Abstract
The use of combination antiretroviral therapy (cART) to prevent HIV mother-to-child transmission during pregnancy and delivery is generally considered safe. However, vigilant assessment of potential risks of these agents remains warranted. Epigenetic changes including DNA methylation are considered potential mechanisms linking the in utero environment with long-term health outcomes. Few studies have examined the epigenetic effects of prenatal exposure to pharmaceutical agents, including antiretroviral therapies, on children. In this study, we examined the methylation status of the LINE-1 and ALU-Yb8 repetitive elements as markers of global DNA methylation alteration in peripheral blood mononuclear cells obtained from newborns participating in the Pediatric HIV/AIDS Cohort Study SMARTT cohort of HIV-exposed, cART-exposed uninfected infants compared to a historical cohort of HIV-exposed, antiretroviral-unexposed infants from the Women and Infants Transmission Study Cohort. In linear regression models controlling for potential confounders, we found the adjusted mean difference of AluYb8 methylation of the cART-exposed compared to the -unexposed was −0.568 (95% CI: −1.023, −0.149) and for LINE-1 methylation was −1.359 (95% CI: −1.860, −0.857). Among those exposed to cART, subjects treated with atazanavir (ATV), compared to those on other treatments, had less AluYb8 methylation (−0.524, 95% CI: −0.025, −1.024). Overall, these results suggest a small but statistically significant reduction in the methylation of these repetitive elements in an HIV-exposed, cART-exposed cohort compared to an HIV-exposed, cART-unexposed historic cohort. The potential long-term implications of these differences are worthy of further examination.
Keywords: anti-retroviral, atazanavir, developmental origins, DNA methylation, HIV, newborn, pediatrics, pharmaceuticals, prenatal, repetitive elements
Introduction
The use of combination antiretroviral therapy (cART) during pregnancy has resulted in a dramatic reduction in HIV mother-to-child transmission.1-3 The use of cART is generally considered safe, with the benefits of HIV prevention far exceeding potential safety concerns from in utero exposure to these drugs.1,4-7 However, there is still a continued need to monitor HIV-exposed, uninfected infants for potential adverse effects from in utero and postnatal exposure to these therapies.4,8-11 There is conflicting evidence demonstrating associations between antiretroviral use during pregnancy and birth defects, reduced birth weight, altered growth trajectories, neurological and neurodevelopmental deficits, and mitochondrial abnormalities.5,12,13 Little is known about the mechanisms by which specific antiretrovirals can lead to these postnatal and even childhood effects.
Epigenetic changes may play an important role as mediators of the effects of the intrauterine environment on long-term health outcomes in offspring, including neurodevelopmental deficits, behavior, metabolic disorders, and obesity.14-22 Epigenetic modifications are broadly defined as the mitotically heritable chemical or structural changes to DNA, which are involved in the regulation of gene expression without alteration of the underlying DNA sequence. The most widely studied of the epigenetic modifications is DNA methylation. This is particularly true in human population-based studies, as the marker is readily accessible and stable for measurement from biologic specimens, and requires much smaller sample amounts and more lenient sample collection procedures than would the examination of other types of epigenetic modification. In human populations, DNA methylation has been studied at specific genes, generally in their promoter regions, as a marker of functional alteration, and at the “global” level, as an overall measure of the epigenetic status of a sample. Global methylation is considered a passive dosimeter, susceptible to alteration by various environmental insults.23 Studies of global methylation make use of analyses of repetitive element regions of the genome, such as long interspersed nuclear elements (LINEs) and small interspersed nuclear elements (SINEs, represented by Alu elements), which are generally highly methylated and can be used to assess variation in the overall DNA methylation. Reductions in methylation of these elements have been associated with various exposures including cigarette smoke and environmental toxicants, as well as with disease outcomes in retrospective and now prospective studies.24-27 As these markers have been heavily studied, and may represent global measures of DNA methylation status, we have focused our analyses on the LINE-1 and AluYb8 repetitive elements, representing members of both the LINE and SINE families.
Although the functional consequences of variation in methylation of LINE and SINE repetitive elements remains a topic of debate, reduction in LINE and SINE repetitive element methylation may indicate potential epigenetic effects of an exposure.28 To date, no studies have directly examined the epigenetic consequences of exposure to antiretrovirals in utero or in early life. However, some investigators have suggested that antiretrovirals exposure in utero is associated with mitochondrial function that became apparent postnatally.29 Altered mitochondrial function is linked to altered DNA methylation profiles.30-32 The association of antiretrovirals with mitochondrial anomalies9,33-35 and metabolic dysfunction36 suggests that antiretroviral drugs may also be linked to changes in DNA methylation. Thus, the purpose of the current study was to examine the distribution of global DNA methylation in peripheral blood samples collected near birth and to examine the relationship of methylation patterns and exposure to antiretroviral drugs in utero among HIV-exposed, cART-exposed uninfected infants compared to a historical cohort of HIV-exposed, antiretroviral-unexposed infants.
Results
Characteristics of the study participants
The demographic characteristics of the 2 cohorts, cART-exposed (PHACS SMARTT), and antiretroviral-unexposed cohort (WITS) were similar (Table 1). Unexposed infants were slightly more likely to be male. The proportions of infants weighting less than 2500 g and considered preterm (<37 weeks) did not differ significantly between the cohorts, consistent with the matching design used. A higher proportion of mothers from the unexposed population had HIV RNA levels ≥400 copies/ml. Unexposed subjects also had mothers with less education and lower household incomes. In addition, greater proportions of mothers from the unexposed group reported use of alcohol, tobacco, cocaine or crack, heroin, and marijuana during pregnancy.
