010; 29(24):3465-76; PMID:20418906; http://dx.doi.org/ 10.1038/onc.2010.101 [DOI] [PubMed] [Google Scholar]
- 90. Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, Eberhart CG. The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: a potential mechanism of therapeutic resistance. Clin Cancer Res 2010; 16(24):6060-70; PMID:21169257; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sanalkumar R, Indulekha CL, Divya TS, Divya MS, Anto RJ, Vinod B, Vidyanand S, Jagatha B, Venugopal S, James J. ATF2 maintains a subset of neural progenitors through CBF1/Notch independent Hes-1 expression and synergistically activates the expression of Hes-1 in Notch-dependent neural progenitors. J Neurochem 2010; 113(4):807-18; PMID:20067572 [DOI] [PubMed] [Google Scholar]
- 92. Stockhausen MT, Sjölund J, Axelson H. Regulation of the Notch target gene Hes-1 by TGFα induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res 2005; 310(1):218-28; PMID:16120441; http://dx.doi.org/ 10.1016/j.yexcr.2005.07.011 [DOI] [PubMed] [Google Scholar]
- 93. Ulasov IV, Nandi S, Dey M, Sonabend AM, Lesniak MS. Inhibition of sonic hedgehog and notch pathways enhances sensitivity of cd133+ glioma stem cells to temozolomide therapy. Mol Med 2011; 17(1-2):103; PMID:20957337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol Therap 2014; 141(2):140-9; PMID:24076266; http://dx.doi.org/ 10.1016/j.pharmthera.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 2005; 435(7044):959-63; PMID:15959515 [DOI] [PubMed] [Google Scholar]