Table 1:
Demographic Characteristics of the Study Participants by Cohort
| Characteristic | ART-exposed (SMARTT, N = 295) | ART-unexposed (WITS, N = 150) | Total (N = 445) | P-Value* | |
|---|---|---|---|---|---|
| Gender | Male | 140 (47%) | 84 (56%) | 224 (50%) | 0.088 |
| Female | 155 (53%) | 66 (44%) | 221 (50%) | ||
| Birth Weight (g) | >=2500 | 256 (87%) | 126 (89%) | 382 (87%) | 0.564 |
| <2500 | 39 (13%) | 16 (11%) | 55 (13%) | ||
| Missing | 0 | 8 | 8 | ||
| Gestational Age (Weeks) | >= 37 | 241 (82%) | 130 (87%) | 371 (83%) | 0.183 |
| < 37 | 54 (18%) | 20 (13%) | 74 (17%) | ||
| Last CD4 Count Prior to Delivery | >=350 | 214 (74%) | 104 (77%) | 318 (75%) | 0.440 |
| <350 | 77 (26%) | 31 (23%) | 108 (25%) | ||
| Missing | 4 | 15 | 19 | ||
| Last Maternal HIV RNA Prior to Delivery | <400 | 236 (83%) | 21 (16%) | 257 (61%) | <0.001 |
| >=400 | 48 (17%) | 114 (84%) | 162 (39%) | ||
| Missing | 11 | 15 | 26 | ||
| Caregiver Education | Less than High School | 111 (38%) | 86 (58%) | 197 (45%) | <0.001 |
| At least High School | 181 (62%) | 62 (42%) | 243 (55%) | ||
| Missing | 3 | 2 | 5 | ||
| African American/Black | No | 135 (46%) | 76 (51%) | 211 (47%) | 0.327 |
| Yes | 160 (54%) | 74 (49%) | 234 (53%) | ||
| Household Income | <= 20k | 216 (77%) | 123 (91%) | 339 (82%) | <0.001 |
| >20k | 63 (23%) | 12 (9%) | 75 (18%) | ||
| Missing | 16 | 15 | 31 | ||
| Alcohol During Pregnancy | No | 268 (91%) | 82 (55%) | 350 (79%) | <0.001 |
| Yes | 25 (9%) | 68 (45%) | 93 (21%) | ||
| Missing | 2 | 0 | 2 | ||
| Tobacco During Pregnancy | No | 238 (81%) | 72 (48%) | 310 (70%) | <0.001 |
| Yes | 55 (19%) | 78 (52%) | 133 (30%) | ||
| Missing | 2 | 0 | 2 | ||
| Cocaine/Crack During Pregnancy | No | 283 (97%) | 92 (61%) | 375 (85%) | <0.001 |
| Yes | 10 (3%) | 58 (39%) | 68 (15%) | ||
| Missing | 2 | 0 | 2 | ||
| Heroin During Pregnancy | No | 290 (99%) | 117 (78%) | 407 (92%) | <0.001 |
| Yes | 3 (1%) | 33 (22%) | 36 (8%) | ||
| Missing | 2 | 0 | 2 | ||
| Marijuana During Pregnancy | No | 268 (91%) | 125 (83%) | 393 (89%) | 0.010 |
| Yes | 25 (9%) | 25 (17%) | 50 (11%) | ||
| Missing | 2 | 0 | 2 |
Chi-Square Test
LINE-1 and AluYb8 DNA Methylation in Newborn Peripheral Blood
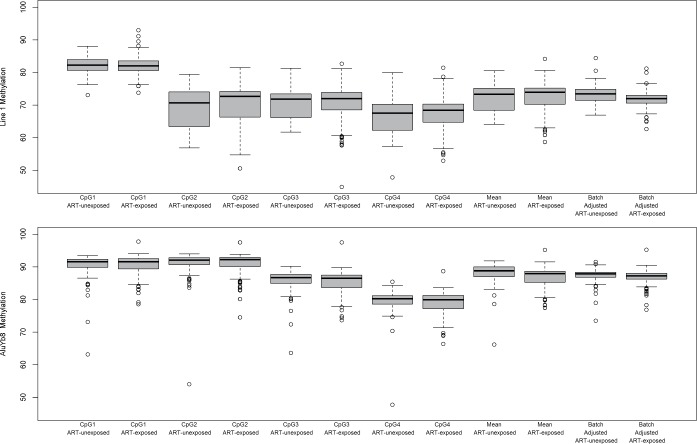
Figure 1 shows the distribution of methylation for each CpG site within the LINE-1 and AluYb8 regions for cART-exposed and -unexposed subjects and the means across the regions with and without batch adjustment. For both LINE-1 and AluYb8, the extent of methylation varied considerably across CpGs. The mean batch adjusted LINE-1 methylation was 71.73 and 73.20 for cART-exposed and -unexposed subjects, respectively, while AluYb8 was also lower in cART-exposed subjects (87.08) compared to -unexposed (87.46).
Figure 1.
Distributions of (A) LINE-1 and (B) AluYb8 methylation extent at individual CpG sites, and the mean across the sequenced regions with and without batch adjustment
Association of cART exposure with DNA methylation of LINE-1 and AluYb8
The results of both unadjusted and fully adjusted models are presented in Table 2. In unadjusted, bivariate analyses, AluYb8 methylation was significantly lower in cART-exposed subjects compared to -unexposed subjects (P = 0.05), and was significantly higher in infants of black race compared to other races (P = 0.05). These results were maintained in the fully adjusted model. AluYb8 methylation was lower in cART-exposed subjects, compared to -unexposed subjects by −0.568 (95% CI: −1.023, −0.149; P = 0.009), in a model adjusted for race, caregiver educational status, household income, birth weight, preterm birth status, sex, and maternal tobacco use during pregnancy. In this adjusted model, black race, compared to all other races, was associated with higher methylation of AluYb8 of 0.4 (P = 0.034).
Table 2:
Association between LINE-1/AluYb8 Mean Methylation and Exposure to ARVs in Unadjusted and Adjusted Linear Regression Models
| Variable | Diff (LCI, UCI) | P-value | aDiff (LCI, UCI)* | aP-value* |
|---|---|---|---|---|
| ART-Exposed vs. Unexposed | ||||
| AluYb8 | −0.382 (−0.761, −0.004) | 0.048 | −0.586 (−1.023, −0.149) | 0.009 |
| LINE-1 | −1.471 (−1.933, −1.008) | <0.001 | −1.359 (−1.860, −0.857) | <0.001 |
| Race Black vs. All Other | ||||
| AluYb8 | 0.363 (0.007, 0.718) | 0.046 | 0.399 (0.029, 0.768) | 0.034 |
| LINE-1 | 0.184 (−0.270, 0.637) | 0.43 | 0.366 (−0.062, 0.793) | 0.094 |
| Caregiver Education < HS vs. ≥ HS | ||||
| AluYb8 | 0.112 (−0.251, 0.474) | 0.55 | 0.135 (−0.257, 0.528) | 0.50 |
| LINE-1 | 0.469 (0.013, 0.925) | 0.044 | 0.116 (−0.338, 0.569) | 0.62 |
| Household Income < $20 k vs. ≥ $20 k | ||||
| AluYb8 | 0.088 (−0.389, 0.565) | 0.72 | 0.111 (−0.389, 0.611) | 0.66 |
| LINE-1 | −0.497 (−1.099, 0.104) | 0.11 | −0.258 (−0.843, 0.326) | 0.39 |
| Birth Wt ≥ 2500 g vs. < 2500 g | ||||
| AluYb8 | 0.291 (−0.245, 0.827) | 0.29 | 0.326 (−0.298, 0.951) | 0.31 |
| LINE-1 | 0.526 (−0.148, 1.199) | 0.13 | 0.511 (−0.210, 1.231) | 0.16 |
| Preterm Birth vs. Term (≥ 37 wks) | ||||
| AluYb8 | −0.153 (−0.632, 0.326) | 0.53 | −0.043 (−0.605, 0.520) | 0.88 |
| LINE-1 | −0.316 (−0.915, 0.283) | 0.30 | −0.296 (−0.940, 0.348) | 0.37 |
| Male vs. Female | ||||
| AluYb8 | 0.314 (−0.041, 0.670) | 0.083 | 0.318 (−0.055, 0.690) | 0.095 |
| LINE-1 | 1.178 (0.739, 1.617) | <0.001 | 1.010 (0.579, 1.442) | <0.001 |
| Tobacco During Pregnancy vs. No Tobacco Use | ||||
| AluYb8 | −0.118 (−0.506, 0.271) | 0.55 | −0.293 (−0.725, 0.139) | 0.18 |
| LINE-1 | 0.765 (0.277, 1.253) | 0.002 | 0.154 (−0.346, 0.655) | 0.55 |
Diff = average difference; (LCI,UCI) = 95% confidence interval; aDiff = adjusted average difference; aP-value = P-value from the adjusted model; HS = High School.
Model adjusted for maternal race, caregiver education, household income, infant birth weight, preterm birth, infant sex, and maternal tobacco use during pregnancy.
LINE-1 methylation was also significantly lower in the cART-exposed subjects compared to -unexposed subjects (P < 0.001) in unadjusted models. These bivariate analyses also suggested that caregiver education, male sex, and maternal tobacco use during pregnancy were associated with increased LINE-1 methylation (P = 0.04, P < 0.001, P = 0.002, respectively). In the fully adjusted model, LINE-1 methylation was lower by greater than one percent (Difference= −1.36, 95% CI: −1.86, −0.86; P < 0.001). Male sex was the only other predictor significantly and independently associated with LINE-1 methylation in this adjusted model (Difference= 1.01, 95% CI: 0.58, 1.44; P<0.001) although the estimate was attenuated.
We performed sensitivity analyses controlling for maternal substance use during pregnancy among those with available data for both LINE-1 and AluYb8 methylation (Table S2). Inclusion of these variables did not substantially alter the effect estimates for ART exposure on methylation.
Comparing those exposed to a specific cART treatment regimen to those unexposed to any treatment in WITS (Table S3), we observed similar findings as the primary analysis: lower DNA methylation at LINE-1 and AluYb8 with each of the potential regimens of treatment compared to the unexposed subjects. However, in a limited number of subjects exposed to non-nucleoside reverse transcription inhibitors (NNRTIs) and the protease inhibitor nelfinavir, the reduction in AluYb8 methylation was not significant, although the estimates were consistent with a reduction in methylation.
Among those exposed to cART (SMARTT subjects only), we estimated the effect of specific regimens that may be driving the observed differences in methylation (Table S4). Only those subjects treated with atazanavir (ATV) compared to those on other treatments had significantly lower AluYb8 methylation [aDiff = −0.524, 95% CI: (−0.025, −1.024), P-value = 0.04] in fully adjusted models (Table 3).
Table 3.
Adjusted and Unadjusted Linear Regression Analyses of AluYb8 and LINE-1 comparing atazanavir (ATV) use During Pregnancy to Other Regimens. Data on ART Exposures were Taken from in the SMARTT Cohort.
| Repetitive Element Methylation | N | Diff (LCI, UCI) | P-value | aDiff (LCI, UCI)* | aP-value* | |
|---|---|---|---|---|---|---|
| LINE-1 | ||||||
| Other Regimens | 210 | Ref | — | Ref | — | |
| ATV Exposed | 71 | 0.130 (0.738, −0.479) | 0.68 | −0.087 (0.498, −0.671) | 0.77 | |
| AluYb8 | ||||||
| Other Regimens | 207 | Ref | — | Ref | — | |
| ATV Exposed | 71 | −0.414 (0.083, −0.910) | 0.10 | −0.524 (−0.025, −1.024) | 0.040 |
Diff = average difference; (LCI,UCI) = 95% confidence interval; aDiff = adjusted average difference, aP-value = P-value from the adjusted model.
Model adjusted for maternal race, caregiver education, household income, infant birth weight, preterm birth, infant sex, and maternal tobacco use during pregnancy
Discussion
We identified small but significant differences in the extent of repetitive element DNA methylation of the LINE-1 and AluYb8 elements in the peripheral blood of newborns based on exposure to prophylactic antiretroviral therapies in utero. Specifically, infants exposed to antiretrovirals had 1.4% lower LINE-1 methylation and 0.6% lower AluYb8 methylation compared to infants not exposed to these therapies. These findings suggest a reduction of DNA methylation related to antiretroviral therapy exposure in utero. However, questions remain as to the stability of these changes over time and on the clinical impact of such differences in the health of these children.
The relatively subtle differences we have observed in this examination are consistent in size and, in some cases, in direction with prior studies of various in utero environmental and nutritional factors and methylation of these repetitive elements in newborn cord or peripheral blood. One study of infants in Bangladesh exposed to arsenic in utero through maternal drinking water found that increases in infant cord blood arsenic were related to a reduction of Alu methylation of 0.45%, and increases in maternal urinary arsenic measure during pregnancy with a reduction of LINE-1 methylation of 0.31%, but only among female infants.37 Another study found the opposite: increased LINE-1 methylation of 1.36% (95% CI: 0.52, 2.21%) comparing the highest quartile of maternal drinking water arsenic to the lowest.38 However, a third study found no differences in LINE-1 methylation in infant cord blood associated with various measures of internal dose of arsenic exposure.39 In a study of newborn infants in Mexico, maternal patella lead burden in the highest quartile was associated with a reduction in LINE-1 methylation of greater than 1% compared to the lowest quartile, while maternal tibial lead burden in the highest quartile was associated with a reduction in ALU methylation of nearly 0.5% compared to the lowest quartile of exposure in adjusted models.40 Reduced Alu methylation of 0.37% was also identified in cord blood of newborns with maternal exposures to dichlorodiphenyl trichloroethane insecticide in a cohort of Mexican-American children in a rural area of California.41
Dietary supplements including folic acid supplementation after the 12th week of pregnancy was associated with a reduction of cord blood LINE-1 methylation of 0.34%,42 choline levels during the periconceptual period were associated with a reduction of LINE-1 cord blood methylation of 0.1% in male infants only within the Boston-based Project Viva cohort,43 but DHA supplementation of maternal smokers led to an increase of LINE-1 methylation of 0.77%.44 Only a single prior study has examined the association of a pharmaceutical agent during gestation with global DNA methylation, specifically examining the use of anti-epileptic drugs.45 Using a measure of DNA methylation based on the average extent of methylation profiled on the Illumina Infinium 27K array platform, the authors identified a reduction in global methylation associated with maternal anti-epileptic drug use during gestation; longer durations of exposures led to greater reductions in methylation, and a number of specific genes were targeted for altered methylation associated with anti-epileptic exposures.45
Within the SMARTT study of antiretroviral-exposed subjects, we identified a specific effect of atazanavir compared to other antiretroviral exposure on a reduction in AluYb8 methylation. This drug can pass into the fetal circulation through the placenta and has been potentially linked to some cases of hyperbilirubinemia in newborns, although without further sequelae.46 In the SMARTT study, atazanavir was associated with elevated odds of congenital anomalies.47 In vitro studies have suggested that atazanavir as well as other protease inhibitors can alter the expression of key metabolic genes in hepatocytes,48 inflammatory regulators in macrophages,49 and mediators of adipocyte formation and autophagy in preadipocytes.50,51 It is of interest that this effect appears specific to AluYb8 and not LINE-1, and may suggest differences in the potential mechanism through which this drug leads to these differences, or in differences in the susceptibility of these repetitive genomic regions for alteration by different agents. Recent epigenomic analyses suggest that the de-methylation and re-methylation of these repetitive elements occurs early in embryogenesis. Hypomethylation of the SINE and LINE repeats is observed in cells at the cleavage and blastocyst stages of development. It is then restored in the SINE elements in early human embryonic stem cells, but only partially in LINE elements, which become fully hypermethylated only in differentiated somatic cells.52 This could also suggest important effects of the timing of exposure, which we were unable to fully interrogate in this study, but would be of great interest and could aid in providing insights into the mechanisms of action and potential toxicity of these agents.
Pregnancy conditions and outcomes also have been related to altered global methylation extent measured at the repetitive elements in newborn blood samples. Gestational diabetes has been associated with a one percent decrease in Alu methylation, but a 2 percent increase in methylation at LINE-1.53 Preterm birth has also been associated with a reduction of 0.73% in LINE-1 methylation, while birth weights under 2500 g or greater than 4000 g were also associated with reductions of 0.82% and 0.43% in LINE-1 methylation, respectively, compared to infants born between 2500–4000g.54 These latter results could suggest potential growth related implications to alterations in global DNA methylation, although the cross-sectional nature of those examinations cannot definitively determine that relationship. We did not observe relationships between these markers of repetitive element DNA methylation and birth outcomes such as birth weight or gestational age in the SMARTT cohort subjects (data not shown), and we did not observe independent effects of birth weight or preterm birth status on repetitive element methylation (Table 3), although our current study design had limited power for this examination. Further studies are necessary to examine the birth and long-term implications of the differences in methylation.
This study has a number of limitations. The most important limitation is our reliance on a historical cohort as a comparison group of antiretroviral unexposed individuals. Yet, this allows us to uniquely and appropriately control for the HIV exposure, which would occur in the SMARTT subjects as well, thereby eliminating the potential impact of this confounder. Given the conclusive data on the efficacy for the prevention of maternal to child transmission of HIV through the use of antiretrovirals during pregnancy, there is no ethical means by which to study these effects in a contemporary cohort. We aimed to appropriately address additional confounders that could bias the results due to the differences in the cohorts, but acknowledge that uncontrolled confounding could still be influencing our results. We do not believe there would be technical reasons based on the age of samples that would influence the extent of DNA methylation being assessed. The addition or loss of the methyl-group to cytosine requires active enzymatic processes, which are unlikely occurring differentially in the samples, which were all stored at −80°C. The examination of the specific regimens, effects within the SMARTT subjects also led to a large number of comparisons, and so our findings may be due to chance related to the introduction of increased Type I error. Further study of the specific effects of these regimens in larger samples would be needed to validate these relationships. Finally, we acknowledge that we are examining generalized biomarkers within a mixed population of cells that make up PBMCs. Our findings may be confounded by subtype; therefore, future work which can address this confounding through isolation of specific cellular subtypes would be warranted.
Important future work should aim to examine the clinical relevance of this altered methylation, in terms of childhood health outcomes such as growth, metabolic health, and immune system function. It would also be of interest to understand how the timing of exposures during pregnancy could impact epigenetic variation, as this may provide more detailed insight into the mechanisms through which these drugs could be impacting the epigenome. In addition, it will be important to examine the long-term stability of this variation, as it will be important to know if these decrements are maintained or propagated, or are potentially reversed over time The true functional implications of methylation of these repetitive elements in not clear, and it would be of interest to examine more gene-specific effects, targeting candidate genes related to known health effects of these agents, or performing genome-wide discovery studies to uncover potentially novel genes or pathways which may be altered through these exposure.
Although we have identified potential epigenetic variability with exposure to anti-retrovirals in utero, there remain significant questions regarding the clinical implications of these findings. As such, the overwhelming evidence of the benefit of the use of these agents in preventing maternal to fetal HIV transmission far outweighs these effects. However, further research is needed to assess the long-term implications of in utero antiretroviral drug exposure in order to identify the most efficacious and safe treatment and preventative antiretroviral regimens.
Patients and Methods
Study Populations. The HIV-exposed, antiretroviral exposed population was drawn from participants in the Surveillance Monitoring for Antiretroviral Therapy Toxicities (SMARTT) study of the Pediatrics HIV/AIDS Cohort Study (PHACS) network.55 The Dynamic Cohort of SMARTT enrolled newborns and their mothers between 22 weeks gestation and 1 week after birth at 22 participating PHACS network sites in the United States including Puerto Rico starting in 2007. A more thorough description of the particular antiretroviral regimens being examined is provided as supplementary methods.
The HIV-exposed, antiretroviral unexposed population was drawn from the Women and Infants Transmission Study (WITS), which enrolled newborns and their mothers between 1989 and 2003 at 6 clinical sites in the United States.56 Institutional review board approval was obtained at all participating sites and at the Harvard School of Public Health, which serves as the coordinating center.
Subjects from WITS (n = 150) were selected from those infants with peripheral blood samples collected prior to the third day of life. SMARTT subjects with peripheral blood samples collected prior to the third day of life were matched in a 2 to one ratio (n = 300) to the WITS subjects. SMARTT subjects were matched on gestational age to minimize the difference in the percentage of preterm delivery between the SMARTT and WITS. Exposure to specific antiretrovirals was defined as any exposure during pregnancy. These sample sizes were chosen based on a priori power simulations to identify a difference of methylation of 0.025.
DNA Methylation Analysis. Genomic DNA was isolated from peripheral blood mononuclear cell pellets using the Qiamp Blood Minikit (Qiagen). The resulting genomic DNA underwent sodium bisulfite modification utilizing the EZ DNA Methylation Kit (Zymo Research) in 96 sample batches. The extent of methylation of cytosines followed guanines (CpGs) of the LINE-1 and AluYb8 repetitive regions (presenting LINE and SINE repetitive elements, respectively) was examined with a quantitative pyrosequencing approach following previously described methods.57-59 These assays allow for the assessment of the percent of DNA methylation at 4 specific CpG sites in the LINE-1 region, and 6 CpG sites within the AluYb8 region.
Statistical methods. Demographic characteristics were compared between those with vs. without antiretroviral exposure using chi-square tests. Mean LINE-1 and AluYb8 methylation at individual CpG sites in those with vs. without cART exposure, and adjusted means adjusting for batch effects were calculated. Batch effects were adjusted using a 2-stage procedure.60 To assure that our normalization procedure adequately controlled for batch effects, we performed additional analyses that included batch in the model. Those analyses demonstrated appropriate control for these effects (Table S1) and so the models reported thereafter in this report did not include the batch effect as a covariate (Table 3), and were based on the adjusted means across all interrogated CpG sites. Linear regression models were used to estimate associations of cART exposure with mean LINE-1 and AluYb8 methylation while adjusting for potential confounders. Potential confounders were chosen a priori and included race, caregiver education, household income, birth weight, preterm delivery, gender, and tobacco use. Sensitivity analyses were performed by adjusting for other substance use exposure during pregnancy including marijuana use, cocaine/crack use, heroin and alcohol use. We further examined if any specific antiretrovirals were associated with reduction in DNA methylation. Linear regression models were constructed comparing specific antiretroviral therapy regimens compared to all other regimens within the cART-exposed subjects only. In addition, models were constructed comparing those subjects on a specific cART regimen compared to unexposed subjects.
Analyses with P-values < 0.05 were considered to be statistically significant, and confidence intervals (CI) are reported as 95% CI. SAS 9.2 and R version 2.15.1 were used for the analyses.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed. The conclusions and opinions expressed in this article are those of the authors and do not necessarily reflect those of the National Institutes of Health or US. Department of Health and Human Services or the US EPA.
Acknowledgments
We thank the children and families for their participation in PHACS, and the individuals and institutions involved in the conduct of PHACS.
The following institutions, clinical site investigators and staff participated in conducting PHACS SMARTT in 2014, in alphabetical order: Ann & Robert H. Lurie Children's Hospital of Chicago: Ram Yogev, Margaret Ann Sanders, Kathleen Malee, Scott Hunter; Baylor College of Medicine: William Shearer, Mary Paul, Norma Cooper, Lynnette Harris; Bronx Lebanon Hospital Center: Murli Purswani, Emma Stuard, Anna Cintron; Children's Diagnostic & Treatment Center: Ana Puga, Dia Cooley, Patricia Garvie, James Blood; New York University School of Medicine: William Borkowsky, Sandra Deygoo, Helen Rozelman; Rutgers - New Jersey Medical School: Arry Dieudonne, Linda Bettica, Susan Adubato; St. Jude Children's Research Hospital: Katherine Knapp, Kim Allison, Megan Wilkins; San Juan Hospital/Department of Pediatrics: Midnela Acevedo-Flores, Lourdes Angeli-Nieves, Vivian Olivera; SUNY Downstate Medical Center: Hermann Mendez, Ava Dennie, Susan Bewley; Tulane University Health Sciences Center: Chi Dola, Robert Maupin, Karen Craig, Patricia Sirois; University of Alabama, Birmingham: Marilyn Crain, Newana Beatty, Dan Marullo; University of California, San Diego: Stephen Spector, Jean Manning, Sharon Nichols; University of Colorado Denver Health Sciences Center: Elizabeth McFarland, Jenna Wallace, Carrie Chambers, Christine Reed; University of Florida/Jacksonville: Mobeen Rathore, Kristi Stowers, Ann Usitalo; University of Illinois, Chicago: Kenneth Rich, Lourdes Richardson, Renee Smith; University of Miami: Gwendolyn Scott, Claudia Florez, Elizabeth Willen; University of Southern California: Toni Frederick, Mariam Davtyan, Maribel Mejia; University of Puerto Rico Medical Center: Zoe Rodriguez, Ibet Heyer, Nydia Scalley Trifilio.
Funding
The study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development with co-funding from the National Institute on Drug Abuse, the National Institute of Allergy and Infectious Diseases, the Office of AIDS Research, the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, the National Institute on Deafness and Other Communication Disorders, the National Heart Lung and Blood Institute, the National Institute of Dental and Craniofacial Research, and the National Institute on Alcohol Abuse and Alcoholism, through cooperative agreements with the Harvard T.H. Chan School of Public Health (HD052102) (Principal Investigator: George Seage; Project Director: Julie Alperen) and the Tulane University School of Medicine (HD052104) (Principal Investigator: Russell Van Dyke; Co-Principal Investigator: Kenneth Rich; Project Director: Patrick Davis). Also through the National Institute of Environmental Health Sciences and US EPA through NIH-NIEHS P01 ES022832 and by US EPA grant RD83544201. Data management services were provided by Frontier Science and Technology Research Foundation (PI: Suzanne Siminski), and regulatory services and logistical support were provided by Westat, Inc. (PtdIns: Julie Davidson).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Connor EM, Sperling RS, Gelber R, Kiselev P, Scott G, O'Sullivan MJ, VanDyke R, Bey M, Shearer W, Jacobson RL, et al.. Reduction of Maternal-Infant Transmission of Human Immunodeficiency Virus Type 1 with Zidovudine Treatment. Pediatric Aids Clinical Trials Group Protocol 076 Study Group. N Engl J Med 1994; 331:1173–80; PMID:7935654; http://dx.doi.org/ 10.1056/NEJM199411033311801 [DOI] [PubMed] [Google Scholar]
- 2.Hurst SA, Appelgren KE, Kourtis AP. Prevention of Mother-to-Child Transmission of Hiv Type 1: The Role of Neonatal and Infant Prophylaxis. Expert Rev Anti Infect Ther 2015; 13:169–81; PMID:25578882; http://dx.doi.org/ 10.1586/14787210.2015.999667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens J, Lyall H. Mother to Child Transmission of Hiv: What Works and How Much Is Enough? J Infect 2014; 69 Suppl 1:S56–62; PMID:25438711; http://dx.doi.org/ 10.1016/j.jinf.2014.07.018 [DOI] [PubMed] [Google Scholar]
- 4.Ciaranello AL, Seage GR 3rd, Freedberg KA, Weinstein MC, Lockman S, Walensky RP. Antiretroviral Drugs for Preventing Mother-to-Child Transmission of Hiv in Sub-Saharan Africa: Balancing Efficacy and Infant Toxicity. AIDS 2008; 22:2359–69; PMID:18981776; http://dx.doi.org/ 10.1097/QAD.0b013e3283189bd7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culnane M, Fowler M, Lee SS, McSherry G, Brady M, O'Donnell K, Mofenson L, Gortmaker SL, Shapiro DE, Scott G, et al.. Lack of Long-Term Effects of in Utero Exposure to Zidovudine among Uninfected Children Born to Hiv-Infected Women. Pediatric Aids Clinical Trials Group Protocol 219/076 Teams. JAMA 1999; 281:151–7; PMID:9917118; http://dx.doi.org/ 10.1001/jama.281.2.151 [DOI] [PubMed] [Google Scholar]
- 6.Ahmed S, Kim MH, Abrams EJ. Risks and Benefits of Lifelong Antiretroviral Treatment for Pregnant and Breastfeeding Women: A Review of the Evidence for the Option B+ Approach. Curr Opin HIV AIDS 2013; 8:474–89; PMID:23925003; http://dx.doi.org/ 10.1097/COH.0b013e328363a8f2 [DOI] [PubMed] [Google Scholar]
- 7.Brogly SB, Ylitalo N, Mofenson LM, Oleske J, Van Dyke R, Crain MJ, Abzug MJ, Brady M, Jean-Philippe P, Hughes MD, et al.. In Utero Nucleoside Reverse Transcriptase Inhibitor Exposure and Signs of Possible Mitochondrial Dysfunction in Hiv-Uninfected Children. AIDS 2007; 21:929–38; PMID:17457086; http://dx.doi.org/ 10.1097/QAD.0b013e3280d5a786 [DOI] [PubMed] [Google Scholar]
- 8.Stek AM. Antiretroviral Treatment in Pregnancy. Curr Opin HIV AIDS 2008; 3:155–60; PMID:19372959; http://dx.doi.org/ 10.1097/COH.0b013e3282f50bfe [DOI] [PubMed] [Google Scholar]
- 9.Blanche S, Tardieu M, Benhammou V, Warszawski J, Rustin P. Mitochondrial Dysfunction Following Perinatal Exposure to Nucleoside Analogues. AIDS 2006; 20:1685–90; PMID:16931932; http://dx.doi.org/ 10.1097/01.aids.0000242814.42344.77 [DOI] [PubMed] [Google Scholar]
- 10.Cossarizza A, Troiano L, Mussini C. Mitochondria and Hiv Infection: The First Decade. J Biol Regul Homeost Agents 2002; 16:18–24; PMID:12003168 [PubMed] [Google Scholar]
- 11.Barret B, Tardieu M, Rustin P, Lacroix C, Chabrol B, Desguerre I, Dollfus C, Mayaux MJ, Blanche S, French Perinatal Cohort Study G . Persistent Mitochondrial Dysfunction in Hiv-1-Exposed but Uninfected Infants: Clinical Screening in a Large Prospective Cohort. AIDS 2003; 17:1769–85; PMID:12891063; http://dx.doi.org/ 10.1097/00002030-200308150-00006 [DOI] [PubMed] [Google Scholar]
- 12.Heidari S, Mofenson L, Cotton MF, Marlink R, Cahn P, Katabira E. Antiretroviral Drugs for Preventing Mother-to-Child Transmission of Hiv: A Review of Potential Effects on Hiv-Exposed but Uninfected Children. J Acquir Immune Defic Syndr 2011; 57:290–6; PMID:21602695; http://dx.doi.org/ 10.1097/QAI.0b013e318221c56a [DOI] [PubMed] [Google Scholar]
- 13.Mofenson LM, Munderi P. Safety of Antiretroviral Prophylaxis of Perinatal Transmission for Hiv-Infected Pregnant Women and Their Infants. J Acquir Immune Defic Syndr 2002; 30:200–15; PMID:12045684; http://dx.doi.org/ 10.1097/00042560-200206010-00010 [DOI] [PubMed] [Google Scholar]
- 14.Cutfield WS, Hofman PL, Mitchell M, Morison IM. Could Epigenetics Play a Role in the Developmental Origins of Health and Disease? Pediatr Res 2007; 61:68R–75R; PMID:17413843; http://dx.doi.org/ 10.1203/pdr.0b013e318045764c [DOI] [PubMed] [Google Scholar]
- 15.Kiser DP, Rivero O, Lesch KP. Annual Research Review: The (Epi)Genetics of Neurodevelopmental Disorders in the Era of Whole-Genome Sequencing - Unveiling the Dark Matter. J Child Psychol Psychiatry 2015; 56:278–95; PMID:25677560; http://dx.doi.org/ 10.1111/jcpp.12392 [DOI] [PubMed] [Google Scholar]
- 16.Isles AR. Neural and Behavioral Epigenetics; What It Is, and What Is Hype. Genes Brain Behav 2015; 14:64–72; PMID:25346298; http://dx.doi.org/ 10.1111/gbb.12184 [DOI] [PubMed] [Google Scholar]
- 17.Desplats PA. Perinatal Programming of Neurodevelopment: Epigenetic Mechanisms and the Prenatal Shaping of the Brain. Adv Neurobiol 2015; 10:335–61; PMID:25287548 [DOI] [PubMed] [Google Scholar]
- 18.Lester BM, Marsit CJ, Conradt E, Bromer C, Padbury JF. Behavioral Epigenetics and the Developmental Origins of Child Mental Health Disorders. J Dev Orig Health Dis 2012; 3:395–408; PMID:25084292; http://dx.doi.org/ 10.1017/S2040174412000426 [DOI] [PubMed] [Google Scholar]
- 19.Lester BM, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa CW, Marsit CJ, Maze I, Meaney MJ, Monteggia LM, et al.. Behavioral Epigenetics. Ann N Y Acad Sci 2011; 1226:14–33; PMID:21615751; http://dx.doi.org/ 10.1111/j.1749-6632.2011.06037.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai M, Jellyman JK, Ross MG. Epigenomics, Gestational Programming and Risk of Metabolic Syndrome. Int J Obes (Lond) 2015; 39(4):633–41; PMID:25640766 [DOI] [PubMed] [Google Scholar]
- 21.Ruchat SM, Hivert MF, Bouchard L. Epigenetic Programming of Obesity and Diabetes by in Utero Exposure to Gestational Diabetes Mellitus. Nutr Rev 2013; 71 Suppl 1:S88–94; PMID:24147929; http://dx.doi.org/ 10.1111/nure.12057 [DOI] [PubMed] [Google Scholar]
- 22.Heerwagen MJ, Miller MR, Barbour LA, Friedman JE. Maternal Obesity and Fetal Metabolic Programming: A Fertile Epigenetic Soil. Am J Physiol Regul Integr Comp Physiol 2010; 299:R711–22; PMID:20631295; http://dx.doi.org/ 10.1152/ajpregu.00310.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz WA, Steinhoff C, Florl AR. Methylation of Endogenous Human Retroelements in Health and Disease. Curr Top Microbiol Immunol 2006; 310:211–50; PMID:16909913 [DOI] [PubMed] [Google Scholar]
- 24.Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, Sparrow D, Vokonas P, Schwartz J. Repetitive Element DNA Methylation and Circulating Endothelial and Inflammation Markers in the Va Normative Aging Study. Epigenetics 2010; 5:222–8; PMID:20305373; http://dx.doi.org/ 10.4161/epi.5.3.11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. Ischemic Heart Disease and Stroke in Relation to Blood DNA Methylation. Epidemiology 2010; 21:819–28; PMID:20805753; http://dx.doi.org/ 10.1097/EDE.0b013e3181f20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in Genomic DNA Methylation through Aging in a Cohort of Elderly Subjects. Mech Ageing Dev 2009; 130:234–9; PMID:19150625; http://dx.doi.org/ 10.1016/j.mad.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, et al.. Implications of Line1 Methylation for Bladder Cancer Risk in Women. Clin Cancer Res 2010; 16:1682–9; PMID:20179218; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson HH, Marsit CJ, Kelsey KT. Global Methylation in Exposure Biology and Translational Medical Science. Environ Health Perspect 2011; 119:1528–33; PMID:21669556; http://dx.doi.org/ 10.1289/ehp.1103423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanche S, Tardieu M, Rustin P, Slama A, Barret B, Firtion G, Ciraru-Vigneron N, Lacroix C, Rouzioux C, Mandelbrot L, et al.. Persistent Mitochondrial Dysfunction and Perinatal Exposure to Antiretroviral Nucleoside Analogues. Lancet 1999; 354:1084–9; PMID:10509500; http://dx.doi.org/ 10.1016/S0140-6736(99)07219-0 [DOI] [PubMed] [Google Scholar]
- 30.Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, et al.. DNA Methylation, Isocitrate Dehydrogenase Mutation, and Survival in Glioma. J Natl Cancer Inst 2011; 103:143–53; PMID:21163902; http://dx.doi.org/ 10.1093/jnci/djq497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al.. Identification of a Cpg Island Methylator Phenotype That Defines a Distinct Subgroup of Glioma. Cancer Cell 2010; 17:510–22; PMID:20399149; http://dx.doi.org/ 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace DC, Fan W. Energetics, Epigenetics, Mitochondrial Genetics. Mitochondrion 2010; 10:12–31; PMID:19796712; http://dx.doi.org/ 10.1016/j.mito.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiMauro S, Schon EA. Mitochondrial DNA Mutations in Human Disease. Am J Med Genet 2001; 106:18–26; PMID:11579421; http://dx.doi.org/ 10.1002/ajmg.1392 [DOI] [PubMed] [Google Scholar]
- 34.Funk MJ, Belinson SE, Pimenta JM, Morsheimer M, Gibbons DC. Mitochondrial Disorders among Infants Exposed to Hiv and Antiretroviral Therapy. Drug Saf 2007; 30:845–59; PMID:17867723; http://dx.doi.org/ 10.2165/00002018-200730100-00004 [DOI] [PubMed] [Google Scholar]
- 35.Jitratkosol MH, Sattha B, Maan EJ, Gadawski I, Harrigan PR, Forbes JC, Alimenti A, van Schalkwyk J, Money DM, Cote HC, et al.. Blood Mitochondrial DNA Mutations in Hiv-Infected Women and Their Infants Exposed to Haart During Pregnancy. AIDS 2012; 26:675–83; PMID:22436539; http://dx.doi.org/ 10.1097/QAD.0b013e32835142eb [DOI] [PubMed] [Google Scholar]
- 36.Jao J, Abrams EJ. Metabolic Complications of in Utero Maternal Hiv and Antiretroviral Exposure in Hiv-Exposed Infants. Pediatr Infect Dis J 2014; 33:734–40; PMID:24378947; http://dx.doi.org/ 10.1097/INF.0000000000000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilsner JR, Hall MN, Liu X, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Yunus M, Rahman M, Graziano JH, et al.. Influence of Prenatal Arsenic Exposure and Newborn Sex on Global Methylation of Cord Blood DNA. PLoS One 2012; 7:e37147; PMID:22662134; http://dx.doi.org/ 10.1371/journal.pone.0037147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Hsueh YM, Wright RO, et al.. Prenatal Arsenic Exposure and DNA Methylation in Maternal and Umbilical Cord Blood Leukocytes. Environ Health Perspect 2012; 120:1061–6; PMID:22466225; http://dx.doi.org/ 10.1289/ehp.1104173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, Ruchirawat M. Effects of Arsenic Exposure on DNA Methylation in Cord Blood Samples from Newborn Babies and in a Human Lymphoblast Cell Line. Environ Health 2012; 11:31; PMID:22551203; http://dx.doi.org/ 10.1186/1476-069X-11-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, et al.. Influence of Prenatal Lead Exposure on Genomic Methylation of Cord Blood DNA. Environ Health Perspect 2009; 117:1466–71; PMID:19750115; http://dx.doi.org/ 10.1289/ehp.0800497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huen K, Yousefi P, Bradman A, Yan L, Harley KG, Kogut K, Eskenazi B, Holland N. Effects of Age, Sex, and Persistent Organic Pollutants on DNA Methylation in Children. Environ Mol Mutagen 2014; 55:209–22; PMID:24375655; http://dx.doi.org/ 10.1002/em.21845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haggarty P, Hoad G, Campbell DM, Horgan GW, Piyathilake C, McNeill G. Folate in Pregnancy and Imprinted Gene and Repeat Element Methylation in the Offspring. Am J Clin Nutr 2013; 97:94–9; PMID:23151531; http://dx.doi.org/ 10.3945/ajcn.112.042572 [DOI] [PubMed] [Google Scholar]
- 43.Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, Tarantini L, Gillman M. Gestational Intake of Methyl Donors and Global Line-1 DNA Methylation in Maternal and Cord Blood: Prospective Results from a Folate-Replete Population. Epigenetics 2012; 7:253–60; PMID:22430801; http://dx.doi.org/ 10.4161/epi.7.3.19082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HS, Barraza-Villarreal A, Hernandez-Vargas H, Sly PD, Biessy C, Ramakrishnan U, Romieu I, Herceg Z. Modulation of DNA Methylation States and Infant Immune System by Dietary Supplementation with Omega-3 Pufa During Pregnancy in an Intervention Study. Am J Clin Nutr 2013; 98:480–7; PMID:23761484; http://dx.doi.org/ 10.3945/ajcn.112.052241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AK, Conneely KN, Newport DJ, Kilaru V, Schroeder JW, Pennell PB, Knight BT, Cubells JC, Stowe ZN, Brennan PA. Prenatal Antiepileptic Exposure Associates with Neonatal DNA Methylation Differences. Epigenetics 2012; 7:458–63; PMID:22419127; http://dx.doi.org/ 10.4161/epi.19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandelbrot L, Mazy F, Floch-Tudal C, Meier F, Azria E, Crenn-Hebert C, Treluyer JM, Herinomenzanahary E, Ferreira C, Peytavin G. Atazanavir in Pregnancy: Impact on Neonatal Hyperbilirubinemia. Eur J Obstet Gynecol Reprod Biol 2011; 157:18–21; PMID:21492993; http://dx.doi.org/ 10.1016/j.ejogrb.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 47.Williams PL, Crain MJ, Yildirim C, Hazra R, Van Dyke RB, Rich K, Read JS, Stuard E, Rathore M, Mendez HA, et al.. Congenital Anomalies and in Utero Antiretroviral Exposure in Human Immunodeficiency Virus-Exposed Uninfected Infants. JAMA Pediatr 2015; 169:48–55; PMID:25383770; http://dx.doi.org/ 10.1001/jamapediatrics.2014.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, Gurley EC, Jarujaron S, Ding H, Fang Y, Xu Z, Pandak WM Jr., Hylemon PB. Hiv Protease Inhibitors Activate the Unfolded Protein Response and Disrupt Lipid Metabolism in Primary Hepatocytes. Am J Physiol Gastrointest Liver Physiol 2006; 291:G1071–80; PMID:16861219; http://dx.doi.org/ 10.1152/ajpgi.00182.2006 [DOI] [PubMed] [Google Scholar]
- 49.Zhou H, Jarujaron S, Gurley EC, Chen L, Ding H, Studer E, Pandak WM Jr., Hu W, Zou T, Wang JY, et al.. Hiv Protease Inhibitors Increase Tnf-Alpha and Il-6 Expression in Macrophages: Involvement of the Rna-Binding Protein Hur. Atherosclerosis 2007; 195:e134–43; PMID:17531241; http://dx.doi.org/ 10.1016/j.atherosclerosis.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 50.Caso G, Mileva I, McNurlan MA, Mynarcik DC, Darras F, Gelato MC. Effect of Ritonavir and Atazanavir on Human Subcutaneous Preadipocyte Proliferation and Differentiation. Antiviral Res 2010; 86:137–43; PMID:20153378; http://dx.doi.org/ 10.1016/j.antiviral.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibellini L, De Biasi S, Pinti M, Nasi M, Riccio M, Carnevale G, Cavallini GM, Sala de Oyanguren FJ, O'Connor JE, Mussini C, et al.. The Protease Inhibitor Atazanavir Triggers Autophagy and Mitophagy in Human Preadipocytes. AIDS 2012; 26:2017–26; PMID:22948272; http://dx.doi.org/ 10.1097/QAD.0b013e328359b8be [DOI] [PubMed] [Google Scholar]
- 52.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA Methylation Dynamics of the Human Preimplantation Embryo. Nature 2014; 511:611–5; PMID:25079558; http://dx.doi.org/ 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El Hajj N, Pliushch G, Schneider E, Dittrich M, Muller T, Korenkov M, Aretz M, Zechner U, Lehnen H, Haaf T. Metabolic Programming of Mest DNA Methylation by Intrauterine Exposure to Gestational Diabetes Mellitus. Diabetes 2013; 62:1320–8; PMID:23209187; http://dx.doi.org/ 10.2337/db12-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michels KB, Harris HR, Barault L. Birthweight, Maternal Weight Trajectories and Global DNA Methylation of Line-1 Repetitive Elements. PLoS One 2011; 6:e25254; PMID:21980406; http://dx.doi.org/ 10.1371/journal.pone.0025254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griner R, Williams PL, Read JS, Seage GR 3rd, Crain M, Yogev R, Hazra R, Rich K, Pediatric HIVACS . In utero and postnatal exposure to antiretrovirals among Hiv-Exposed but Uninfected Children in the United States. AIDS Patient Care STDS 2011; 25:385–94; PMID:21992592; http://dx.doi.org/ 10.1089/apc.2011.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez EM, Mendez H, Rich K, Sheon A, Fox H, Green K, Diaz C, Brambilla D, Mofenson L. Maternal Drug Use in Perinatal Hiv Studies. The Women and Infants Transmission Study. Ann N Y Acad Sci 1993; 693:245–8; PMID:8267268; http://dx.doi.org/ 10.1111/j.1749-6632.1993.tb26272.x [DOI] [PubMed] [Google Scholar]
- 57.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al.. Changes in DNA Methylation Patterns in Subjects Exposed to Low-Dose Benzene. Cancer Res 2007; 67:876–80; PMID:17283117; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-2995 [DOI] [PubMed] [Google Scholar]
- 58.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A Simple Method for Estimating Global DNA Methylation Using Bisulfite Pcr of Repetitive DNA Elements. Nucleic Acids Res 2004; 32:e38; PMID:14973332; http://dx.doi.org/ 10.1093/nar/gnh032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, Gagne LA, Banister CE, Padbury JF, Marsit CJ. In Utero Exposures, Infant Growth, and DNA Methylation of Repetitive Elements and Developmentally Related Genes in Human Placenta. Environ Health Perspect 2012; 120:296–302; PMID:22005006; http://dx.doi.org/ 10.1289/ehp.1103927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giordan M. A Two-Stage Procedure for the Removal of Batch Effect in Microarray Studies. Statistics in Bioscience 2014; 6.1:73–84; http://dx.doi.org/ 10.1007/s12561-013-9081-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.